Elucidation of the Correlation between Heme Distortion and Tertiary Structure of the Heme-Binding Pocket Using a Convolutional Neural Network
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collation on Heme Proteins and Dataset Preparation for Deep Learning
2.2. CNN Model
- 1.
- A subset was split into validation and test datasets at a ratio of 0.2:0.8.
- 2.
- The model was trained using the remaining four subsets (training set) for 300 epochs. (In the training process, a network is trained to reduce the loss between the predicted and observed values by using an optimizer. The number of epochs indicates the number of times that training is carried out for the entire training dataset.)
- 3.
- The model with the minimum value of loss, calculated as the mean-square error, in the validation dataset was selected.
- 4.
- The resulting model was validated on the test dataset; prediction was performed by using the resulting model on the test dataset.
2.3. Clustering and Principal Component Analyses of Heme-Binding Pockets
2.4. Alignment of Amino Acid Sequences of Heme Proteins
3. Results and Discussion
3.1. Prediction of Heme Distortion from the Tertiary Structure of the Heme-Binding Pocket Using a CNN Model
3.2. Differences in the Importance of Information Included in Subsets of Input Data
3.3. Similarity of the Structure of Heme-Binding Pockets and Hemes
3.4. Similarity of the Structures of Heme-Binding Pockets between Protein Chains with Similar Amino Acid Sequences
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Poulos, T.L. The Janus Nature of Heme. Nat. Prod. Rep. 2007, 24, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Louie, G.V.; Brayer, G.D. High-Resolution Refinement of Yeast Iso-1-Cytochrome c and Comparisons with Other Eukaryotic Cytochromes C. J. Mol. Biol. 1990, 214, 527–555. [Google Scholar] [CrossRef]
- Shaik, S.; Kumar, D.; de Visser, S.P.; Altun, A.; Thiel, W. Theoretical Perspective on the Structure and Mechanism of Cytochrome P450 Enzymes. Chem. Rev. 2005, 105, 2279–2328. [Google Scholar] [CrossRef] [PubMed]
- Ostermeier, C. Cytochrome c Oxidase. Curr. Opin. Struct. Biol. 1996, 6, 460–466. [Google Scholar] [CrossRef]
- Perutz, M.F.; Rossmann, M.G.; Cullis, A.F.; Muirhead, H.; Will, G.; North, A.C.T. Structure of Hæmoglobin: A Three-Dimensional Fourier Synthesis at 5.5-Å. Resolution, Obtained by X-Ray Analysis. Nature 1960, 185, 416–422. [Google Scholar] [CrossRef]
- Kendrew, J.C.; Dickerson, R.E.; Strandberg, B.E.; Hart, R.G.; Davies, D.R.; Phillips, D.C.; Shore, V.C. Structure of Myoglobin: A Three-Dimensional Fourier Synthesis at 2 Å. Resolution. Nature 1960, 185, 422–427. [Google Scholar] [CrossRef]
- Faller, M.; Matsunaga, M.; Yin, S.; Loo, J.A.; Guo, F. Heme Is Involved in MicroRNA Processing. Nat. Struct. Mol. Biol. 2007, 14, 23–29. [Google Scholar] [CrossRef]
- Sun, J.; Hoshino, H.; Takaku, K.; Nakajima, O.; Muto, A.; Suzuki, H.; Tashiro, S.; Takahashi, S.; Shibahara, S.; Alam, J.; et al. Hemoprotein Bach1 Regulates Enhancer Availability of Heme Oxygenase-1 Gene. EMBO J. 2002, 21, 5216–5224. [Google Scholar] [CrossRef]
- Liu, H.-L.; Zhou, H.-N.; Xing, W.-M.; Zhao, J.-F.; Li, S.-X.; Huang, J.-F.; Bi, R.-C. 2.6 Å Resolution Crystal Structure of the Bacterioferritin from Azotobacter Vinelandii. FEBS Lett. 2004, 573, 93–98. [Google Scholar] [CrossRef]
- Bateman, T.J.; Shah, M.; Ho, T.P.; Shin, H.E.; Pan, C.; Harris, G.; Fegan, J.E.; Islam, E.A.; Ahn, S.K.; Hooda, Y.; et al. A Slam-Dependent Hemophore Contributes to Heme Acquisition in the Bacterial Pathogen Acinetobacter Baumannii. Nat. Commun. 2021, 12, 6270. [Google Scholar] [CrossRef]
- Reedy, C.J.; Elvekrog, M.M.; Gibney, B.R. Development of a Heme Protein Structure Electrochemical Function Database. Nucleic Acids Res. 2007, 36, D307–D313. [Google Scholar] [CrossRef] [PubMed]
- Kondo, H.X.; Kanematsu, Y.; Masumoto, G.; Takano, Y. PyDISH: Database and Analysis Tools for Heme Porphyrin Distortion in Heme Proteins. Database 2020, 2020, baaa066. [Google Scholar] [CrossRef] [PubMed]
- Rydberg, P.; Sigfridsson, E.; Ryde, U. On the Role of the Axial Ligand in Heme Proteins: A Theoretical Study. J. Biol. Inorg. Chem. 2004, 9, 203–223. [Google Scholar] [CrossRef] [PubMed]
- Walker, F.A. Magnetic Spectroscopic (EPR, ESEEM, Mossbauer, MCD and NMR) Studies of Low-Spin Ferriheme Centers and Their Corresponding Heme Proteins. Coord. Chem. Rev. 1999, 185–186, 471–534. [Google Scholar] [CrossRef]
- Takano, Y.; Nakamura, H. Density Functional Study of Roles of Porphyrin Ring in Electronic Structures of Heme. Int. J. Quantum Chem. 2009, 109, 3583–3591. [Google Scholar] [CrossRef]
- Takano, Y.; Kondo, H.X.; Kanematsu, Y.; Imada, Y. Computational Study of Distortion Effect of Fe-Porphyrin Found as a Biological Active Site. Jpn. J. Appl. Phys. 2020, 59, 010502. [Google Scholar] [CrossRef]
- Jentzen, W.; Song, X.Z.; Shelnutt, J.A. Structural Characterization of Synthetic and Protein-Bound Porphyrins in Terms of the Lowest-Frequency Normal Coordinates of the Macrocycle. J. Phys. Chem. B 1997, 101, 1684–1699. [Google Scholar] [CrossRef]
- Bikiel, D.E.; Forti, F.; Boechi, L.; Nardini, M.; Luque, F.J.; Martí, M.A.; Estrin, D.A. Role of Heme Distortion on Oxygen Affinity in Heme Proteins: The Protoglobin Case. J. Phys. Chem. B 2010, 114, 8536–8543. [Google Scholar] [CrossRef]
- Sun, Y.; Benabbas, A.; Zeng, W.; Kleingardner, J.G.; Bren, K.L.; Champion, P.M. Investigations of Heme Distortion, Low-Frequency Vibrational Excitations, and Electron Transfer in Cytochrome C. Proc. Natl. Acad. Sci. USA 2014, 111, 6570–6575. [Google Scholar] [CrossRef]
- Imada, Y.; Nakamura, H.; Takano, Y. Density Functional Study of Porphyrin Distortion Effects on Redox Potential of Heme. J. Comput. Chem. 2018, 39, 143–150. [Google Scholar] [CrossRef]
- Kanematsu, Y.; Kondo, H.X.; Imada, Y.; Takano, Y. Statistical and Quantum-Chemical Analysis of the Effect of Heme Porphyrin Distortion in Heme Proteins: Differences between Oxidoreductases and Oxygen Carrier Proteins. Chem. Phys. Lett. 2018, 710, 108–112. [Google Scholar] [CrossRef]
- Kondo, H.X.; Takano, Y. Analysis of Fluctuation in the Heme-Binding Pocket and Heme Distortion in Hemoglobin and Myoglobin. Life 2022, 12, 210. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Bonkovsky, H.L.; Guo, J. Structural Analysis of Heme Proteins: Implications for Design and Prediction. BMC Struct. Biol. 2011, 11, 13. [Google Scholar] [CrossRef]
- Kondo, H.X.; Kanematsu, Y.; Takano, Y. Structure of Heme-Binding Pocket in Heme Protein Is Generally Rigid and Can Be Predicted by AlphaFold2. Chem. Lett. 2022, 51, 704–708. [Google Scholar] [CrossRef]
- Sacquin-Mora, S.; Lavery, R. Investigating the Local Flexibility of Functional Residues in Hemoproteins. Biophys. J. 2006, 90, 2706–2717. [Google Scholar] [CrossRef] [PubMed]
- Kondo, H.X.; Fujii, M.; Tanioka, T.; Kanematsu, Y.; Yoshida, T.; Takano, Y. Global Analysis of Heme Proteins Elucidates the Correlation between Heme Distortion and the Heme-Binding Pocket. J. Chem. Inf. Model. 2022, 62, 775–784. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Advances in Neural Information Processing Systems; Pereira, F., Burges, C.J., Bottou, L., Weinberger, K.Q., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2012; Volume 25. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015. [Google Scholar] [CrossRef]
- Kinjo, A.R.; Suzuki, H.; Yamashita, R.; Ikegawa, Y.; Kudou, T.; Igarashi, R.; Kengaku, Y.; Cho, H.; Standley, D.M.; Nakagawa, A.; et al. Protein Data Bank Japan (PDBj): Maintaining a Structural Data Archive and Resource Description Framework Format. Nucleic Acids Res. 2012, 40, D453–D460. [Google Scholar] [CrossRef]
- Kinjo, A.R.; Yamashita, R.; Nakamura, H. PDBj Mine: Design and Implementation of Relational Database Interface for Protein Data Bank Japan. Database 2010, 2010, baq021. [Google Scholar] [CrossRef][Green Version]
- Hamelryck, T.; Manderick, B. PDB File Parser and Structure Class Implemented in Python. Bioinformatics 2003, 19, 2308–2310. [Google Scholar] [CrossRef]
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely Available Python Tools for Computational Molecular Biology and Bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- McGibbon, R.T.; Beauchamp, K.A.; Harrigan, M.P.; Klein, C.; Swails, J.M.; Hernández, C.X.; Schwantes, C.R.; Wang, L.P.; Lane, T.J.; Pande, V.S. MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophys. J. 2015, 109, 1528–1532. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Dunbrack, R.L. PISCES: A Protein Sequence Culling Server. Bioinformatics 2003, 19, 1589–1591. [Google Scholar] [CrossRef] [PubMed]
- Adamo, C.; Barone, V. Toward Reliable Density Functional Methods without Adjustable Parameters: The PBE0 Model. J. Chem. Phys. 1999, 110, 6158–6170. [Google Scholar] [CrossRef]
- Ditchfield, R.; Hehre, W.J.; Pople, J.A. Self-Consistent Molecular-Orbital Methods. IX. An Extended Gaussian-Type Basis for Molecular-Orbital Studies of Organic Molecules. J. Chem. Phys. 1971, 54, 724–728. [Google Scholar] [CrossRef]
- Hariharan, P.C.; Pople, J.A. The Influence of Polarization Functions on Molecular Orbital Hydrogenation Energies. Theor. Chim. Acta 1973, 28, 213–222. [Google Scholar] [CrossRef]
- Rassolov, V.A.; Pople, J.A.; Ratner, M.A.; Windus, T.L. 6-31G* Basis Set for Atoms K through Zn. J. Chem. Phys. 1998, 109, 1223–1229. [Google Scholar] [CrossRef]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Advances in Neural Information Processing Systems 32; Wallach, H., Larochelle, H., Beygelzimer, A., Alché-Buc, F., Fox, E., Garnett, R., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2019; pp. 8024–8035. [Google Scholar]
- Kingma, P.D.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014. [Google Scholar] [CrossRef]
- Wagner, J.R.; Sørensen, J.; Hensley, N.; Wong, C.; Zhu, C.; Perison, T.; Amaro, R.E. POVME 3.0: Software for Mapping Binding Pocket Flexibility. J. Chem. Theory Comput. 2017, 13, 4584–4592. [Google Scholar] [CrossRef]
- Case, D.A.; Ben-Shalom, I.Y.; Brozell, S.; Cerutti, D.S.; Cheatham, T.E.; Cruzeiro, V.W.D., III; Darden, T.A.; Duke, R.E.; Ghoreishi, D.; Giambasu, G.; et al. AMBER 2019; University of California: San Francisco, CA, USA, 2019. [Google Scholar]
- Jolliffe, I.T. Principal Component Analysis, Second Edition. Encycl. Stat. Behav. Sci. 2002, 30, 487. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-Hit: A Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Shelnutt, J.A.; Song, X.Z.; Ma, J.G.; Jia, S.L.; Jentzen, W.; Medforth, C.J. Nonplanar Porphyrins and Their Significance in Proteins. Chem. Soc. Rev. 1998, 27, 31–42. [Google Scholar] [CrossRef]
- Bolognesi, M.; Onesti, S.; Gatti, G.; Coda, A.; Ascenzi, P.; Brunori, M. Aplysia Limacina Myoglobin. J. Mol. Biol. 1989, 205, 529–544. [Google Scholar] [CrossRef]
- Li, H.; Shimizu, H.; Flinspach, M.; Jamal, J.; Yang, W.; Xian, M.; Cai, T.; Wen, E.Z.; Jia, Q.; Wang, P.G.; et al. The Novel Binding Mode of N-Alkyl-N’-Hydroxyguanidine to Neuronal Nitric Oxide Synthase Provides Mechanistic Insights into NO Biosynthesis. Biochemistry 2002, 41, 13868–13875. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Wang, Y.; Lovell, S.; Kumar, R.; Ruvinsky, A.M.; Battaile, K.P.; Vakser, I.A.; Rivera, M. The Structure of the BfrB–Bfd Complex Reveals Protein–Protein Interactions Enabling Iron Release from Bacterioferritin. J. Am. Chem. Soc. 2012, 134, 13470–13481. [Google Scholar] [CrossRef] [PubMed]
- Hui, H.L.; Kavanaugh, J.S.; Doyle, M.L.; Wierzba, A.; Rogers, P.H.; Arnone, A.; Holt, J.M.; Ackers, G.K.; Noble, R.W. Structural and Functional Properties of Human Hemoglobins Reassembled after Synthesis in Escherichia Coli. Biochemistry 1999, 38, 1040–1049. [Google Scholar] [CrossRef]
- Kavanaugh, J.S.; Rogers, P.H.; Arnone, A. High-Resolution x-Ray Study of Deoxy Recombinant Human Hemoglobins Synthesized from Beta-Globins Having Mutated Amino Termini. Biochemistry 1992, 31, 8640–8647. [Google Scholar] [CrossRef]
- Wang, Y.; Yao, H.; Cheng, Y.; Lovell, S.; Battaile, K.P.; Midaugh, C.R.; Rivera, M. Characterization of the Bacterioferritin/Bacterioferritin Associated Ferredoxin Protein–Protein Interaction in Solution and Determination of Binding Energy Hot Spots. Biochemistry 2015, 54, 6162–6175. [Google Scholar] [CrossRef]
- Tsukihara, T.; Shimokata, K.; Katayama, Y.; Shimada, H.; Muramoto, K.; Aoyama, H.; Mochizuki, M.; Shinzawa-Itoh, K.; Yamashita, E.; Yao, M.; et al. The Low-Spin Heme of Cytochrome c Oxidase as the Driving Element of the Proton-Pumping Process. Proc. Natl. Acad. Sci. USA 2003, 100, 15304–15309. [Google Scholar] [CrossRef]
- LaCount, M.W.; Zhang, E.; Chen, Y.P.; Han, K.; Whitton, M.M.; Lincoln, D.E.; Woodin, S.A.; Lebioda, L. The Crystal Structure and Amino Acid Sequence of Dehaloperoxidase from Amphitrite Ornata Indicate Common Ancestry with Globins. J. Biol. Chem. 2000, 275, 18712–18716. [Google Scholar] [CrossRef]
- Chen, Z.; de Serrano, V.; Betts, L.; Franzen, S. Distal Histidine Conformational Flexibility in Dehaloperoxidase from Amphitrite Ornata. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009, 65, 34–40. [Google Scholar] [CrossRef]
- Polyakov, K.M.; Boyko, K.M.; Tikhonova, T.V.; Slutsky, A.; Antipov, A.N.; Zvyagilskaya, R.A.; Popov, A.N.; Bourenkov, G.P.; Lamzin, V.S.; Popov, V.O. High-Resolution Structural Analysis of a Novel Octaheme Cytochrome c Nitrite Reductase from the Haloalkaliphilic Bacterium Thioalkalivibrio Nitratireducens. J. Mol. Biol. 2009, 389, 846–862. [Google Scholar] [CrossRef] [PubMed]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Žídek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly Accurate Protein Structure Prediction for the Human Proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef] [PubMed]
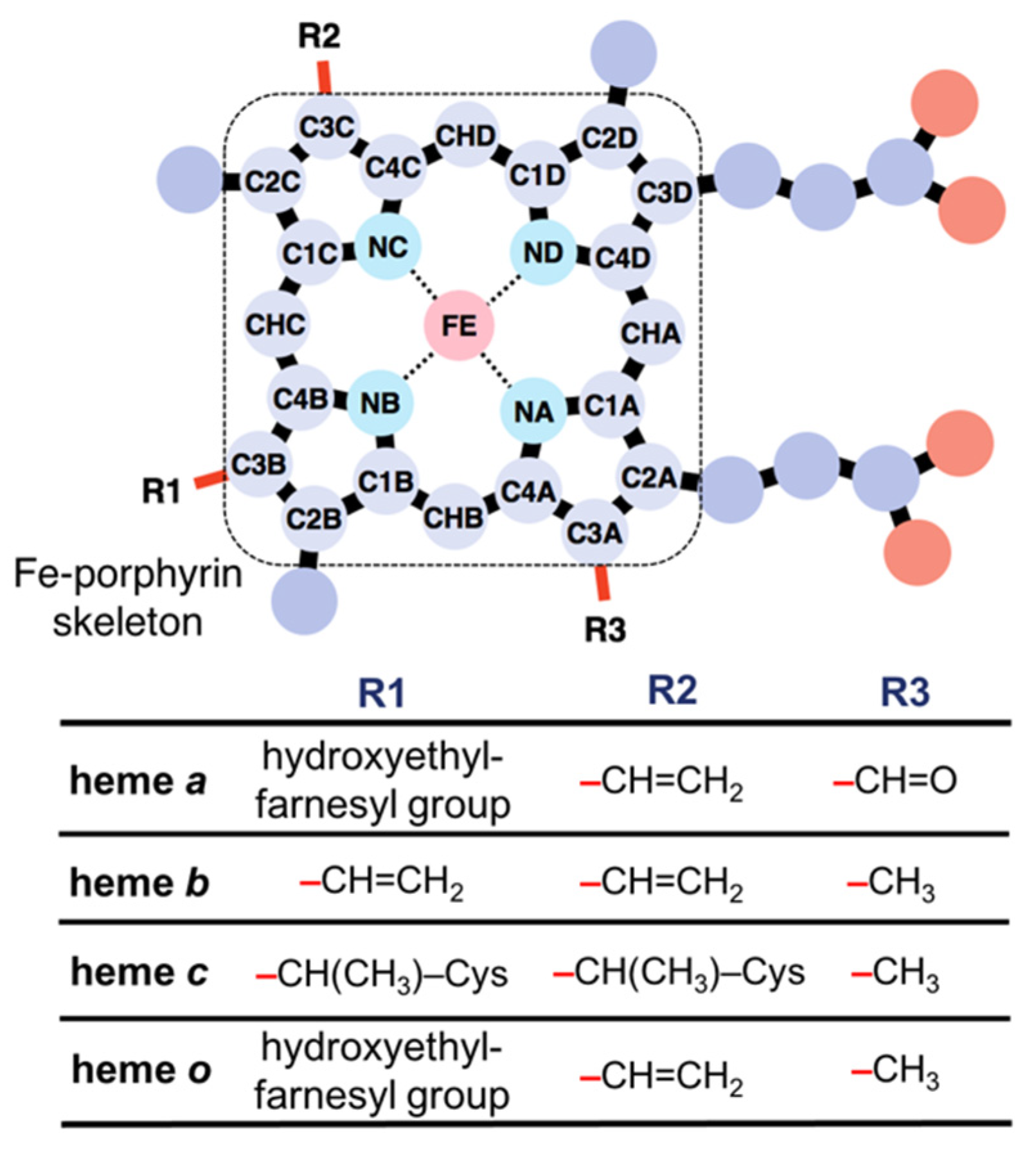


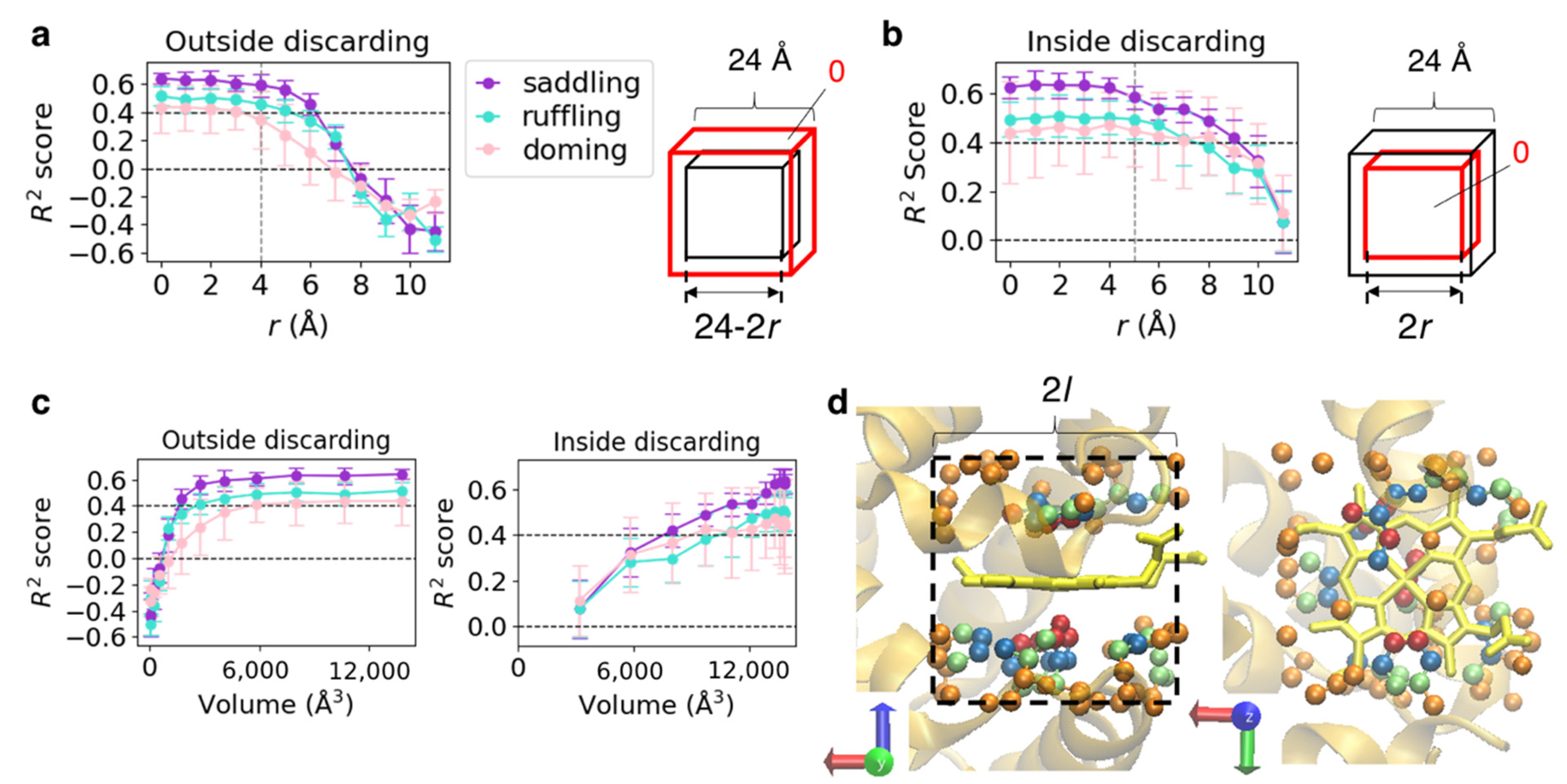
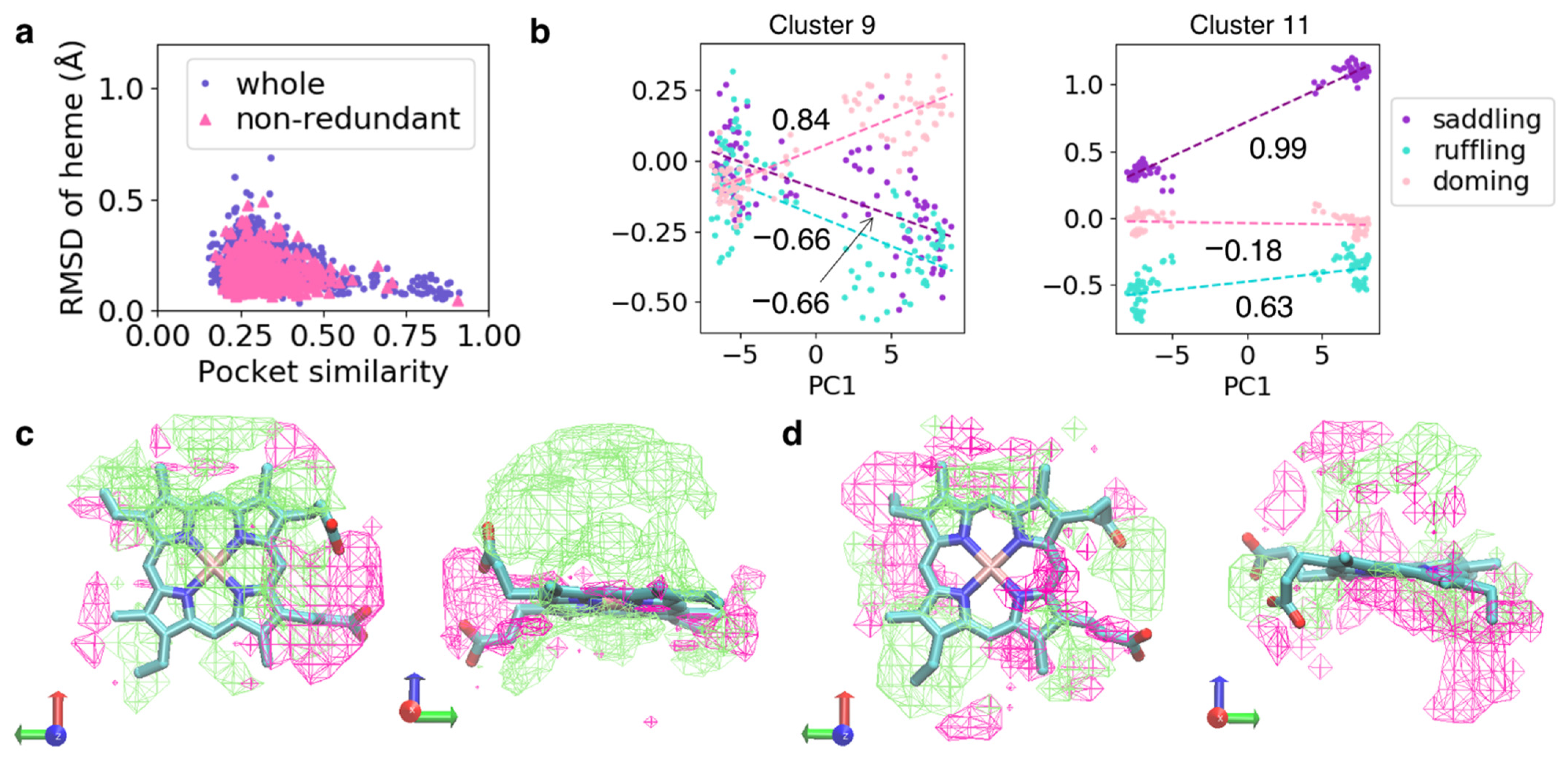

| Layer | Function | Filter (Kernel) | Output Dimension (Channel × Depth × Width × Height) |
|---|---|---|---|
| 1 | Conv3d | 2 × 2 × 2 with 0-padding | 64 × 21 × 21 × 21 |
| 2 | Conv3d | 2 × 2 × 2 with 0-padding | 128 × 22 × 22 × 22 |
| 3 | BatchNorm3d | - | 128 × 22 × 22 × 22 |
| 4 | Conv3d | 2 × 2 × 2 without padding | 128 × 21 × 21 × 21 |
| 5 | ReLU | - | 128 × 21 × 21 × 21 |
| 6 | BatchNorm3d | - | 128 × 21 × 21 × 21 |
| 7 | MaxPool3d | 2 × 2 × 2 stride: 2 × 2 × 2 | 128 × 10 × 10 × 10 |
| 8 | Full connection | - | 128,000 |
| 9 | Linear | - | 128 |
| 10 | ReLU | - | 128 |
| 11 | Dropout | 0.4 | 128 |
| 12 | Linear | - | 64 |
| 13 | BatchNorm1d | - | 64 |
| 14 | ReLU | - | 64 |
| 15 | Linear | - | 1 (or 12) |
| Saddling | Ruffling | Doming | |
|---|---|---|---|
| R2 score (max., min.) | 0.62 ± 0.05 (0.70, 0.55) | 0.50 ± 0.09 (0.65, 0.39) | 0.46 ± 0.15 (0.70, 0.25) |
| RMSE † (min., max.) | 0.21 ± 0.02 (0.20, 0.24) | 0.31 ± 0.04 (0.25, 0.37) | 0.16 ± 0.03 (0.11, 0.20) |
| Heme Type | Saddling | Ruffling | Doming |
|---|---|---|---|
| heme c (85.8 ± 2.7) † | 0.20 ± 0.01 | 0.22 ± 0.02 | 0.11 ± 0.01 |
| heme b (64.2 ± 3.0) | 0.22 ± 0.02 | 0.41 ± 0.07 | 0.22 ± 0.06 |
| Saddling | Ruffling | Doming | |
|---|---|---|---|
| R2 score (max., min.) | 0.63 ± 0.07 (0.72, 0.53) | 0.39 ± 0.10 (0.52, 0.24) | 0.43 ± 0.17 (0.68, 0.17) |
| RMSE † (min., max.) | 0.21 ± 0.02 (0.19, 0.25) | 0.34 ± 0.02 (0.31, 0.37) | 0.16 ± 0.03 (0.12, 0.21) |
| Cluster Index | Sample Number | Protein Name | |
|---|---|---|---|
| 1 | 407 (407) † | 7.55 | Nitric-oxide synthase |
| 2 | 146 (146) | 8.43 | Hemoglobin (beta chain) |
| 3 | 133 (95) | 5.46 | Bacterioferritin |
| 4 | 103 (103) | 7.72 | Hemoglobin (alpha chain) |
| 5 | 99 (99) | 8.18 | Nitric oxide synthase |
| 6 | 64 (81) | 8.94 | Cytochrome c oxidase subunit 1 |
| 7 | 55 (55) | 11.14 | Dehaloperoxidase |
| 8 | 50 (50) | 6.41 | Nitric oxide synthase oxygenase |
| 9 | 47 (47) | 9.84 | Cytochrome c |
| 10 | 46 (321) | 14.46 | Eight-heme nitrite reductase |
| whole dataset | 3843 | 17.27 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kondo, H.X.; Iizuka, H.; Masumoto, G.; Kabaya, Y.; Kanematsu, Y.; Takano, Y. Elucidation of the Correlation between Heme Distortion and Tertiary Structure of the Heme-Binding Pocket Using a Convolutional Neural Network. Biomolecules 2022, 12, 1172. https://doi.org/10.3390/biom12091172
Kondo HX, Iizuka H, Masumoto G, Kabaya Y, Kanematsu Y, Takano Y. Elucidation of the Correlation between Heme Distortion and Tertiary Structure of the Heme-Binding Pocket Using a Convolutional Neural Network. Biomolecules. 2022; 12(9):1172. https://doi.org/10.3390/biom12091172
Chicago/Turabian StyleKondo, Hiroko X., Hiroyuki Iizuka, Gen Masumoto, Yuichi Kabaya, Yusuke Kanematsu, and Yu Takano. 2022. "Elucidation of the Correlation between Heme Distortion and Tertiary Structure of the Heme-Binding Pocket Using a Convolutional Neural Network" Biomolecules 12, no. 9: 1172. https://doi.org/10.3390/biom12091172
APA StyleKondo, H. X., Iizuka, H., Masumoto, G., Kabaya, Y., Kanematsu, Y., & Takano, Y. (2022). Elucidation of the Correlation between Heme Distortion and Tertiary Structure of the Heme-Binding Pocket Using a Convolutional Neural Network. Biomolecules, 12(9), 1172. https://doi.org/10.3390/biom12091172







