Revisiting Concurrent Radiation Therapy, Temozolomide, and the Histone Deacetylase Inhibitor Valproic Acid for Patients with Glioblastoma—Proteomic Alteration and Comparison Analysis with the Standard-of-Care Chemoirradiation
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. SomaLogic SOMAScan® Assays
2.3. Data Process
2.4. Class Comparisons
2.5. Gene Set Enrichment Analysis
2.6. Survival Analysis
3. Results
3.1. Clinical Features Comparison between VPA and Non-VPA Cohort
3.2. Clinical Factors Survival and Progression-Free Survival Analysis
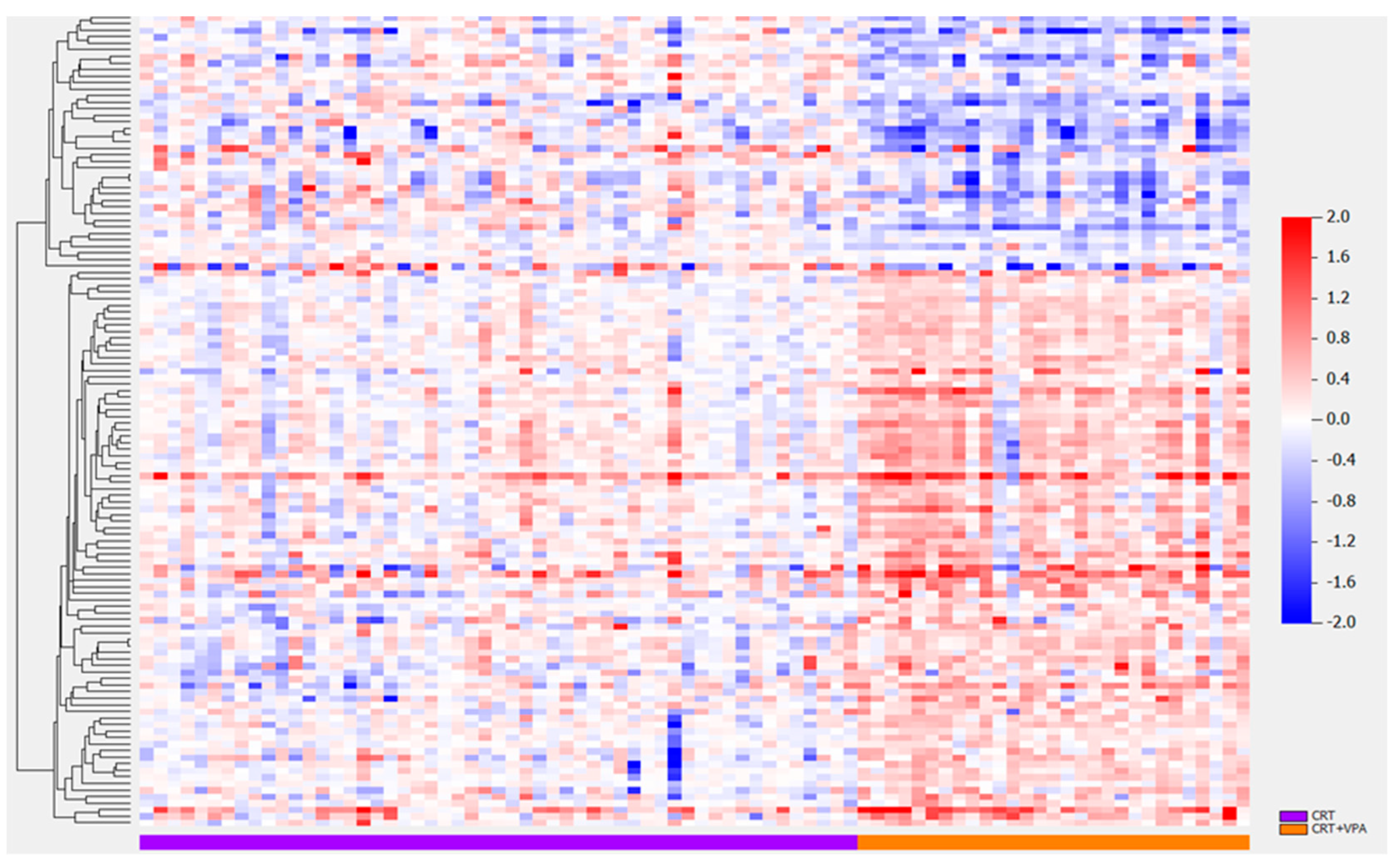
3.3. Differentially Expressed Proteins VPA vs. No VPA
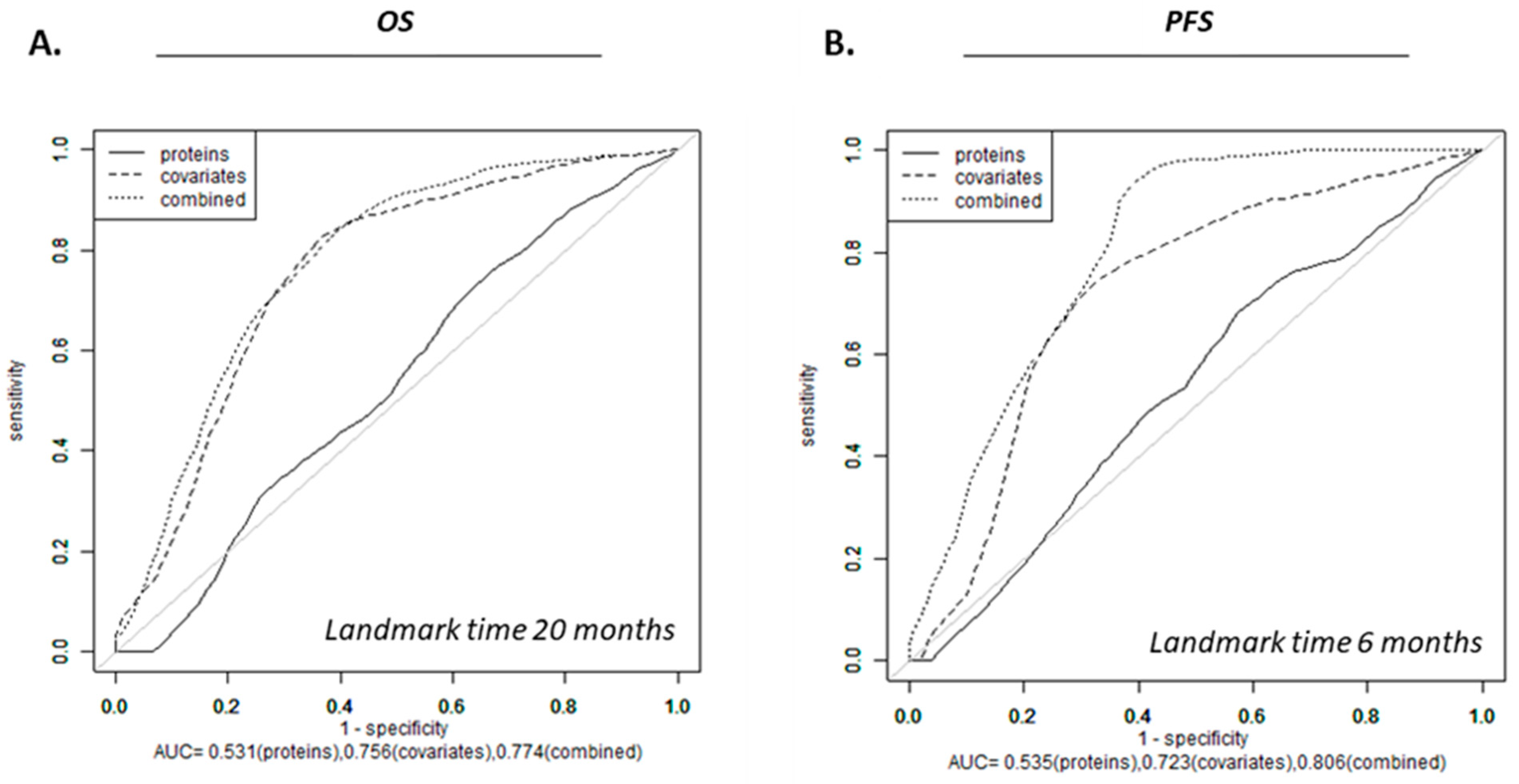
3.4. Survival Models for OS and PFS
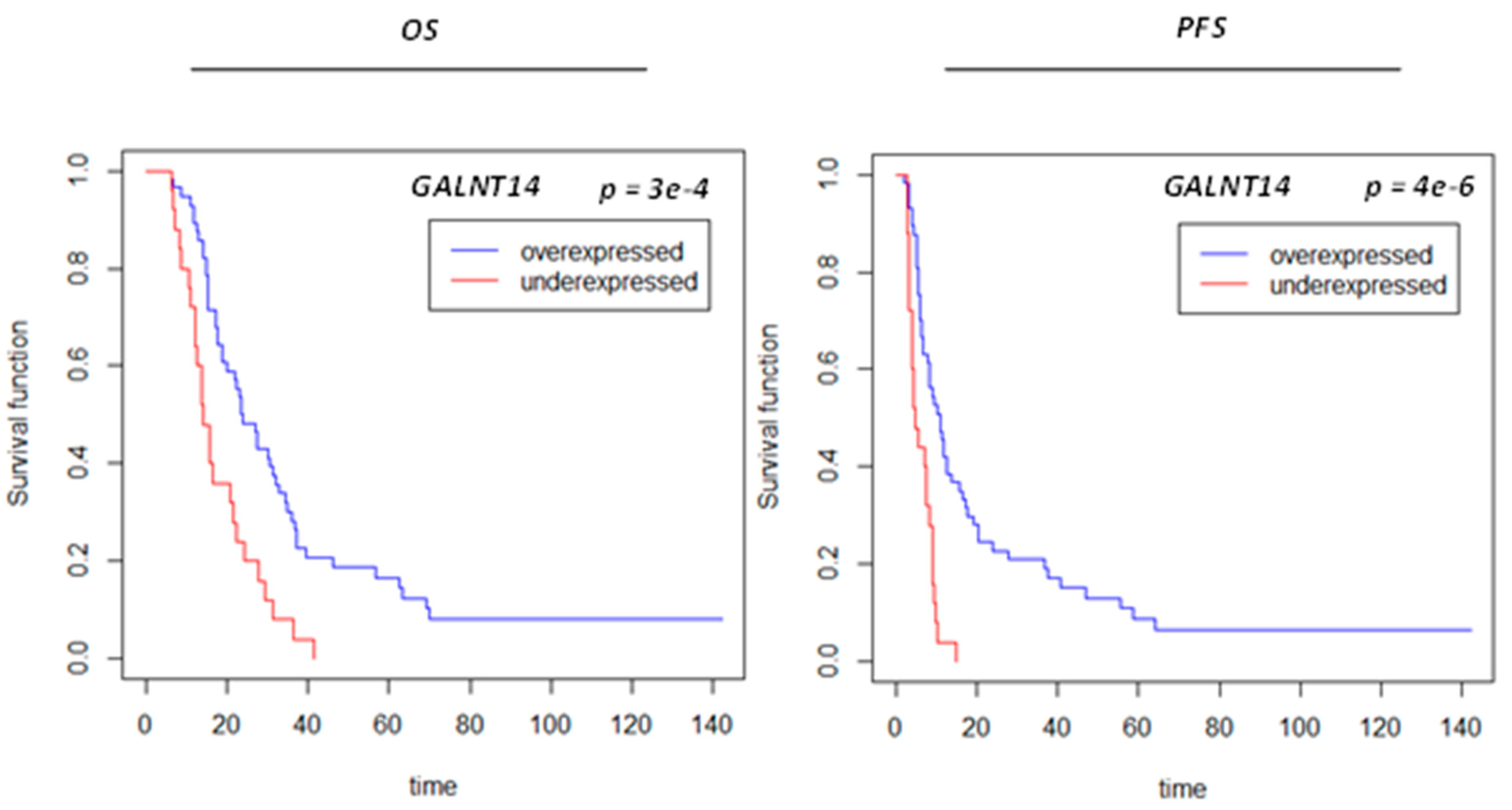
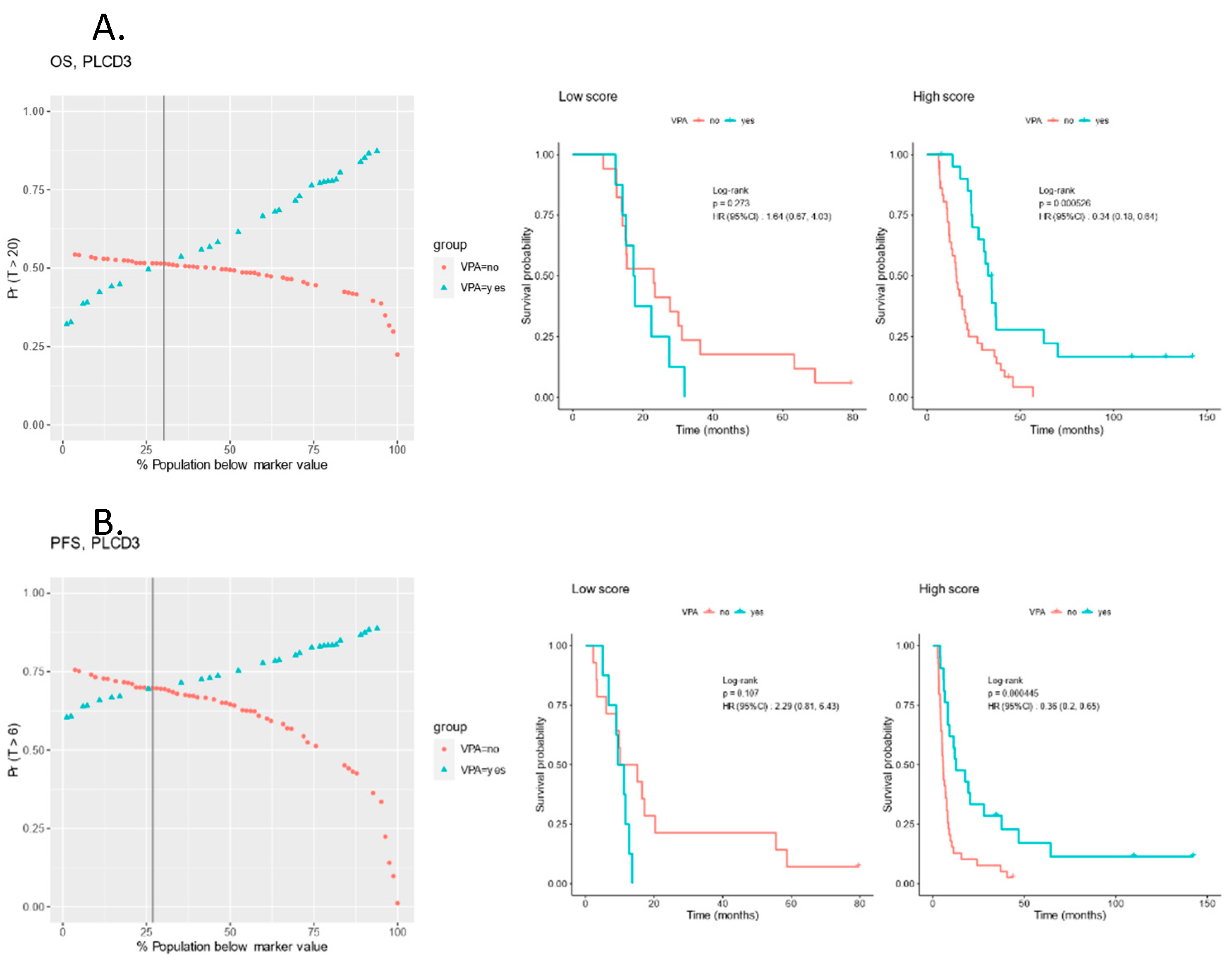
3.5. Protein Signals Predictive of VPA Response
3.6. Pathway Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CRT | Concurrent Chemoirradiation |
| EMT | Epithelial–Mesenchymal Transition |
| GBM | Glioblastoma |
| GTV T1 | Gross Tumor Volume on T1 Gadolinium-Enhanced MRI Sequence |
| GTV T2 | Gross Tumor Volume on T2 FLAIR Signal Sequence |
| MGMT | O6-Methylguanine-DNA Methyltransferase |
| OS | Overall survival |
| PFS | Progression-Free Survival |
| RPA | Recursive Partitioning Analysis |
| RT | Radiation Therapy |
| TMZ | Temozolomide |
| VPA | Valproic Acid |
| WHO | World Health Organisation |
References
- Ostrom, Q.T.; Patil, N.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2013–2017. Neuro-Oncol. 2020, 22, iv1–iv96. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Mathen, P.; Rowe, L.; Mackey, M.; Smart, D.; Tofilon, P.; Camphausen, K. Radiosensitizers in the temozolomide era for newly diagnosed glioblastoma. Neuro-Oncol. Pract. 2020, 7, 268–276. [Google Scholar] [CrossRef]
- Cornago, M.; Garcia-Alberich, C.; Blasco-Angulo, N.; Vall-Llaura, N.; Nager, M.; Herreros, J.; Comella, J.X.; Sanchis, D.; Llovera, M. Histone deacetylase inhibitors promote glioma cell death by G2 checkpoint abrogation leading to mitotic catastrophe. Cell Death Dis. 2014, 5, e1435. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Xu, Y.; Wang, H.; Niu, J.; Hou, H.; Jiang, Y. Histone deacetylase inhibitor, valproic acid, radiosensitizes the C6 glioma cell line in vitro. Oncol. Lett. 2014, 7, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, S.; Frijlink, E.; Kroonen, J.; Spliet, W.G.M.; van Hecke, W.; Seute, T.; Snijders, T.J.; Robe, P.A. Effects of valproic acid on histone deacetylase inhibition in vitro and in glioblastoma patient samples. Neuro-Oncol. Adv. 2019, 1, vdz025. [Google Scholar] [CrossRef]
- Kerkhof, M.; Dielemans, J.C.; van Breemen, M.S.; Zwinkels, H.; Walchenbach, R.; Taphoorn, M.J.; Vecht, C.J. Effect of valproic acid on seizure control and on survival in patients with glioblastoma multiforme. Neuro-Oncol. 2013, 15, 961–967. [Google Scholar] [CrossRef]
- Guthrie, G.D.; Eljamel, S. Impact of particular antiepileptic drugs on the survival of patients with glioblastoma multiforme. J. Neurosurg. 2013, 118, 859–865. [Google Scholar] [CrossRef]
- Redjal, N.; Reinshagen, C.; Le, A.; Walcott, B.P.; McDonnell, E.; Dietrich, J.; Nahed, B.V. Valproic acid, compared to other antiepileptic drugs, is associated with improved overall and progression-free survival in glioblastoma but worse outcome in grade II/III gliomas treated with temozolomide. J. Neuro-Oncol. 2016, 127, 505–514. [Google Scholar] [CrossRef]
- Weller, M.; Gorlia, T.; Cairncross, J.G.; van den Bent, M.J.; Mason, W.; Belanger, K.; Brandes, A.A.; Bogdahn, U.; Macdonald, D.R.; Forsyth, P.; et al. Prolonged survival with valproic acid use in the EORTC/NCIC temozolomide trial for glioblastoma. Neurology 2011, 77, 1156–1164. [Google Scholar] [CrossRef] [PubMed]
- Lu, V.M.; Texakalidis, P.; McDonald, K.L.; Mekary, R.A.; Smith, T.R. The survival effect of valproic acid in glioblastoma and its current trend: A systematic review and meta-analysis. Clin. Neurol. Neurosurg. 2018, 174, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.J.; Yang, Y.H.; Lee, I.Y.; Chen, P.C.; Yang, J.T.; Wang, T.C.; Lin, M.H.; Yang, W.H.; Cheng, C.Y.; Chen, K.T.; et al. Effect of valproic acid on overall survival in patients with high-grade gliomas undergoing temozolomide: A nationwide population-based cohort study in Taiwan. Medicine 2020, 99, e21147. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Guan, S.; Yang, X.; Sun, S.; Huang, B.; Li, X. Administration of Valproic Acid Improves the Survival of Patients with Glioma Treated with Postoperative Radiotherapy. Oncol. Res. Treat. 2022, 45, 650–659. [Google Scholar] [CrossRef]
- Fisher, C.; Broderick, W. Sodium valproate or valproate semisodium: Is there a difference in the treatment of bipolar disorder? Psychiatr. Bull. 2003, 27, 446–448. [Google Scholar] [CrossRef]
- Krauze, A.V.; Megan, M.; Theresa, C.Z.; Peter, M.; Shih, J.H.; Tofilon, P.J.; Rowe, L.; Gilbert, M.; Camphausen, K. The addition of Valproic acid to concurrent radiation therapy and temozolomide improves patient outcome: A Correlative analysis of RTOG 0525, SEER and a Phase II NCI trial. Cancer Stud. Ther. 2020, 5. [Google Scholar] [CrossRef][Green Version]
- Krauze, A.V.; Mackey, M.; Rowe, L.; Chang, M.G.; Holdford, D.J.; Cooley, T.; Shih, J.; Tofilon, P.J.; Camphausen, K. Late toxicity in long-term survivors from a phase 2 study of concurrent radiation therapy, temozolomide and valproic acid for newly diagnosed glioblastoma. Neuro-Oncol. Pract. 2018, 5, 246–250. [Google Scholar] [CrossRef]
- Krauze, A.V.; Myrehaug, S.D.; Chang, M.G.; Holdford, D.J.; Smith, S.; Shih, J.; Tofilon, P.J.; Fine, H.A.; Camphausen, K. A Phase 2 Study of Concurrent Radiation Therapy, Temozolomide, and the Histone Deacetylase Inhibitor Valproic Acid for Patients With Glioblastoma. Int. J. Radiat. Oncol. Biol. Phys. 2015, 92, 986–992. [Google Scholar] [CrossRef]
- Su, J.M.; Murray, J.C.; McNall-Knapp, R.Y.; Bowers, D.C.; Shah, S.; Adesina, A.M.; Paulino, A.C.; Jo, E.; Mo, Q.; Baxter, P.A.; et al. A phase 2 study of valproic acid and radiation, followed by maintenance valproic acid and bevacizumab in children with newly diagnosed diffuse intrinsic pontine glioma or high-grade glioma. Pediatr. Blood Cancer 2020, 67, e28283. [Google Scholar] [CrossRef]
- Yuan, Y.; Xiang, W.; Qing, M.; Yanhui, L.; Jiewen, L.; Yunhe, M. Survival analysis for valproic acid use in adult glioblastoma multiforme: A meta-analysis of individual patient data and a systematic review. Seizure 2014, 23, 830–835. [Google Scholar] [CrossRef]
- Ochiai, S.; Nomoto, Y.; Yamashita, Y.; Watanabe, Y.; Toyomasu, Y.; Kawamura, T.; Takada, A.; Ii, N.; Kobayashi, S.; Sakuma, H. Roles of Valproic Acid in Improving Radiation Therapy for Glioblastoma: A Review of Literature Focusing on Clinical Evidence. Asian Pac. J. Cancer Prev. 2016, 17, 463–466. [Google Scholar] [CrossRef]
- Han, W.; Guan, W. Valproic Acid: A Promising Therapeutic Agent in Glioma Treatment. Front. Oncol. 2021, 11, 687362. [Google Scholar] [CrossRef]
- Osuka, S.; Takano, S.; Watanabe, S.; Ishikawa, E.; Yamamoto, T.; Matsumura, A. Valproic acid inhibits angiogenesis in vitro and glioma angiogenesis in vivo in the brain. Neurol. Med.-Chir. 2012, 52, 186–193. [Google Scholar] [CrossRef]
- Hoja, S.; Schulze, M.; Rehli, M.; Proescholdt, M.; Herold-Mende, C.; Hau, P.; Riemenschneider, M.J. Molecular dissection of the valproic acid effects on glioma cells. Oncotarget 2016, 7, 62989–63002. [Google Scholar] [CrossRef][Green Version]
- Han, W.; Yu, F.; Cao, J.; Dong, B.; Guan, W.; Shi, J. Valproic Acid Enhanced Apoptosis by Promoting Autophagy Via Akt/mTOR Signaling in Glioma. Cell Transpl. 2020, 29, 963689720981878. [Google Scholar] [CrossRef]
- Yang, Z.Y.; Wang, X.H. Valproic Acid Inhibits Glioma and Its Mechanisms. J. Healthc. Eng. 2022, 2022, 4985781. [Google Scholar] [CrossRef] [PubMed]
- Ryu, C.H.; Yoon, W.S.; Park, K.Y.; Kim, S.M.; Lim, J.Y.; Woo, J.S.; Jeong, C.H.; Hou, Y.; Jeun, S.S. Valproic acid downregulates the expression of MGMT and sensitizes temozolomide-resistant glioma cells. J. Biomed. Biotechnol. 2012, 2012, 987495. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.C.; Wei, K.C.; Chen, P.Y.; Huang, C.Y.; Chen, K.T.; Lin, Y.J.; Cheng, H.W.; Chen, Y.R.; Wang, H.T. Valproic Acid Enhanced Temozolomide-Induced Anticancer Activity in Human Glioma Through the p53-PUMA Apoptosis Pathway. Front. Oncol. 2021, 11, 722754. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, S.; Yuan, X.; Hu, Z.; Li, H.; Wu, M.; Yuan, J.; Zhao, Z.; Su, J.; Wang, X.; et al. Valproic Acid Promotes Human Glioma U87 Cells Apoptosis and Inhibits Glycogen Synthase Kinase-3beta Through ERK/Akt Signaling. Cell. Physiol. Biochem. 2016, 39, 2173–2185. [Google Scholar] [CrossRef]
- Riva, G.; Cilibrasi, C.; Bazzoni, R.; Cadamuro, M.; Negroni, C.; Butta, V.; Strazzabosco, M.; Dalpra, L.; Lavitrano, M.; Bentivegna, A. Valproic Acid Inhibits Proliferation and Reduces Invasiveness in Glioma Stem Cells Through Wnt/beta Catenin Signalling Activation. Genes 2018, 9, 522. [Google Scholar] [CrossRef]
- Riva, G.; Butta, V.; Cilibrasi, C.; Baronchelli, S.; Redaelli, S.; Dalpra, L.; Lavitrano, M.; Bentivegna, A. Epigenetic targeting of glioma stem cells: Short-term and long-term treatments with valproic acid modulate DNA methylation and differentiation behavior, but not temozolomide sensitivity. Oncol. Rep. 2016, 35, 2811–2824. [Google Scholar] [CrossRef]
- Happold, C.; Gorlia, T.; Chinot, O.; Gilbert, M.R.; Nabors, L.B.; Wick, W.; Pugh, S.L.; Hegi, M.; Cloughesy, T.; Roth, P.; et al. Does Valproic Acid or Levetiracetam Improve Survival in Glioblastoma? A Pooled Analysis of Prospective Clinical Trials in Newly Diagnosed Glioblastoma. J. Clin. Oncol. 2016, 34, 731–739. [Google Scholar] [CrossRef]
- Berendsen, S.; Varkila, M.; Kroonen, J.; Seute, T.; Snijders, T.J.; Kauw, F.; Spliet, W.G.; Willems, M.; Poulet, C.; Broekman, M.L.; et al. Prognostic relevance of epilepsy at presentation in glioblastoma patients. Neuro-Oncol. 2016, 18, 700–706. [Google Scholar] [CrossRef]
- Knudsen-Baas, K.M.; Storstein, A.M.; Zarabla, A.; Maialetti, A.; Giannarelli, D.; Beghi, E.; Maschio, M. Antiseizure medication in patients with Glioblastoma- a collaborative cohort study. Seizure 2021, 87, 107–113. [Google Scholar] [CrossRef]
- Krauze, A.V.; Sierk, M.; Nguyen, T.; Chen, Q.; Yan, C.; Hu, Y.; Jiang, W.; Tasci, E.; Cooley Zgela, T.; Sproull, M.; et al. Glioblastoma survival is associated with distinct proteomic alteration signatures post chemoirradiation in a large-scale proteomic panel. Front. Oncol. 2023, 13, 1127645. [Google Scholar] [CrossRef]
- Gold, L.; Walker, J.J.; Wilcox, S.K.; Williams, S. Advances in human proteomics at high scale with the SOMAscan proteomics platform. New Biotechnol. 2012, 29, 543–549. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Mirimanoff, R.O.; Gorlia, T.; Mason, W.; Van den Bent, M.J.; Kortmann, R.D.; Fisher, B.; Reni, M.; Brandes, A.A.; Curschmann, J.; Villa, S.; et al. Radiotherapy and temozolomide for newly diagnosed glioblastoma: Recursive partitioning analysis of the EORTC 26981/22981-NCIC CE3 phase III randomized trial. J. Clin. Oncol. 2006, 24, 2563–2569. [Google Scholar] [CrossRef] [PubMed]
- Hodapp, N. The ICRU Report 83: Prescribing, recording and reporting photon-beam intensity-modulated radiation therapy (IMRT). Strahlenther. Onkol. 2012, 188, 97–99. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat.Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Simon, R.; Lam, A.; Li, M.C.; Ngan, M.; Menenzes, S.; Zhao, Y. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform. 2007, 3, 11–17. [Google Scholar] [CrossRef]
- Dobbin, K.K.; Simon, R.M. Sample size planning for developing classifiers using high-dimensional DNA microarray data. Biostatistics 2006, 8, 101–117. [Google Scholar] [CrossRef] [PubMed]
- Dobbin, K.K.; Zhao, Y.; Simon, R.M. How large a training set is needed to develop a classifier for microarray data? Clin. Cancer Res. 2008, 14, 108–114. [Google Scholar] [CrossRef] [PubMed]
- R Core Team (2023). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. Available online: https://www.R-project.org/ (accessed on 22 September 2023).
- Tibshirani, R. The lasso method for variable selection in the Cox model. Stat. Med. 1997, 16, 385–395. [Google Scholar] [CrossRef]
- Simon, N.; Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Cox’s Proportional Hazards Model via Coordinate Descent. J. Stat. Softw. 2011, 39, 1–13. [Google Scholar] [CrossRef]
- Simon, R.M.; Subramanian, J.; Li, M.C.; Menezes, S. Using cross-validation to evaluate predictive accuracy of survival risk classifiers based on high-dimensional data. Brief. Bioinform. 2011, 12, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Janes, H.; Pepe, M.S.; Bossuyt, P.M.; Barlow, W.E. Measuring the performance of markers for guiding treatment decisions. Ann. Intern. Med. 2011, 154, 253–259. [Google Scholar] [CrossRef]
- Stupp, R.; Taillibert, S.; Kanner, A.; Read, W.; Steinberg, D.M.; Lhermitte, B.; Toms, S.; Idbaih, A.; Ahluwalia, M.S.; Fink, K.; et al. Effect of Tumor-Treating Fields Plus Maintenance Temozolomide vs Maintenance Temozolomide Alone on Survival in Patients With Glioblastoma: A Randomized Clinical Trial. JAMA 2017, 318, 2306–2316. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Hegi, M.E.; Gorlia, T.; Erridge, S.C.; Perry, J.; Hong, Y.K.; Aldape, K.D.; Lhermitte, B.; Pietsch, T.; Grujicic, D.; et al. Cilengitide combined with standard treatment for patients with newly diagnosed glioblastoma with methylated MGMT promoter (CENTRIC EORTC 26071-22072 study): A multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2014, 15, 1100–1108. [Google Scholar] [CrossRef]
- Weller, M.; Butowski, N.; Tran, D.D.; Recht, L.D.; Lim, M.; Hirte, H.; Ashby, L.; Mechtler, L.; Goldlust, S.A.; Iwamoto, F.; et al. Rindopepimut with temozolomide for patients with newly diagnosed, EGFRvIII-expressing glioblastoma (ACT IV): A randomised, double-blind, international phase 3 trial. Lancet Oncol. 2017, 18, 1373–1385. [Google Scholar] [CrossRef]
- Kim, H.K.; Choi, I.J.; Kim, C.G.; Kim, H.S.; Oshima, A.; Yamada, Y.; Arao, T.; Nishio, K.; Michalowski, A.; Green, J.E. Three-gene predictor of clinical outcome for gastric cancer patients treated with chemotherapy. Pharmacogenomics J. 2012, 12, 119–127. [Google Scholar] [CrossRef]
- Simon, R.; Radmacher, M.D.; Dobbin, K. Design of studies using DNA microarrays. Genet. Epidemiol. 2002, 23, 21–36. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.R.; Yeh, C.T. GALNT14: An Emerging Marker Capable of Predicting Therapeutic Outcomes in Multiple Cancers. Int. J. Mol. Sci. 2020, 21, 1491. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.R.; Hoessli, D.C.; Fang, M. N-acetylgalactosaminyltransferases in cancer. Oncotarget 2016, 7, 54067–54081. [Google Scholar] [CrossRef] [PubMed]
- Wagner, K.W.; Punnoose, E.A.; Januario, T.; Lawrence, D.A.; Pitti, R.M.; Lancaster, K.; Lee, D.; von Goetz, M.; Yee, S.F.; Totpal, K.; et al. Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat. Med. 2007, 13, 1070–1077. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Xing, H.; Kim, T.M.; Jung, Y.; Huang, W.; Yang, H.W.; Song, S.; Park, P.J.; Carroll, R.S.; Johnson, M.D. Numb regulates glioma stem cell fate and growth by altering epidermal growth factor receptor and Skp1-Cullin-F-box ubiquitin ligase activity. Stem Cells 2012, 30, 1313–1326. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.B.; Karpova, A.; Gritsenko, M.A.; Kyle, J.E.; Cao, S.; Li, Y.; Rykunov, D.; Colaprico, A.; Rothstein, J.H.; Hong, R.; et al. Proteogenomic and metabolomic characterization of human glioblastoma. Cancer Cell 2021, 39, 509–528.e520. [Google Scholar] [CrossRef] [PubMed]
- Pridham, K.J.; Varghese, R.T.; Sheng, Z. The Role of Class IA Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunits in Glioblastoma. Front. Oncol. 2017, 7, 312. [Google Scholar] [CrossRef]
- Franco, C.; Kausar, S.; Silva, M.F.B.; Guedes, R.C.; Falcao, A.O.; Brito, M.A. Multi-Targeting Approach in Glioblastoma Using Computer-Assisted Drug Discovery Tools to Overcome the Blood-Brain Barrier and Target EGFR/PI3Kp110β Signaling. Cancers 2022, 14, 3506. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Wang, W.F.; Xie, S.; Zhang, X.L.; Qi, W.F.; Zhou, X.P.; Hu, J.X.; Shi, Q.; Yu, R.T. β-transducin repeat-containing E3 ubiquitin protein ligase inhibits migration, invasion and proliferation of glioma cells. Oncol. Lett. 2017, 14, 3131–3135. [Google Scholar] [CrossRef]
- Wang, H.; Pan, J.Q.; Luo, L.; Ning, X.J.; Ye, Z.P.; Yu, Z.; Li, W.S. NF-κB induces miR-148a to sustain TGF-β/Smad signaling activation in glioblastoma. Mol. Cancer 2015, 14, 2. [Google Scholar] [CrossRef]
- Thompson, L.L.; Baergen, A.K.; Lichtensztejn, Z.; McManus, K.J. Reduced SKP1 Expression Induces Chromosome Instability through Aberrant Cyclin E1 Protein Turnover. Cancers 2020, 12, 531. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Gao, P.; Dai, X.; Ye, L.; Wang, Z.; Cheng, H. New prognostic biomarker CMTM3 in low grade glioma and its immune infiltration. Ann. Transl. Med. 2022, 10, 206. [Google Scholar] [CrossRef] [PubMed]
- Diggins, N.L.; Webb, D.J. APPL1 is a multifunctional endosomal signaling adaptor protein. Biochem. Soc. Trans. 2017, 45, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Korbecki, J.; Kojder, K.; Simińska, D.; Bohatyrewicz, R.; Gutowska, I.; Chlubek, D.; Baranowska-Bosiacka, I. CC Chemokines in a Tumor: A Review of Pro-Cancer and Anti-Cancer Properties of the Ligands of Receptors CCR1, CCR2, CCR3, and CCR4. Int. J. Mol. Sci. 2020, 21, 8412. [Google Scholar] [CrossRef]
- Autelitano, F.; Loyaux, D.; Roudières, S.; Déon, C.; Guette, F.; Fabre, P.; Ping, Q.; Wang, S.; Auvergne, R.; Badarinarayana, V.; et al. Identification of novel tumor-associated cell surface sialoglycoproteins in human glioblastoma tumors using quantitative proteomics. PLoS ONE 2014, 9, e110316. [Google Scholar] [CrossRef]
- de Vega, S.; Kondo, A.; Suzuki, M.; Arai, H.; Jiapaer, S.; Sabit, H.; Nakada, M.; Ikeuchi, T.; Ishijima, M.; Arikawa-Hirasawa, E.; et al. Fibulin-7 is overexpressed in glioblastomas and modulates glioblastoma neovascularization through interaction with angiopoietin-1. Int. J. Cancer 2019, 145, 2157–2169. [Google Scholar] [CrossRef]
- Thotala, D.; Karvas, R.M.; Engelbach, J.A.; Garbow, J.R.; Hallahan, A.N.; DeWees, T.A.; Laszlo, A.; Hallahan, D.E. Valproic acid enhances the efficacy of radiation therapy by protecting normal hippocampal neurons and sensitizing malignant glioblastoma cells. Oncotarget 2015, 6, 35004–35022. [Google Scholar] [CrossRef]
- Raja, E.; Komuro, A.; Tanabe, R.; Sakai, S.; Ino, Y.; Saito, N.; Todo, T.; Morikawa, M.; Aburatani, H.; Koinuma, D.; et al. Bone morphogenetic protein signaling mediated by ALK-2 and DLX2 regulates apoptosis in glioma-initiating cells. Oncogene 2017, 36, 4963–4974. [Google Scholar] [CrossRef]
- Talwadekar, M.; Fernandes, S.; Kale, V.; Limaye, L. Valproic acid enhances the neural differentiation of human placenta derived-mesenchymal stem cells in vitro. J. Tissue Eng. Regen. Med. 2017, 11, 3111–3123. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Tasci, E.; Zhuge, Y.; Camphausen, K.; Krauze, A.V. Bias and Class Imbalance in Oncologic Data-Towards Inclusive and Transferrable AI in Large Scale Oncology Data Sets. Cancers 2022, 14, 2897. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Ge, M.; Xia, L.; Shan, G.; Qian, J. CTSV (cathepsin V) promotes bladder cancer progression by increasing NF-κB activity. Bioengineered 2022, 13, 10180–10190. [Google Scholar] [CrossRef]
- Mitrović, A.; Senjor, E.; Jukić, M.; Bolčina, L.; Prunk, M.; Proj, M.; Nanut, M.P.; Gobec, S.; Kosa, J. New inhibitors of cathepsin V impair tumor cell proliferation and elastin degradation and increase immune cell cytotoxicity. Comput. Struct. Biotechnol. J. 2022, 20, 4667–4687. [Google Scholar] [CrossRef]
- Gao, L.; Xiong, D.-D.; Yang, X.; Li, J.-D.; He, R.-Q.; Huang, Z.-G.; Lai, Z.F.; Liu, L.-M.; Luo, J.-Y.; Du, X.F.; et al. The expression characteristics and clinical significance of ACP6, a potential target of nitidine chloride, in hepatocellular carcinoma. BMC Cancer 2022, 22, 1244. [Google Scholar] [CrossRef]
- Berezovsky, A.D.; Poisson, L.M.; Cherba, D.; Webb, C.P.; Transou, A.D.; Lemke, N.W.; Hong, X.; Hasselbach, L.A.; Irtenkauf, S.M.; Mikkelsen, T.; et al. Sox2 promotes malignancy in glioblastoma by regulating plasticity and astrocytic differentiation. Neoplasia 2014, 16, 193–206. [Google Scholar] [CrossRef][Green Version]
- Ehata, S.; Miyazono, K. Bone Morphogenetic Protein Signaling in Cancer; Some Topics in the Recent 10 Years. Front. Cell Dev. Biol. 2022, 10, 883523. [Google Scholar] [CrossRef]
- Sachdeva, R.; Wu, M.; Johnson, K.; Kim, H.; Celebre, A.; Shahzad, U.; Graham, M.S.; Kessler, J.A.; Chuang, J.H.; Karamchandani, K.; et al. BMP signaling mediates glioma stem cell quiescence and confers treatment resistance in glioblastoma. Sci. Rep. 2019, 9, 14569. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.-Q.; Ming, S.-L.; Wu, H.-T.; Zeng, L.; Ba, G.; Li, J.; Lu, W.-F.; Han, J.; Du, Q.-J.; Sun, M.-M.; et al. Myostatin knockout induces apoptosis in human cervical cancer cells via elevated reactive oxygen species generation. Redox Biol. 2018, 19, 412–428. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Li, Z.; Fu, J.; Zhou, R. Growth and differentiation factor 15 regulates PD-L1 expression in glioblastoma. Cancer Manag. Res. 2019, 11, 2653–2661. [Google Scholar] [CrossRef]
- Mir, M.A.; Pandith, A.A.; Mansoor, S.; Baba, S.M.; Makhdoomi, R.; Ain, Q.-U.; Anwar, I.; Para, S.A.; Bhat, A.H.; Koul, A.M.; et al. Differential expression of SLITRK6 gene as a potential therapeutic target for urothelial carcinoma in particular upper tract cancer. Gene 2023, 878, 147583. [Google Scholar] [CrossRef]
- Aruga, J.; Yokota, N.; Mikoshiba, K. Human SLITRK family genes: Genomic organization and expression profiling in normal brain and brain tumor tissue. Gene 2003, 315, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Kammerer, S.; Roth, R.B.; Reneland, R.; Marnellos, G.; Hoyal, C.R.; Markward, N.J.; Ebner, F.; Kiechle, M.; Schwarz-Boeger, U.; Griffiths, L.R.; et al. Large-scale association study identifies ICAM gene region as breast and prostate cancer susceptibility locus. Cancer Res. 2004, 64, 8906–8910. [Google Scholar] [CrossRef]
- Shen, C.K.; Shen, C.-K.; Huang, B.-R.; Yeh, W.-L.; Chen, C.-W.; Liu, Y.-S.; Lai, S.-W.; Tseng, W.P.; Lu, D.-Y.; Tsai, C.-F. Regulatory effects of IL-1β in the interaction of GBM and tumor-associated monocyte through VCAM-1 and ICAM-1. Eur. J. Pharmacol. 2021, 905, 174216. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, X.; Zou, H.; Dai, Z.; Feng, S.; Zhang, M.; Xiao, G.; Liu, Z.; Cheng, Q. The Basic Characteristics of the Pentraxin Family and Their Functions in Tumor Progression. Front. Immunol. 2020, 11, 1757. [Google Scholar] [CrossRef]
- Gómez de San José, N.; Massa, F.; Halbgebauer, S.; Oeckl, P.; Steinacker, P.; Otto, M. Neuronal pentraxins as biomarkers of synaptic activity: From physiological functions to pathological changes in neurodegeneration. J. Neural. Transm. 2022, 129, 207–230. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Shin, D.Y.; Eiseman, M.; Yallowitz, A.R.; Li, N.; Lalani, S.; Li, Z.; Cung, M.; Bok, S.; Debnath, S.; et al. SLITRK5 is a negative regulator of hedgehog signaling in osteoblasts. Nat. Commun. 2021, 12, 4611. [Google Scholar] [CrossRef]
- Hiz, S.; Kiliç, S.; Bademci, G.; Karakulak, T.; Erdoğan, A.; Özden, B.; Eresen, C.; Erdal, E.; Yiş, U.; Tekin, M.; et al. VARS1 mutations associated with neurodevelopmental disorder are located on a short amino acid stretch of the anticodon-binding domain. Turk. J. Biol. 2022, 46, 458–464. [Google Scholar] [CrossRef]
- Arnoult, N.; Correia, A.; Ma, J.; Merlo, A.; Garcia-Gomez, S.; Maric, M.; Tognetti, M.; Benner, C.W.; Boulton, S.J.; Saghatelian, A.; et al. Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN. Nature 2017, 549, 548–552. [Google Scholar] [CrossRef]
- Ahmed, E.A.; Rosemann, M.; Scherthan, H. NHEJ Contributes to the Fast Repair of Radiation-induced DNA Double-strand Breaks at Late Prophase I Telomeres. Health Phys. 2018, 115, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Jia, L.; Yang, A.G. Roles of HLA-G/KIR2DL4 in Breast Cancer Immune Microenvironment. Front. Immunol. 2022, 13, 791975. [Google Scholar] [CrossRef]
- Fabian, K.P.; Hodge, J.W. The emerging role of off-the-shelf engineered natural killer cells in targeted cancer immunotherapy. Mol. Ther. Oncolytics 2021, 23, 266–276. [Google Scholar] [CrossRef] [PubMed]
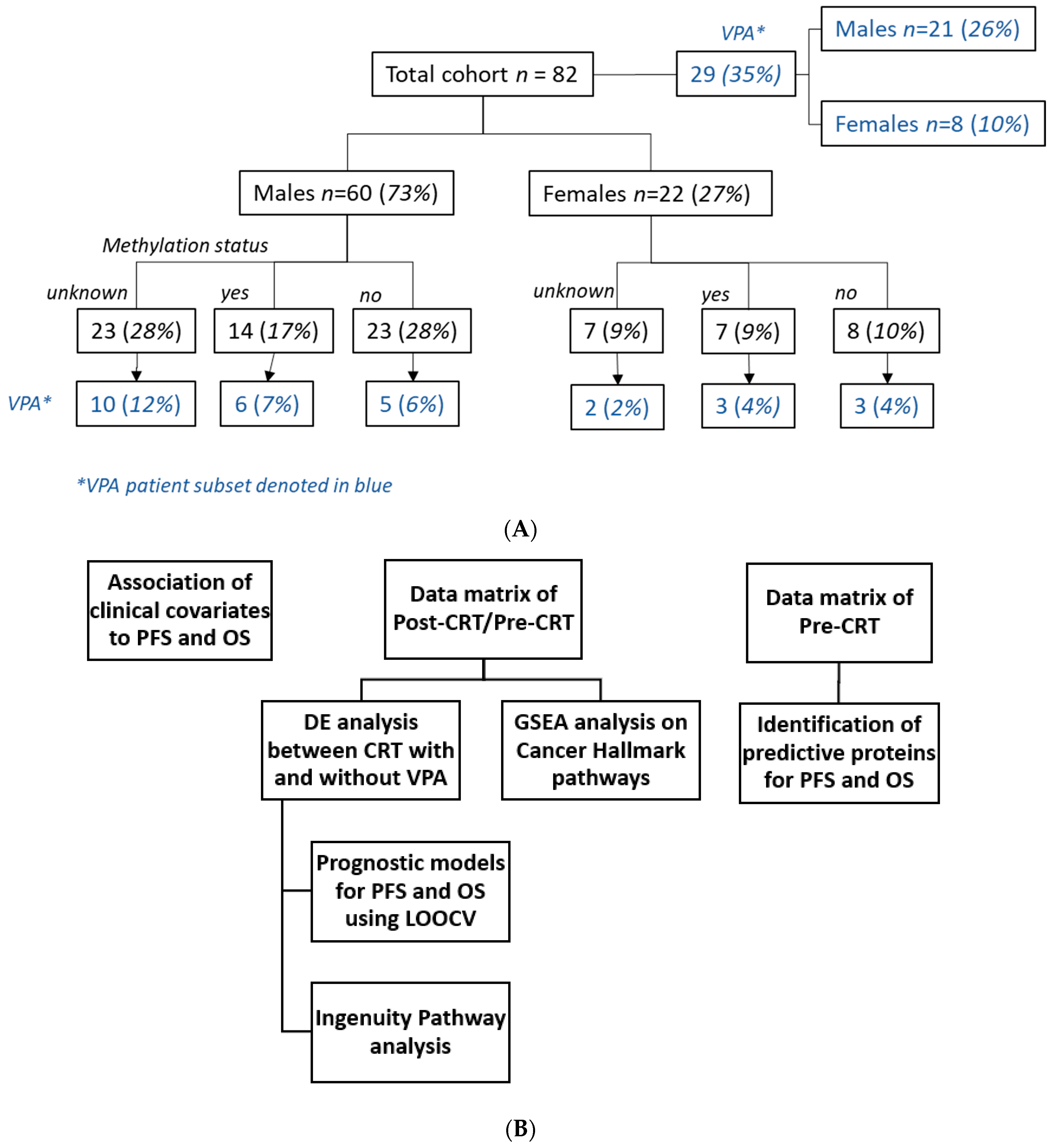


| VPA Administration | No | Yes | p-Value |
|---|---|---|---|
| Total cohort n = 82 (%) | 53 (64.6) | 29 (35.4) | |
| Age (mean (SD)) | 58.06 (10.54) | 52.34 (9.60) | 0.018 |
| Age Range | 31–70 | 31–71 | |
| Gender = Male (%) | 39 (73.6) | 21 (72.4) | 1 |
| Location | |||
| Periventricular or Cortical = Periventricular (%) | 22 (41.5) | 6 (20.7) | 0.097 |
| Hemisphere | 0.614 | ||
| Left | 23 (43.4) | 15 (51.7) | |
| Right | 29 (54.7) | 14 (48.3) | |
| Both | 1 (1.9) | 0 (0.0) | |
| Extent of Resection | 0.04 | ||
| GTR | 15 (28.3) | 16 (55.2) | |
| STR | 31 (58.5) | 12 (41.4) | |
| Biopsy | 7 (13.2) | 1 (3.4) | |
| MGMT status | 0.361 | ||
| methylated | 12 (22.6) | 9 (31.0) | |
| unmethylated | 23 (43.4) | 8 (27.6) | |
| unknown | 18 (34.0) | 12 (41.4) | |
| KPS | 0.014 | ||
| 60–80 | 14 (26.4) | 3 (10.3) | |
| 90 | 24 (45.3) | 12 (41.4) | |
| 100 | 10 (18.9) | 14 (48.3) | |
| Unknown | 5 (9.4) | 0 (0.0) | |
| RPA | 0.019 | ||
| 3 | 5 (9.4) | 9 (31.0) | |
| 4 | 29 (54.7) | 17 (58.6) | |
| 5 | 16 (30.2) | 3 (10.3) | |
| Unknown | 3 (5.7) | 0 (0.0) | |
| RT volumes | |||
| GTV T1 (cc) | 0.437 | ||
| <20 cc | 13 (24.5) | 11 (37.9) | |
| 20–40 cc | 19 (35.8) | 9 (31.0) | |
| >40 cc | 21 (39.6) | 9 (31.0) | |
| GTV T2 (cc) | 0.152 | ||
| <10 cc | 8 (15.1) | 0 (0.0) | |
| 10–50 cc | 13 (24.5) | 8 (27.6) | |
| 50–100 cc | 15 (28.3) | 8 (27.6) | |
| >100 cc | 17 (32.1) | 13 (44.8) | |
| RT Technique | <0.001 | ||
| 3D | 15 (28.3) | 9 (31.0) | |
| IMRT | 18 (34.0) | 20 (69.0) | |
| Arc | 20 (37.7) | 0 (0.0) |
| Class Comparison | Class 0 | Class 1 | Adjusted by | # of Significant Proteins (p < 0.001) |
|---|---|---|---|---|
| VPA | VPA = 0 (53) | VPA = 1 (29) | 124 | |
| VPA | VPA = 0 (53) | VPA = 1 (29) | age (>65) | 109 |
| VPA | VPA = 0 (53) | VPA = 1 (29) | sex | 128 |
| VPA | VPA = 0 (53) | VPA = 1 (29) | GTV-T1 (median) | 127 |
| VPA | VPA = 0 (53) | VPA = 1 (29) | Resection type | 134 |
| VPA | VPA = 0 (53) | VPA = 1 (29) | KPS | 110 |
| VPA | VPA = 0 (53) | VPA = 1 (29) | RPA | 108 |
| VPA | VPA = 0 (53) | VPA = 1 (29) | Radiation Technique | 89 |
| VPA | VPA = 0 (53) | VPA = 1 (29) | GTV-V2 (median) | 125 |
| VPA | VPA = 0 (53) | VPA = 1 (29) | Infiltration | 104 |
| VPA | VPA = 0 (52) | VPA = 1 (29) | Hemisphere | 120 |
| VPA | VPA = 0 (51) | VPA = 1 (28) | BMI (median) | 114 |
| VPA | VPA = 0 (53) | VPA = 1 (29) | Location | 83 |
| VPA with known MGMT | VPA = 0 (35) | VPA = 1 (17) | 57 | |
| VPA with known MGMT | VPA = 0 (35) | VPA = 1 (17) | MGMT | 59 |
| Symbol | Name | EntrezID | Fold-Change | Mean log2(COT/PRE) | OS p-Value | OS FDR | OS HR | PFS p-Value | PFS FDR | PFS HR |
|---|---|---|---|---|---|---|---|---|---|---|
| CCL17 | C-C motif chemokine ligand 17 | 6361 | 1.620 | −0.004 | 0.003 | 0.198 | 1.513 | 0.024 | 0.232 | 1.363 |
| GALNT14 | polypeptide N-acetylgalactosaminyltransferase 14 | 79623 | 0.780 | 0.186 | 0.003 | 0.198 | 0.407 | 0.004 | 0.232 | 0.388 |
| CTSV | chathepsin V | 1515 | 1.160 | −0.088 | 0.009 | 0.285 | 3.647 | 0.024 | 0.232 | 3.031 |
| ACP6 | acid phosphatase 6, lysophosphatidic | 51205 | 0.810 | 0.184 | 0.011 | 0.285 | 0.408 | 0.007 | 0.232 | 0.378 |
| BMP6 | bone morphogenetic protein 6 | 654 | 0.850 | 0.134 | 0.012 | 0.285 | 0.344 | 0.027 | 0.240 | 0.369 |
| MSTN | myostatin | 2660 | 0.850 | 0.089 | 0.019 | 0.304 | 0.277 | 0.093 | 0.339 | 0.424 |
| SLITRK6 | SLIT and NTRK like family protein 6 | 84189 | 0.770 | 0.116 | 0.021 | 0.304 | 0.494 | 0.194 | 0.496 | 0.729 |
| ICAM4 | intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) | 3386 | 1.240 | −0.267 | 0.027 | 0.304 | 2.002 | 0.039 | 0.246 | 1.806 |
| NPTX1 | neuronal pentraxin 1 | 4884 | 0.870 | 0.050 | 0.029 | 0.304 | 0.286 | 0.280 | 0.504 | 0.615 |
| SLITRK5 | SLIT and NTRK like family protein 5 | 26050 | 0.720 | 0.229 | 0.031 | 0.304 | 0.542 | 0.013 | 0.232 | 0.475 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krauze, A.V.; Zhao, Y.; Li, M.-C.; Shih, J.; Jiang, W.; Tasci, E.; Cooley Zgela, T.; Sproull, M.; Mackey, M.; Shankavaram, U.; et al. Revisiting Concurrent Radiation Therapy, Temozolomide, and the Histone Deacetylase Inhibitor Valproic Acid for Patients with Glioblastoma—Proteomic Alteration and Comparison Analysis with the Standard-of-Care Chemoirradiation. Biomolecules 2023, 13, 1499. https://doi.org/10.3390/biom13101499
Krauze AV, Zhao Y, Li M-C, Shih J, Jiang W, Tasci E, Cooley Zgela T, Sproull M, Mackey M, Shankavaram U, et al. Revisiting Concurrent Radiation Therapy, Temozolomide, and the Histone Deacetylase Inhibitor Valproic Acid for Patients with Glioblastoma—Proteomic Alteration and Comparison Analysis with the Standard-of-Care Chemoirradiation. Biomolecules. 2023; 13(10):1499. https://doi.org/10.3390/biom13101499
Chicago/Turabian StyleKrauze, Andra V., Yingdong Zhao, Ming-Chung Li, Joanna Shih, Will Jiang, Erdal Tasci, Theresa Cooley Zgela, Mary Sproull, Megan Mackey, Uma Shankavaram, and et al. 2023. "Revisiting Concurrent Radiation Therapy, Temozolomide, and the Histone Deacetylase Inhibitor Valproic Acid for Patients with Glioblastoma—Proteomic Alteration and Comparison Analysis with the Standard-of-Care Chemoirradiation" Biomolecules 13, no. 10: 1499. https://doi.org/10.3390/biom13101499
APA StyleKrauze, A. V., Zhao, Y., Li, M.-C., Shih, J., Jiang, W., Tasci, E., Cooley Zgela, T., Sproull, M., Mackey, M., Shankavaram, U., Tofilon, P., & Camphausen, K. (2023). Revisiting Concurrent Radiation Therapy, Temozolomide, and the Histone Deacetylase Inhibitor Valproic Acid for Patients with Glioblastoma—Proteomic Alteration and Comparison Analysis with the Standard-of-Care Chemoirradiation. Biomolecules, 13(10), 1499. https://doi.org/10.3390/biom13101499







