The Proteasome Activator PA200/PSME4: An Emerging New Player in Health and Disease
Abstract
1. Introduction
2. Expression and Regulation of PA200
3. Structure
4. Function
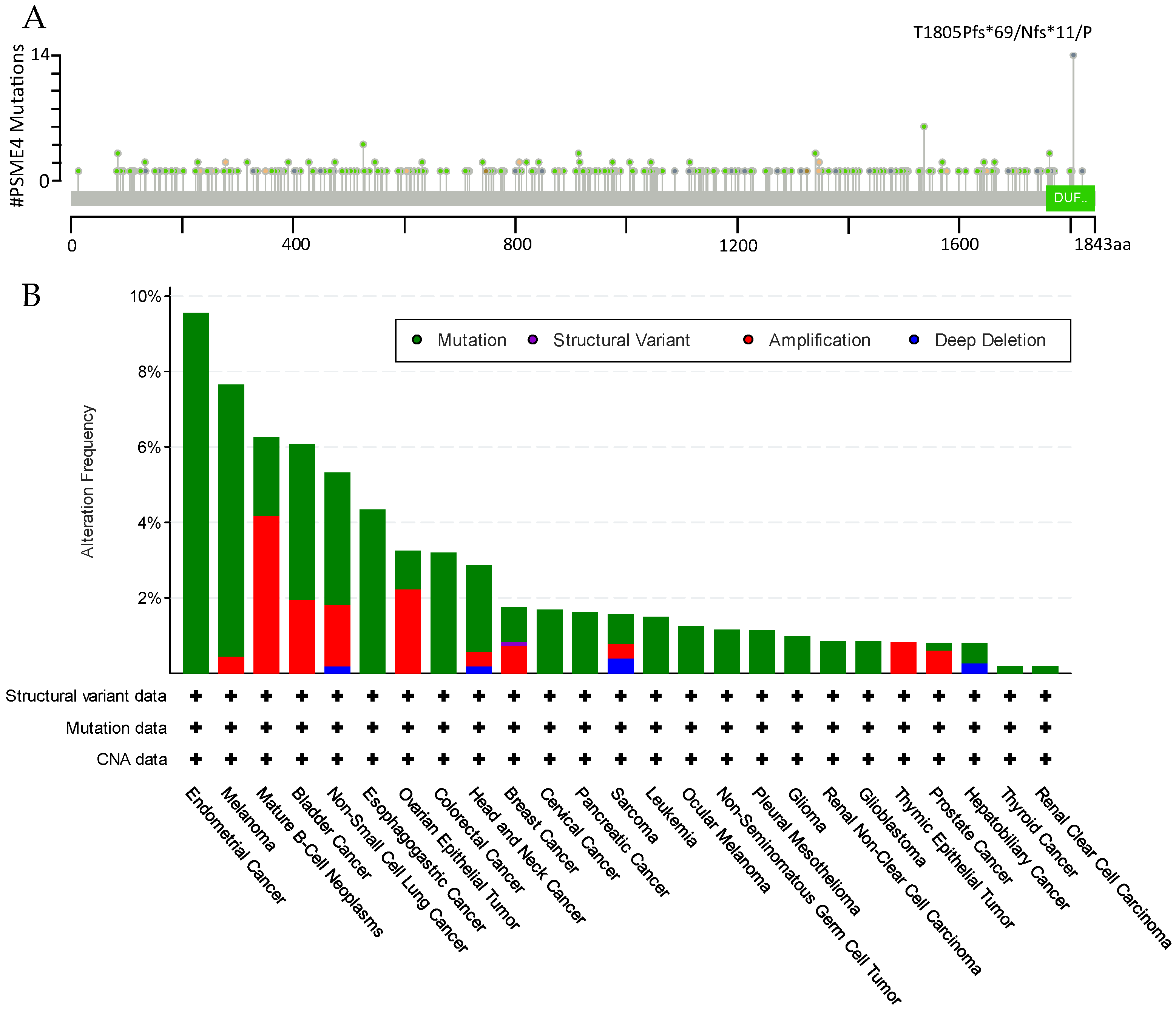
5. Dysregulation in Disease
6. Conclusions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Ustrell, V.; Hoffman, L.; Pratt, G.; Rechsteiner, M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 2002, 21, 3516–3525. [Google Scholar] [CrossRef]
- Fort, P.; Kajava, A.V.; Delsuc, F.; Coux, O. Evolution of proteasome regulators in eukaryotes. Genome Biol. Evol. 2015, 7, 1363–1379. [Google Scholar] [CrossRef] [PubMed]
- Savulescu, A.F.; Glickman, M.H. Proteasome activator 200: The heat is on. Mol. Cell. Proteom. 2011, 10, R110.006890. [Google Scholar] [CrossRef]
- Toste Rego, A.; da Fonseca, P.C.A. Characterization of Fully Recombinant Human 20S and 20S-PA200 Proteasome Complexes. Mol. Cell 2019, 76, 138–147.e5. [Google Scholar] [CrossRef]
- Welk, V.; Meul, T.; Lukas, C.; Kammerl, I.E.; Mulay, S.R.; Schamberger, A.C.; Semren, N.; Fernandez, I.E.; Anders, H.J.; Gunther, A.; et al. Proteasome activator PA200 regulates myofibroblast differentiation. Sci. Rep. 2019, 9, 15224. [Google Scholar] [CrossRef] [PubMed]
- Proteinatlas.org. PSME4 Protein Expression Summary—The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000068878-PSME4 (accessed on 2 June 2022).
- Khor, B.; Bredemeyer, A.L.; Huang, C.Y.; Turnbull, I.R.; Evans, R.; Maggi, L.B., Jr.; White, J.M.; Walker, L.M.; Carnes, K.; Hess, R.A.; et al. Proteasome activator PA200 is required for normal spermatogenesis. Mol. Cell. Biol. 2006, 26, 2999–3007. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Haratake, K.; Miyahara, H.; Chiba, T. Proteasome activators, PA28gamma and PA200, play indispensable roles in male fertility. Sci. Rep. 2016, 6, 23171. [Google Scholar] [CrossRef] [PubMed]
- Guan, H.; Wang, Y.; Yu, T.; Huang, Y.; Li, M.; Saeed, A.; Perculija, V.; Li, D.; Xiao, J.; Wang, D.; et al. Cryo-EM structures of the human PA200 and PA200-20S complex reveal regulation of proteasome gate opening and two PA200 apertures. PLoS Biol. 2020, 18, e3000654. [Google Scholar] [CrossRef]
- Qian, M.X.; Pang, Y.; Liu, C.H.; Haratake, K.; Du, B.Y.; Ji, D.Y.; Wang, G.F.; Zhu, Q.Q.; Song, W.; Yu, Y.; et al. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell 2013, 153, 1012–1024. [Google Scholar] [CrossRef]
- Mandemaker, I.K.; Geijer, M.E.; Kik, I.; Bezstarosti, K.; Rijkers, E.; Raams, A.; Janssens, R.C.; Lans, H.; Hoeijmakers, J.H.; Demmers, J.A.; et al. DNA damage-induced replication stress results in PA200-proteasome-mediated degradation of acetylated histones. EMBO Rep. 2018, 19, e45566. [Google Scholar] [CrossRef]
- Javitt, A.; Shmueli, M.D.; Kramer, M.P.; Kolodziejczyk, A.A.; Cohen, I.J.; Kamer, I.; Litchfield, K.; Bab-Dinitz, E.; Zadok, O.; Neiens, V. The proteasome regulator PSME4 drives immune evasion and abrogates anti-tumor immunity in NSCLC. bioRxiv 2021. [Google Scholar] [CrossRef]
- Fabre, B.; Lambour, T.; Delobel, J.; Amalric, F.; Monsarrat, B.; Burlet-Schiltz, O.; Bousquet-Dubouch, M.P. Subcellular distribution and dynamics of active proteasome complexes unraveled by a workflow combining in vivo complex cross-linking and quantitative proteomics. Mol. Cell. Proteom. 2013, 12, 687–699. [Google Scholar] [CrossRef]
- Blickwedehl, J.; McEvoy, S.; Wong, I.; Kousis, P.; Clements, J.; Elliott, R.; Cresswell, P.; Liang, P.; Bangia, N. Proteasomes and proteasome activator 200 kDa (PA200) accumulate on chromatin in response to ionizing radiation. Radiat. Res. 2007, 167, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Douida, A.; Batista, F.; Robaszkiewicz, A.; Boto, P.; Aladdin, A.; Szenykiv, M.; Czinege, R.; Virag, L.; Tar, K. The proteasome activator PA200 regulates expression of genes involved in cell survival upon selective mitochondrial inhibition in neuroblastoma cells. J. Cell. Mol. Med. 2020, 24, 6716–6730. [Google Scholar] [CrossRef] [PubMed]
- Ensembl PSME4 Human. Available online: https://www.ensembl.org/Homo_sapiens/Gene/Summary?db=core;g=ENSG00000068878;r=2:53864069-53970993 (accessed on 9 June 2022).
- Sha, Z.; Goldberg, A.L. Proteasome-mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits and p97. Curr. Biol. 2014, 24, 1573–1583. [Google Scholar] [CrossRef] [PubMed]
- Meiners, S.; Heyken, D.; Weller, A.; Ludwig, A.; Stangl, K.; Kloetzel, P.M.; Kruger, E. Inhibition of proteasome activity induces concerted expression of proteasome genes and de novo formation of Mammalian proteasomes. J. Biol. Chem. 2003, 278, 21517–21525. [Google Scholar] [CrossRef] [PubMed]
- Welk, V.; Coux, O.; Kleene, V.; Abeza, C.; Trumbach, D.; Eickelberg, O.; Meiners, S. Inhibition of Proteasome Activity Induces Formation of Alternative Proteasome Complexes. J. Biol. Chem. 2016, 291, 13147–13159. [Google Scholar] [CrossRef]
- Jagannathan, S.; Vad, N.; Vallabhapurapu, S.; Vallabhapurapu, S.; Anderson, K.C.; Driscoll, J.J. MiR-29b replacement inhibits proteasomes and disrupts aggresome+autophagosome formation to enhance the antimyeloma benefit of bortezomib. Leukemia 2015, 29, 727–738. [Google Scholar] [CrossRef]
- Blickwedehl, J.; Agarwal, M.; Seong, C.; Pandita, R.K.; Melendy, T.; Sung, P.; Pandita, T.K.; Bangia, N. Role for proteasome activator PA200 and postglutamyl proteasome activity in genomic stability. Proc. Natl. Acad. Sci. USA 2008, 105, 16165–16170. [Google Scholar] [CrossRef]
- Zhao, J.; Makhija, S.; Huang, B.; Cheng, Y. Efficient tagging and purification of endogenous proteins for structural studies by single particle cryo-EM. bioRxiv 2021. [Google Scholar] [CrossRef]
- Fabre, B.; Lambour, T.; Garrigues, L.; Ducoux-Petit, M.; Amalric, F.; Monsarrat, B.; Burlet-Schiltz, O.; Bousquet-Dubouch, M.P. Label-free quantitative proteomics reveals the dynamics of proteasome complexes composition and stoichiometry in a wide range of human cell lines. J. Proteome Res. 2014, 13, 3027–3037. [Google Scholar] [CrossRef]
- Ortega, J.; Heymann, J.B.; Kajava, A.V.; Ustrell, V.; Rechsteiner, M.; Steven, A.C. The axial channel of the 20S proteasome opens upon binding of the PA200 activator. J. Mol. Biol. 2005, 346, 1221–1227. [Google Scholar] [CrossRef] [PubMed]
- Macbeth, M.R.; Schubert, H.L.; Vandemark, A.P.; Lingam, A.T.; Hill, C.P.; Bass, B.L. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 2005, 309, 1534–1539. [Google Scholar] [CrossRef]
- Scaiola, A.; Mangia, F.; Imseng, S.; Boehringer, D.; Berneiser, K.; Shimobayashi, M.; Stuttfeld, E.; Hall, M.N.; Ban, N.; Maier, T. The 3.2-A resolution structure of human mTORC2. Sci. Adv. 2020, 6, eabc1251. [Google Scholar] [CrossRef] [PubMed]
- Okamura, M.; Yamanaka, Y.; Shigemoto, M.; Kitadani, Y.; Kobayashi, Y.; Kambe, T.; Nagao, M.; Kobayashi, I.; Okumura, K.; Masuda, S. Depletion of mRNA export regulator DBP5/DDX19, GLE1 or IPPK that is a key enzyme for the production of IP6, resulting in differentially altered cytoplasmic mRNA expression and specific cell defect. PLoS ONE 2018, 13, e0197165. [Google Scholar] [CrossRef]
- Folkmann, A.W.; Noble, K.N.; Cole, C.N.; Wente, S.R. Dbp5, Gle1-IP6 and Nup159: A working model for mRNP export. Nucleus 2011, 2, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Hanakahi, L.A.; West, S.C. Specific interaction of IP6 with human Ku70/80, the DNA-binding subunit of DNA-PK. EMBO J. 2002, 21, 2038–2044. [Google Scholar] [CrossRef]
- Byrum, J.; Jordan, S.; Safrany, S.T.; Rodgers, W. Visualization of inositol phosphate-dependent mobility of Ku: Depletion of the DNA-PK cofactor InsP6 inhibits Ku mobility. Nucleic Acids Res. 2004, 32, 2776–2784. [Google Scholar] [CrossRef][Green Version]
- Marcum, R.D.; Radhakrishnan, I. Inositol phosphates and core subunits of the Sin3L/Rpd3L histone deacetylase (HDAC) complex up-regulate deacetylase activity. J. Biol. Chem. 2019, 294, 13928–13938. [Google Scholar] [CrossRef]
- Lin, H.; Zhang, X.; Liu, L.; Fu, Q.; Zang, C.; Ding, Y.; Su, Y.; Xu, Z.; He, S.; Yang, X.; et al. Basis for metabolite-dependent Cullin-RING ligase deneddylation by the COP9 signalosome. Proc. Natl. Acad. Sci. USA 2020, 117, 4117–4124. [Google Scholar] [CrossRef]
- Brehm, M.A.; Schenk, T.M.; Zhou, X.; Fanick, W.; Lin, H.; Windhorst, S.; Nalaskowski, M.M.; Kobras, M.; Shears, S.B.; Mayr, G.W. Intracellular localization of human Ins(1,3,4,5,6)P5 2-kinase. Biochem. J. 2007, 408, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Zivkovic, D.; Sanchez Dafun, A.; Menneteau, T.; Schahl, A.; Lise, S.; Kervarrec, C.; Toste Rego, A.; da Fonseca, P.C.A.; Chavent, M.; Pineau, C.; et al. Proteasome complexes experience profound structural and functional rearrangements throughout mammalian spermatogenesis. Proc. Natl. Acad. Sci. USA 2022, 119, e2116826119. [Google Scholar] [CrossRef] [PubMed]
- Bard, J.A.M.; Goodall, E.A.; Greene, E.R.; Jonsson, E.; Dong, K.C.; Martin, A. Structure and Function of the 26S Proteasome. Annu. Rev. Biochem. 2018, 87, 697–724. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.X.; Ma, S.; Han, X.; Luo, Z.Y.; Zhu, Q.Q.; Chiba, T.; Xie, W.; Lin, K.; Qiu, X.B. Proteasome activator PA200 maintains stability of histone marks during transcription and aging. Theranostics 2021, 11, 1458–1472. [Google Scholar] [CrossRef]
- Kim, Y.S.; Kim, M.; Cho, D.I.; Lim, S.Y.; Jun, J.H.; Kim, M.R.; Kang, B.G.; Eom, G.H.; Kang, G.; Yoon, S.; et al. PSME4 Degrades Acetylated YAP1 in the Nucleus of Mesenchymal Stem Cells. Pharmaceutics 2022, 14, 1659. [Google Scholar] [CrossRef]
- Blickwedehl, J.; Olejniczak, S.; Cummings, R.; Sarvaiya, N.; Mantilla, A.; Chanan-Khan, A.; Pandita, T.K.; Schmidt, M.; Thompson, C.B.; Bangia, N. The proteasome activator PA200 regulates tumor cell responsiveness to glutamine and resistance to ionizing radiation. Mol. Cancer Res. 2012, 10, 937–944. [Google Scholar] [CrossRef]
- Wang, F.; Ma, H.; Liang, W.J.; Yang, J.J.; Wang, X.Q.; Shan, M.R.; Chen, Y.; Jia, M.; Yin, Y.L.; Sun, X.Y.; et al. Lovastatin upregulates microRNA-29b to reduce oxidative stress in rats with multiple cardiovascular risk factors. Oncotarget 2017, 8, 9021–9034. [Google Scholar] [CrossRef]
- Douida, A.; Batista, F.; Boto, P.; Regdon, Z.; Robaszkiewicz, A.; Tar, K. Cells Lacking PA200 Adapt to Mitochondrial Dysfunction by Enhancing Glycolysis via Distinct Opa1 Processing. Int. J. Mol. Sci. 2021, 22, 1629. [Google Scholar] [CrossRef]
- cBioPortal. TCGA PanCancer Atlas Studies—PA200/PSME4. Available online: https://bit.ly/3bWk4Cz (accessed on 16 August 2022).
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Wendler, P.; Enenkel, C. Nuclear Transport of Yeast Proteasomes. Front. Mol. Biosci. 2019, 6, 34. [Google Scholar] [CrossRef] [PubMed]
- Macheret, M.; Bhowmick, R.; Sobkowiak, K.; Padayachy, L.; Mailler, J.; Hickson, I.D.; Halazonetis, T.D. High-resolution mapping of mitotic DNA synthesis regions and common fragile sites in the human genome through direct sequencing. Cell Res. 2020, 30, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Huang, H.; Huang, W.; Ji, R.; Chen, J.; Wu, S.; Wang, L.; Huang, T.; Sheng, Y.; Yan, H.; et al. PSME4 Activates mTOR Signaling and Promotes the Malignant Progression of Hepatocellular Carcinoma. Int. J. Gen. Med. 2022, 15, 885–895. [Google Scholar] [CrossRef]
- Tian, T.; Li, X.; Zhang, J. mTOR Signaling in Cancer and mTOR Inhibitors in Solid Tumor Targeting Therapy. Int. J. Mol. Sci. 2019, 20, 755. [Google Scholar] [CrossRef] [PubMed]
- Suraweera, A.; Munch, C.; Hanssum, A.; Bertolotti, A. Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol. Cell 2012, 48, 242–253. [Google Scholar] [CrossRef] [PubMed]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef]
- Yuan, Q.; Zhang, S.; Li, J.; Xiao, J.; Li, X.; Yang, J.; Lu, D.; Wang, Y. Comprehensive analysis of core genes and key pathways in Parkinson’s disease. Am. J. Transl. Res. 2020, 12, 5630–5639. [Google Scholar] [PubMed]
- Bronner, I.F.; Bochdanovits, Z.; Rizzu, P.; Kamphorst, W.; Ravid, R.; van Swieten, J.C.; Heutink, P. Comprehensive mRNA expression profiling distinguishes tauopathies and identifies shared molecular pathways. PLoS ONE 2009, 4, e6826. [Google Scholar] [CrossRef][Green Version]
- Nayler, S.P.; Powell, J.E.; Vanichkina, D.P.; Korn, O.; Wells, C.A.; Kanjhan, R.; Sun, J.; Taft, R.J.; Lavin, M.F.; Wolvetang, E.J. Human iPSC-Derived Cerebellar Neurons from a Patient with Ataxia-Telangiectasia Reveal Disrupted Gene Regulatory Networks. Front. Cell. Neurosci. 2017, 11, 321. [Google Scholar] [CrossRef]
- Minor, M.M.; Hollinger, F.B.; McNees, A.L.; Jung, S.Y.; Jain, A.; Hyser, J.M.; Bissig, K.D.; Slagle, B.L. Hepatitis B Virus HBx Protein Mediates the Degradation of Host Restriction Factors through the Cullin 4 DDB1 E3 Ubiquitin Ligase Complex. Cells 2020, 9, 834. [Google Scholar] [CrossRef]
- Expression Atlas—PSME4. Available online: https://www.ebi.ac.uk/gxa/genes/ensg00000068878?bs=%7B%22homo%20sapiens%22%3A%5B%22ORGANISM_PART%22%5D%7D&ds=%7B%22kingdom%22%3A%5B%22animals%22%5D%7D#differential (accessed on 4 July 2022).
- Kobayashi, S.D.; Braughton, K.R.; Palazzolo-Ballance, A.M.; Kennedy, A.D.; Sampaio, E.; Kristosturyan, E.; Whitney, A.R.; Sturdevant, D.E.; Dorward, D.W.; Holland, S.M.; et al. Rapid neutrophil destruction following phagocytosis of Staphylococcus aureus. J. Innate Immun. 2010, 2, 560–575. [Google Scholar] [CrossRef] [PubMed]
- Yeung, T.L.; Leung, C.S.; Wong, K.K.; Samimi, G.; Thompson, M.S.; Liu, J.; Zaid, T.M.; Ghosh, S.; Birrer, M.J.; Mok, S.C. TGF-beta modulates ovarian cancer invasion by upregulating CAF-derived versican in the tumor microenvironment. Cancer Res. 2013, 73, 5016–5028. [Google Scholar] [CrossRef]
- Kupfer, D.M.; White, V.L.; Strayer, D.L.; Crouch, D.J.; Burian, D. Microarray characterization of gene expression changes in blood during acute ethanol exposure. BMC Med. Genom. 2013, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.L.; Lucas, J.E.; Schroeder, T.; Mori, S.; Wu, J.; Nevins, J.; Dewhirst, M.; West, M.; Chi, J.T. The genomic analysis of lactic acidosis and acidosis response in human cancers. PLoS Genet. 2008, 4, e1000293. [Google Scholar] [CrossRef]
- Yu, Z.; Wei, X.; Liu, L.; Sun, H.; Fang, T.; Wang, L.; Li, Y.; Sui, W.; Wang, K.; He, Y.; et al. Indirubin-3′-monoxime acts as proteasome inhibitor: Therapeutic application in multiple myeloma. EBioMedicine 2022, 78, 103950. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Dong, X.; Jin, J.; He, Y. The Expression Patterns and Prognostic Value of the Proteasome Activator Subunit Gene Family in Gastric Cancer Based on Integrated Analysis. Front. Cell Dev. Biol. 2021, 9, 663001. [Google Scholar] [CrossRef]
- Tong, M.; Chan, K.W.; Bao, J.Y.; Wong, K.Y.; Chen, J.N.; Kwan, P.S.; Tang, K.H.; Fu, L.; Qin, Y.R.; Lok, S.; et al. Rab25 is a tumor suppressor gene with antiangiogenic and anti-invasive activities in esophageal squamous cell carcinoma. Cancer Res. 2012, 72, 6024–6035. [Google Scholar] [CrossRef]
- Maag, J.L.V.; Fisher, O.M.; Levert-Mignon, A.; Kaczorowski, D.C.; Thomas, M.L.; Hussey, D.J.; Watson, D.I.; Wettstein, A.; Bobryshev, Y.V.; Edwards, M.; et al. Novel Aberrations Uncovered in Barrett’s Esophagus and Esophageal Adenocarcinoma Using Whole Transcriptome Sequencing. Mol. Cancer Res. 2017, 15, 1558–1569. [Google Scholar] [CrossRef]
- Farah, C.S.; Dalley, A.J.; Nguyen, P.; Batstone, M.; Kordbacheh, F.; Perry-Keene, J.; Fielding, D. Improved surgical margin definition by narrow band imaging for resection of oral squamous cell carcinoma: A prospective gene expression profiling study. Head Neck 2016, 38, 832–839. [Google Scholar] [CrossRef]
- Sanchez-Palencia, A.; Gomez-Morales, M.; Gomez-Capilla, J.A.; Pedraza, V.; Boyero, L.; Rosell, R.; Farez-Vidal, M.E. Gene expression profiling reveals novel biomarkers in nonsmall cell lung cancer. Int. J. Cancer 2011, 129, 355–364. [Google Scholar] [CrossRef]
- Abdueva, D.; Wing, M.; Schaub, B.; Triche, T.; Davicioni, E. Quantitative expression profiling in formalin-fixed paraffin-embedded samples by affymetrix microarrays. J. Mol. Diagn. 2010, 12, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.; Otu, H.; Spentzos, D.; Kolia, S.; Inan, M.; Beecken, W.D.; Fellbaum, C.; Gu, X.; Joseph, M.; Pantuck, A.J.; et al. Gene signatures of progression and metastasis in renal cell cancer. Clin. Cancer Res. 2005, 11, 5730–5739. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.B.; Salah, Z.; Del Mare, S.; Galasso, M.; Gaudio, E.; Nuovo, G.J.; Lovat, F.; LeBlanc, K.; Palatini, J.; Randall, R.L.; et al. miRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res. 2012, 72, 1865–1877. [Google Scholar] [CrossRef] [PubMed]
- Opoku-Nsiah, K.A.; Gestwicki, J.E. Aim for the core: Suitability of the ubiquitin-independent 20S proteasome as a drug target in neurodegeneration. Transl. Res. 2018, 198, 48–57. [Google Scholar] [CrossRef]
- Woulfe, J. Nuclear bodies in neurodegenerative disease. Biochim. Biophys. Acta 2008, 1783, 2195–2206. [Google Scholar] [CrossRef]
- Abramov, A.Y.; Berezhnov, A.V.; Fedotova, E.I.; Zinchenko, V.P.; Dolgacheva, L.P. Interaction of misfolded proteins and mitochondria in neurodegenerative disorders. Biochem. Soc. Trans. 2017, 45, 1025–1033. [Google Scholar] [CrossRef]
- Chung, C.G.; Lee, H.; Lee, S.B. Mechanisms of protein toxicity in neurodegenerative diseases. Cell. Mol. Life Sci. 2018, 75, 3159–3180. [Google Scholar] [CrossRef]
- Enenkel, C.; Kang, R.W.; Wilfling, F.; Ernst, O.P. Intracellular localization of the proteasome in response to stress conditions. J. Biol. Chem. 2022, 298, 102083. [Google Scholar] [CrossRef]
- Doherty, K.M.; Pride, L.D.; Lukose, J.; Snydsman, B.E.; Charles, R.; Pramanik, A.; Muller, E.G.; Botstein, D.; Moore, C.W. Loss of a 20S proteasome activator in Saccharomyces cerevisiae downregulates genes important for genomic integrity, increases DNA damage, and selectively sensitizes cells to agents with diverse mechanisms of action. G3 2012, 2, 943–959. [Google Scholar] [CrossRef]
- Benmerzoug, S.; Ryffel, B.; Togbe, D.; Quesniaux, V.F.J. Self-DNA Sensing in Lung Inflammatory Diseases. Trends Immunol. 2019, 40, 719–734. [Google Scholar] [CrossRef]
- Barber, G.N. STING-dependent cytosolic DNA sensing pathways. Trends Immunol. 2014, 35, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Kempuraj, D.; Thangavel, R.; Natteru, P.A.; Selvakumar, G.P.; Saeed, D.; Zahoor, H.; Zaheer, S.; Iyer, S.S.; Zaheer, A. Neuroinflammation Induces Neurodegeneration. J. Neurol. Neurosurg. Spine 2016, 1, 1003. [Google Scholar] [PubMed]
- Pickering, A.M.; Davies, K.J. Differential roles of proteasome and immunoproteasome regulators Pa28alphabeta, Pa28gamma and Pa200 in the degradation of oxidized proteins. Arch. Biochem. Biophys. 2012, 523, 181–190. [Google Scholar] [CrossRef]
- Ebstein, F.; Lange, N.; Urban, S.; Seifert, U.; Kruger, E.; Kloetzel, P.M. Maturation of human dendritic cells is accompanied by functional remodelling of the ubiquitin-proteasome system. Int. J. Biochem. Cell Biol. 2009, 41, 1205–1215. [Google Scholar] [CrossRef] [PubMed]
- Newey, A.; Yu, L.; Barber, L.J.; Choudhary, J.S.; Bassani-Sternberg, M.; Gerlinger, M. Multifactorial remodeling of the cancer immunopeptidome by interferon gamma. bioRxiv 2022. [Google Scholar] [CrossRef]
- Pialoux, V.; Mounier, R. Hypoxia-induced oxidative stress in health disorders. Oxid. Med. Cell. Longev. 2012, 2012, 940121. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pickering, A.M.; Koop, A.L.; Teoh, C.Y.; Ermak, G.; Grune, T.; Davies, K.J. The immunoproteasome, the 20S proteasome and the PA28alphabeta proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem. J. 2010, 432, 585–594. [Google Scholar] [CrossRef]
- Abu-El-Rub, E.; Sareen, N.; Yan, W.; Alagarsamy, K.N.; Rafieerad, A.; Srivastava, A.; Desiderio, V.; Dhingra, S. Hypoxia-induced shift in the phenotype of proteasome from 26S toward immunoproteasome triggers loss of immunoprivilege of mesenchymal stem cells. Cell Death Dis. 2020, 11, 419. [Google Scholar] [CrossRef]
- D’Souza, A.J.; Desai, S.D.; Rudner, X.L.; Kelly, M.N.; Ruan, S.; Shellito, J.E. Suppression of the macrophage proteasome by ethanol impairs MHC class I antigen processing and presentation. PLoS ONE 2013, 8, e56890. [Google Scholar] [CrossRef][Green Version]
- Osna, N.A.; Bardag-Gorce, F.; White, R.L.; Weinman, S.A.; Donohue, T.M., Jr.; Kharbanda, K.K. Ethanol and hepatitis C virus suppress peptide-MHC class I presentation in hepatocytes by altering proteasome function. Alcohol. Clin. Exp. Res. 2012, 36, 2028–2035. [Google Scholar] [CrossRef]


| Context | Disease | PA200/PSME4 Expression | Reference |
|---|---|---|---|
| Neurodevelopment and Neurodegeneration | Parkinson’s disease | ↓ | [50] |
| Pick disease | ↓ | [51] | |
| Ataxia-Telangiectasia | ↓ | [52] | |
| Infection | Hepatitis B virus infection | ↓ | [53] |
| Herpes simplex virus G207 infection | ↓ | [54] | |
| Staphylococcus aureus infection | ↓ | [55] | |
| Environmental stresses | Hypoxia | ↓ | [56] |
| Acute ethanol exposure | ↓ | [57] | |
| Lactic acidosis and hypoxia | ↓ | [58] | |
| Cardiovascular diseases | Endothelial dysfunction | ↑ | [39] |
| Cancer | Multiple myeloma | ↑ (loss of miR-29b) | [20] |
| ↑ | [59] | ||
| Gastric cancer | ↑ | [60] | |
| Esophageal squamous cell carcinoma | ↑ | [61] | |
| Esophageal Adenocarcinoma | ↑ | [62] | |
| Oral squamous cell cancer (OSCC) | ↑ | [63] | |
| Hepatocellular carcinoma | ↑ | [46] | |
| Non-small lung cancer | ↑ | [64] | |
| Lung cancer | ↑ | [65] | |
| Transitional cell carcinoma of the kidney | ↑ | [66] | |
| Osteosarcoma | ↓ | [67] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yazgili, A.S.; Ebstein, F.; Meiners, S. The Proteasome Activator PA200/PSME4: An Emerging New Player in Health and Disease. Biomolecules 2022, 12, 1150. https://doi.org/10.3390/biom12081150
Yazgili AS, Ebstein F, Meiners S. The Proteasome Activator PA200/PSME4: An Emerging New Player in Health and Disease. Biomolecules. 2022; 12(8):1150. https://doi.org/10.3390/biom12081150
Chicago/Turabian StyleYazgili, Ayse Seda, Frédéric Ebstein, and Silke Meiners. 2022. "The Proteasome Activator PA200/PSME4: An Emerging New Player in Health and Disease" Biomolecules 12, no. 8: 1150. https://doi.org/10.3390/biom12081150
APA StyleYazgili, A. S., Ebstein, F., & Meiners, S. (2022). The Proteasome Activator PA200/PSME4: An Emerging New Player in Health and Disease. Biomolecules, 12(8), 1150. https://doi.org/10.3390/biom12081150






