SiCoDEA: A Simple, Fast and Complete App for Analyzing the Effect of Individual Drugs and Their Combinations
Abstract
:1. Introduction
2. Materials and Methods
2.1. SiCoDEA Strategy
- Response additivity model. Additionally known as the linear interaction effect, the response additivity model [12] consists of comparing the effect of the combination and the effect obtained by adding that of the individual drugs at the same dose. It is therefore possible to obtain a CI from the ratiowhere EA is the effect of drug A alone, EB is the effect of drug B alone and EAB is the effect of drugs A and B in combination.
- Highest single agent (HSA) model. The highest single agent model (HSA) or Gaddum’s non-interaction model [13] assumes that the expected effect of the combination is equal to the highest effect of the individual drug at the same dose as it has in the combination. Thus, a synergistic combination should produce an additional beneficial effect compared to what individual drugs alone can achieve. The CI is given by the difference between the effect of the combination at a given dose and the highest effect of one of the single drugs at that same dose
- Bliss independence model. The Bliss independence model [14] assumes a stochastic process in which two drugs produce their effect independently. Therefore, the expected effect of the combination can be calculated as the probability of two independent events: where and . The CI will beThe dose–effect-based strategy includes:
- Loewe additivity model. The principle on which the Loewe additivity model [15,16,17] is based is that to calculate the CI it is necessary to compare the doses of the drugs in combination with the doses of the individual drugs necessary to achieve the same effect. In this way, if the dose required for a single drug is lower than that in combination, we will have an antagonistic effect between the two drugs, if, instead, it is higher, the effect will be synergistic. The CI will be calculated aswhere 𝑎 is the dose of drug A in the combination, 𝐴 is the equivalent dose, i.e., the dose of drug A needed to achieve the same effect of the combination, 𝑏 is the dose of drug B in the combination and 𝐵 is the equivalent dose, i.e., the dose of drug B needed to achieve the same effect of the combination.
- Zero Interaction Potency (ZIP) model. The Zero Interaction Potency (ZIP) model [18,19] combines the Loewe model and the Bliss model together if, in combination, the two dose–effect curves do not change. It then uses the same calculation of two independent events as the Bliss model, but using the values calculated through the dose–effect curve as in the Loewe model. The CI will bewhere 𝐸𝐸𝐴 is the expected effect of drug A, and 𝐸𝐸𝐵 is the expected effect of drug B calculated based on the dose–effect curve.
- Chou–Talalay Method [20] (median-effect), which is the most commonly used model based on the median-effect equation, derived from the mass action law principlewhere 𝐷 is the dose of interest, 𝐷𝑚 is the median-effect dose, 𝑓𝑎 is the fraction affected and 𝑚 is the slope.
- Log-logistic with four parameters (log-logistic).
- Log-logistic with three parameters (minimum equal to zero, log-logistic[0]).
- Log-logistic with three parameters (maximum equal to one, log-logistic[1]).
- Log-logistic with two parameters (log-logistic[01]).
2.2. SiCoDEA Validation
3. Results
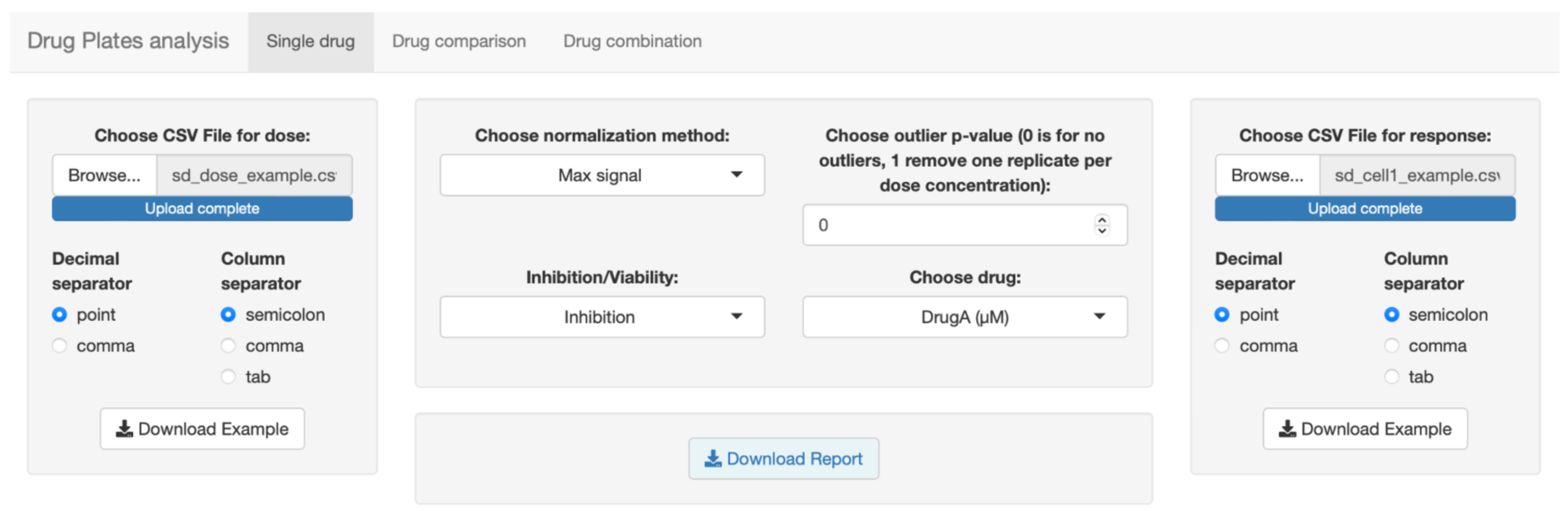
3.1. Implementation and Description of SiCoDEA Functions
3.2. Application of SiCoDEA
4. Discussion
- Single drug analysis. Obtain the dose–response curve for different drugs by uploading a single file. For each drug, curves are displayed for all five models with relative R2 and IC50.
- Drug comparison. Compare the effect of the same drug on different samples, up to a maximum of four, by choosing the most fitting model from the five options.
- Drug combination analysis. Perform a combination analysis in two steps: first, visualizing the dose–response curves for the five models in the two drugs considered; second, based on the R2, choosing the best model to be adopted for the dose–response curve and for the CI. Results are displayed in an isobologram plot and in a heatmap.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Frei, E., 3rd; Karon, M.; Levin, R.H.; Freireich, E.J.; Taylor, R.J.; Hananian, J.; Selawry, O.; Holland, J.F.; Hoogstraten, B.; Wolman, I.J.; et al. The effectiveness of combinations of antileukemic agents in inducing and maintaining remission in children with acute leukemia. Blood 1965, 26, 642–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeVita, V.T., Jr.; Chu, E. A history of cancer chemotherapy. Cancer Res. 2008, 68, 8643–8653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, F.; Zhao, W.; Gong, X.; Ren, S.; Su, C.; Jiang, T.; Zhou, C. Immune-checkpoint inhibitors plus chemotherapy versus chemotherapy as first-line treatment for patients with extensive-stage small cell lung cancer. J. Immunother. Cancer 2020, 8, e001300. [Google Scholar] [CrossRef] [PubMed]
- Mullard, A. 2021 FDA approvals. Nat. Rev. Drug Discov. 2022, 21, 83–88. [Google Scholar] [CrossRef]
- Times, N. Cancer Drug Trials Encounter a Problem Too Few Patients. Available online: https://www.nytimes.com/2017/08/12/health/cancer-drug-trials-encounter-a-problem-too-fewpatients.html (accessed on 20 April 2022).
- Hay, M.; Thomas, D.W.; Craighead, J.L.; Economides, C.; Rosenthal, J. Clinical development success rates for investigational drugs. Nat. Biotechnol. 2014, 32, 40–51. [Google Scholar] [CrossRef]
- Moore, T.J.; Zhang, H.; Anderson, G.; Alexander, G.C. Estimated Costs of Pivotal Trials for Novel Therapeutic Agents Approved by the US Food and Drug Administration, 2015–2016. JAMA Intern. Med. 2018, 178, 1451–1457. [Google Scholar] [CrossRef] [Green Version]
- Foucquier, J.; Guedj, M. Analysis of drug combinations: Current methodological landscape. Pharmacol. Res. Perspect. 2015, 3, e00149. [Google Scholar] [CrossRef]
- Chou, T.C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010, 70, 440–446. [Google Scholar] [CrossRef] [Green Version]
- Zheng, S.; Wang, W.; Aldahdooh, J.; Malyutina, A.; Shadbahr, T.; Tanoli, Z.; Pessia, A.; Tang, J. SynergyFinder Plus: Toward Better Interpretation and Annotation of Drug Combination Screening Datasets. Genom. Proteom. Bioinform. 2022; in press. [Google Scholar] [CrossRef]
- Zhang, T. Drug-Drug Combination Visualization (DDCV): Evaluation of Drug-Drug Interactions using Shiny by RStudio. Available online: https://github.com/xtmgah/DDCV (accessed on 20 April 2022).
- Slinker, B.K. The statistics of synergism. J. Mol. Cell. Cardiol. 1998, 30, 723–731. [Google Scholar] [CrossRef]
- Lehar, J.; Zimmermann, G.R.; Krueger, A.S.; Molnar, R.A.; Ledell, J.T.; Heilbut, A.M.; Short, G.F., 3rd; Giusti, L.C.; Nolan, G.P.; Magid, O.A.; et al. Chemical combination effects predict connectivity in biological systems. Mol. Syst. Biol. 2007, 3, 80. [Google Scholar] [CrossRef] [PubMed]
- Bliss, C.I. The toxicity of poisons applied jointly. Ann. Appl. Biol. 1939, 26, 585–615. [Google Scholar] [CrossRef]
- Loewe, S.; Muischnek, H. Über Kombinationswirkungen. Naunyn-Schmiedebergs Archiv für Experimentelle Pathologie und Pharmakologie 1926, 114, 313–326. [Google Scholar] [CrossRef]
- Loewe, S. Die quantitativen Probleme der Pharmakologie. Ergeb. Physiol. 1928, 27, 47–187. [Google Scholar] [CrossRef]
- Loewe, S. The problem of synergism and antagonism of combined drugs. Arzneimittelforschung 1953, 3, 285–290. [Google Scholar] [PubMed]
- Yadav, B.; Wennerberg, K.; Aittokallio, T.; Tang, J. Searching for Drug Synergy in Complex Dose-Response Landscapes Using an Interaction Potency Model. Comput. Struct. Biotechnol. J. 2015, 13, 504–513. [Google Scholar] [CrossRef] [Green Version]
- Yadav, B.; Wennerberg, K.; Aittokallio, T.; Tang, J. Corrigendum to: Searching for drug synergy in complex dose-response landscapes using an interaction potency model. Comput. Struct. Biotechnol. J. 2017, 15, 387. [Google Scholar] [CrossRef]
- Chou, T.C.; Talalay, P. Quantitative analysis of dose-effect relationships: The combined effects of multiple drugs or enzyme inhibitors. Adv. Enzym. Regul. 1984, 22, 27–55. [Google Scholar] [CrossRef]
- Grubbs, F.E. Sample Criteria for testing outlying observations. Ann. Math. Stat. 1950, 21, 27–58. [Google Scholar] [CrossRef]
- Chang, W.; Cheng, J.; Allaire, J.J.; Xie, Y.; McPherson, J. Shiny: Web Application Framework for R. R Package Version 1.1.0. Available online: https://cran.r-project.org/web/packages/shiny/index.html (accessed on 20 April 2022).
- Attali, D. shinyjs: Easily Improve the User Experience of Your Shiny Apps in Seconds. Available online: https://cran.r-project.org/web/packages/shinyjs/index.html (accessed on 20 April 2022).
- Wickham, H. The Split-Apply-Combine Strategy for Data Analysis. J. Stat. Softw. 2011, 40, 1–29. [Google Scholar] [CrossRef] [Green Version]
- Fox, J.; Weisberg, S. An R Companion to Applied Regression. Available online: https://socialsciences.mcmaster.ca/jfox/Books/Companion/ (accessed on 20 April 2022).
- Ritz, C.; Baty, F.; Streibig, J.C.; Gerhard, D. Dose-Response Analysis Using R. PLoS ONE 2015, 10, e0146021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: Cham, Switzerland, 2009. [Google Scholar]
- Wickham, H. Tidyr: Tidy Messy Data. Available online: https://CRAN.R-project.org/package=tidyr (accessed on 20 April 2022).
- Gregory, R.; Warnes, B.B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; et al. gplots: Various R Programming Tools for Plotting Data. 2020, 3. Available online: https://cran.r-project.org/web/packages/gplots/index.html (accessed on 20 April 2022).
- Komsta, L. Outliers: Tests for outliers. 2022. Available online: https://cran.r-project.org/web/packages/outliers/index.html (accessed on 20 April 2022).
- Wickham, H.; Seidel, D. scales: Scale Functions for Visualization. Available online: https://CRAN.R-project.org/package=scales (accessed on 20 April 2022).
- Ren, K. Rlist: A Toolbox for Non-Tabular Data Manipulation. Available online: https://CRAN.R-project.org/package=rlist (accessed on 20 April 2022).
- Wickham, H.; François, R.; Henry, L.; Müller, K. dplyr: A Grammar of Data Manipulation. Available online: https://CRAN.R-project.org/package=dplyr (accessed on 20 April 2022).
- Quentmeier, H.; Martelli, M.P.; Dirks, W.G.; Bolli, N.; Liso, A.; Macleod, R.A.; Nicoletti, I.; Mannucci, R.; Pucciarini, A.; Bigerna, B.; et al. Cell line OCI/AML3 bears exon-12 NPM gene mutation-A and cytoplasmic expression of nucleophosmin. Leukemia 2005, 19, 1760–1767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiacci, E.; Spanhol-Rosseto, A.; Martelli, M.P.; Pasqualucci, L.; Quentmeier, H.; Grossmann, V.; Drexler, H.G.; Falini, B. The NPM1 wild-type OCI-AML2 and the NPM1-mutated OCI-AML3 cell lines carry DNMT3A mutations. Leukemia 2012, 26, 554–557. [Google Scholar] [CrossRef] [PubMed]
- Martelli, M.P.; Gionfriddo, I.; Mezzasoma, F.; Milano, F.; Pierangeli, S.; Mulas, F.; Pacini, R.; Tabarrini, A.; Pettirossi, V.; Rossi, R.; et al. Arsenic trioxide and all-trans retinoic acid target NPM1 mutant oncoprotein levels and induce apoptosis in NPM1-mutated AML cells. Blood 2015, 125, 3455–3465. [Google Scholar] [CrossRef]
- Falini, B.; Mecucci, C.; Tiacci, E.; Alcalay, M.; Rosati, R.; Pasqualucci, L.; La Starza, R.; Diverio, D.; Colombo, E.; Santucci, A.; et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N. Engl. J. Med. 2005, 352, 254–266. [Google Scholar] [CrossRef]
- Souers, A.J.; Leverson, J.D.; Boghaert, E.R.; Ackler, S.L.; Catron, N.D.; Chen, J.; Dayton, B.D.; Ding, H.; Enschede, S.H.; Fairbrother, W.J.; et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 2013, 19, 202–208. [Google Scholar] [CrossRef]
- Kantarjian, H.M.; Talpaz, M.; Santini, V.; Murgo, A.; Cheson, B.; O’Brien, S.M. Homoharringtonine: History, current research, and future direction. Cancer 2001, 92, 1591–1605. [Google Scholar] [CrossRef]
- Shi, Y.; Ye, J.; Yang, Y.; Zhao, Y.; Shen, H.; Ye, X.; Xie, W. The Basic Research of the Combinatorial Therapy of ABT-199 and Homoharringtonine on Acute Myeloid Leukemia. Front. Oncol. 2021, 11, 692497. [Google Scholar] [CrossRef]
- Mezzasoma, F.; Cardinali, V.; Gionfriddo, I.; Milano, F.; Sciabolacci, S.; Ferrari, A.; Sabino, M.; Silvestri, S.; Donini, S.; Tini, V.; et al. Abstract P195: Omacetaxine mepessucinate plus venetoclax show strong synergistic anti-leukemic activity in nucleophosmin (NPM1)-mutated AML patient-derived xenograft (PDX) models to support a phase 1 clinical trial. Mol. Cancer Ther. 2021, 20, P195. [Google Scholar] [CrossRef]
- Spinozzi, G.; Tini, V.; Adorni, A.; Falini, B.; Martelli, M.P. ARPIR: Automatic RNA-Seq pipelines with interactive report. BMC Bioinform. 2020, 21, 574. [Google Scholar] [CrossRef]
- Spinozzi, G.; Tini, V.; Mincarelli, L.; Falini, B.; Martelli, M.P. A comprehensive RNA-Seq pipeline includes meta-analysis, interactivity and automatic reporting. PeerJ Prepr. 2018, 6, e27317v2. [Google Scholar]
- Calabria, A.; Spinozzi, G.; Benedicenti, F.; Tenderini, E.; Montini, E. adLIMS: A customized open source software that allows bridging clinical and basic molecular research studies. BMC Bioinform. 2015, 16, S5. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| CI Models | Drug–Response Models | Open Source | Report | Single Drug Analysis | Customizable Outlier Analysis | Platform | 3 Drugs Analysis | |
|---|---|---|---|---|---|---|---|---|
| SynergyFinder Plus | 4 | 1 | 🗸 | 🗸 | 🗸 | Win/Mac/Linux | 🗸 | |
| CompuSyn | 1 | 1 | 🗸 | Win | ||||
| DDCV | 1 | 1 | 🗸 | 🗸 | Win/Mac/Linux | |||
| SiCoDEA | 5 | 5 | 🗸 | 🗸 | 🗸 | 🗸 | Win/Mac/Linux |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spinozzi, G.; Tini, V.; Ferrari, A.; Gionfriddo, I.; Ranieri, R.; Milano, F.; Pierangeli, S.; Donnini, S.; Mezzasoma, F.; Silvestri, S.; et al. SiCoDEA: A Simple, Fast and Complete App for Analyzing the Effect of Individual Drugs and Their Combinations. Biomolecules 2022, 12, 904. https://doi.org/10.3390/biom12070904
Spinozzi G, Tini V, Ferrari A, Gionfriddo I, Ranieri R, Milano F, Pierangeli S, Donnini S, Mezzasoma F, Silvestri S, et al. SiCoDEA: A Simple, Fast and Complete App for Analyzing the Effect of Individual Drugs and Their Combinations. Biomolecules. 2022; 12(7):904. https://doi.org/10.3390/biom12070904
Chicago/Turabian StyleSpinozzi, Giulio, Valentina Tini, Alessio Ferrari, Ilaria Gionfriddo, Roberta Ranieri, Francesca Milano, Sara Pierangeli, Serena Donnini, Federica Mezzasoma, Serenella Silvestri, and et al. 2022. "SiCoDEA: A Simple, Fast and Complete App for Analyzing the Effect of Individual Drugs and Their Combinations" Biomolecules 12, no. 7: 904. https://doi.org/10.3390/biom12070904
APA StyleSpinozzi, G., Tini, V., Ferrari, A., Gionfriddo, I., Ranieri, R., Milano, F., Pierangeli, S., Donnini, S., Mezzasoma, F., Silvestri, S., Falini, B., & Martelli, M. P. (2022). SiCoDEA: A Simple, Fast and Complete App for Analyzing the Effect of Individual Drugs and Their Combinations. Biomolecules, 12(7), 904. https://doi.org/10.3390/biom12070904






