Continuous Low-Intensity Ultrasound Preserves Chondrogenesis of Mesenchymal Stromal Cells in the Presence of Cytokines by Inhibiting NFκB Activation
Abstract
1. Introduction
2. Materials and Methods
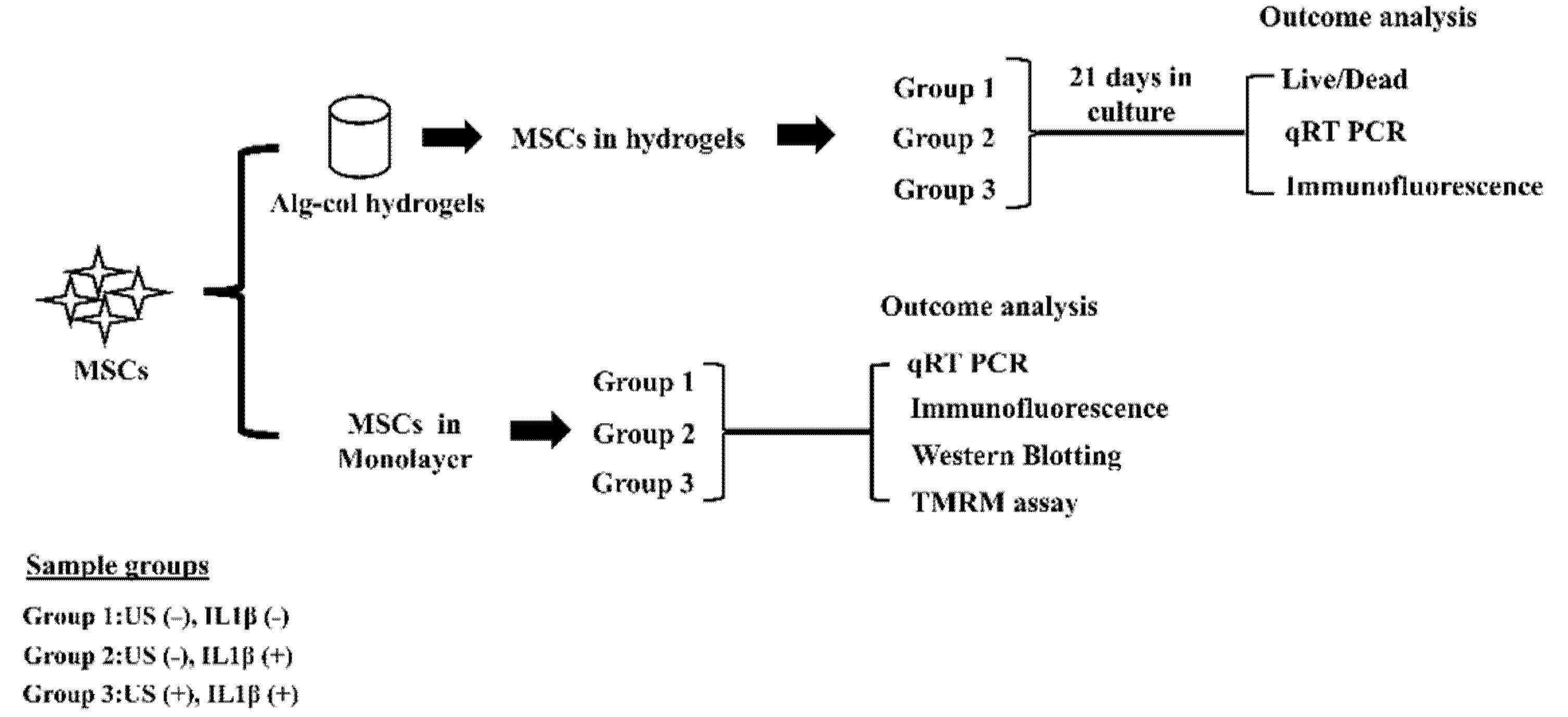
2.1. Cell Culture
2.2. Preparation and Encapsulation of MSCs in 3D Hydrogels
2.3. MSCs Culture in Hydrogels and cLIUS Treatment
2.4. MSCs Culture in Monolayer and cLIUS Treatment
2.5. Cell Viability Assay
2.6. Quantitative Real-Time PCR (qRT-PCR)
2.7. Immunofluorescence Staining
2.8. Protein Isolation and Western Blotting
2.9. Measurement of Mitochondrial Potential Using TMRM Assay
2.10. Statistical Analysis
3. Results
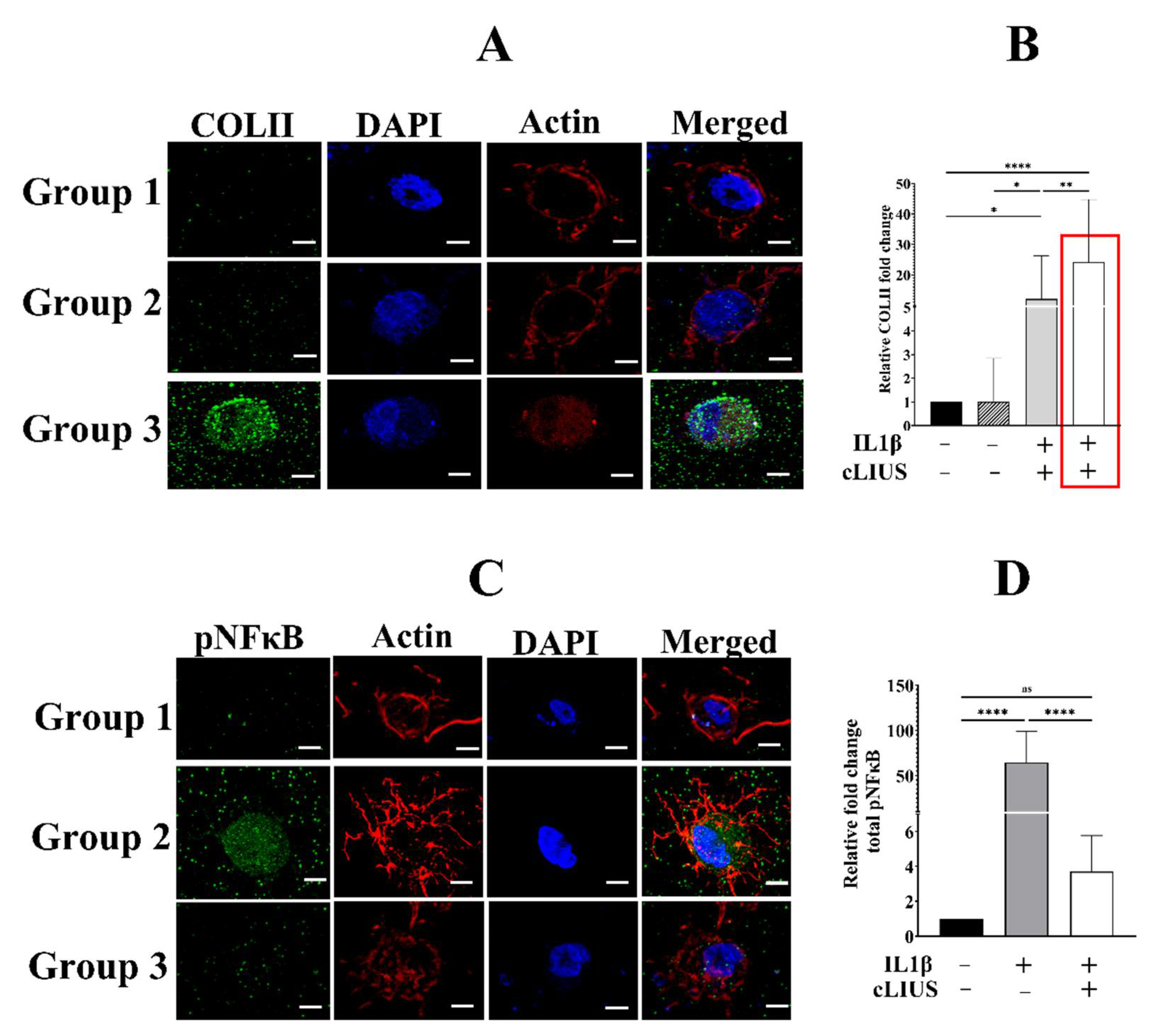
3.1. cLIUS Preserves Collagen II Protein Expression in the Presence of Cytokines
3.2. cLIUS Abrogates pNFκB Nuclear Translocation Induced by IL1β in 3D Scaffolds
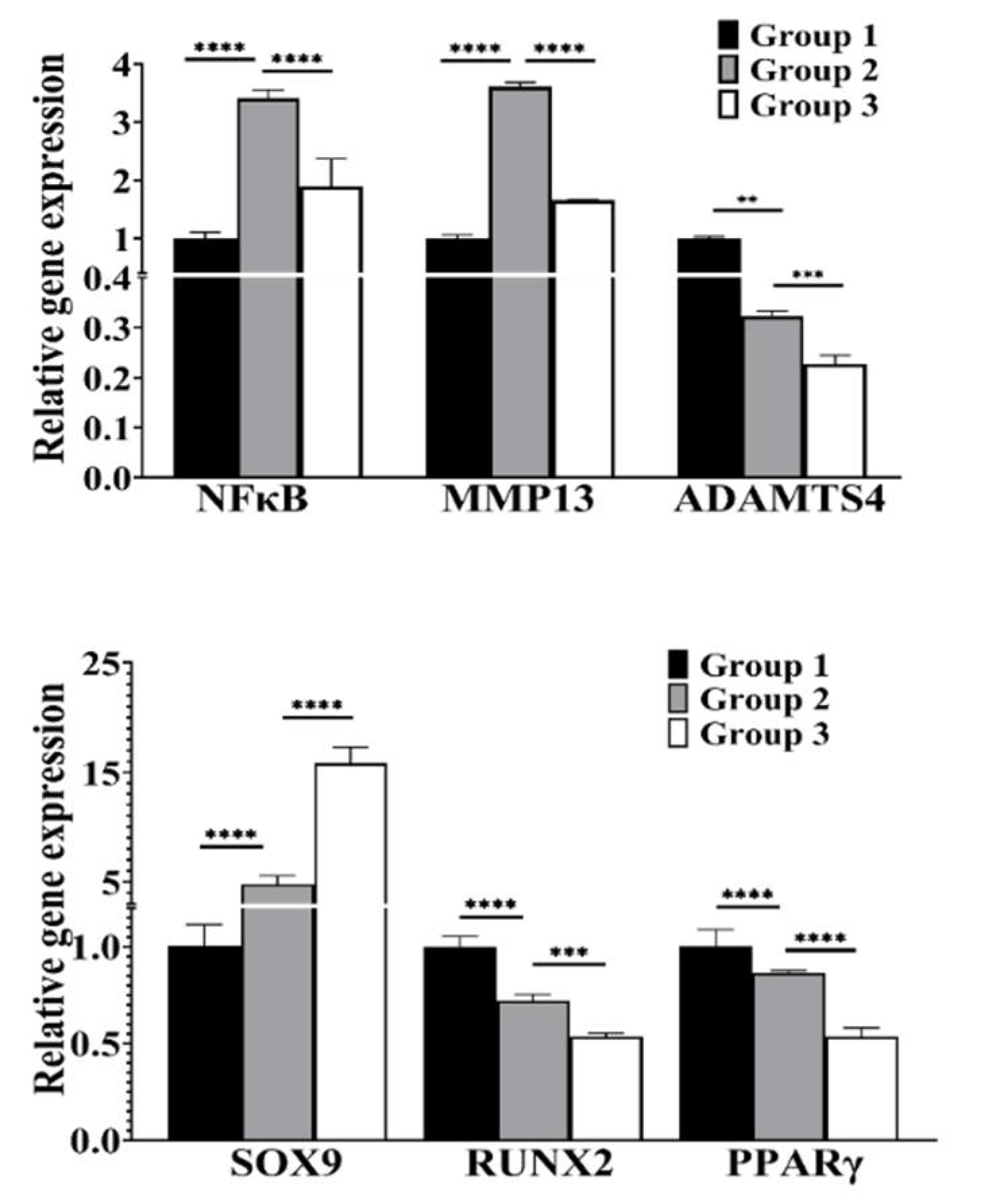
3.3. cLIUS Attenuates NFκB Expression and Upregulates SOX9 Gene Expression in the Presence of IL1β
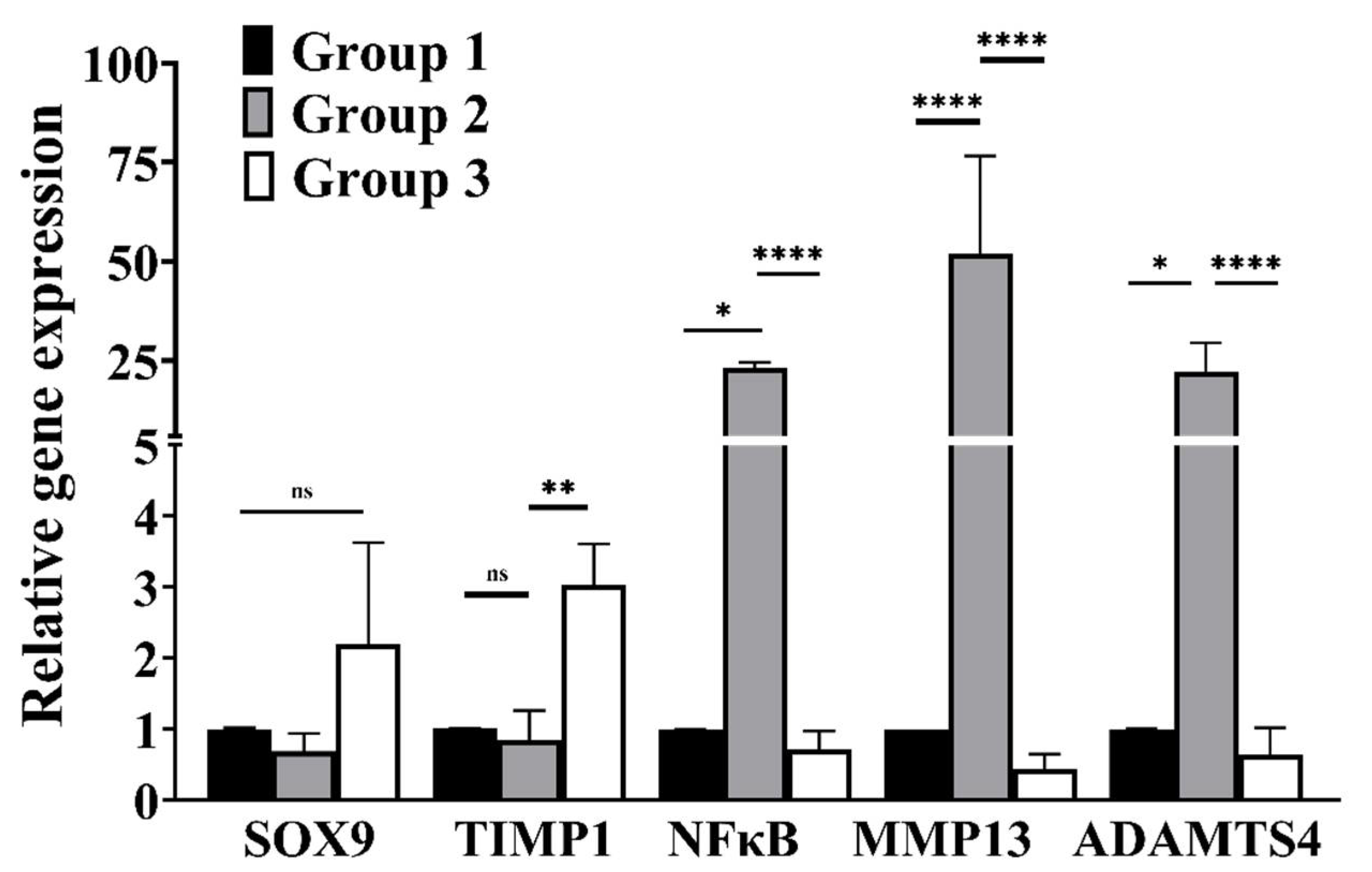
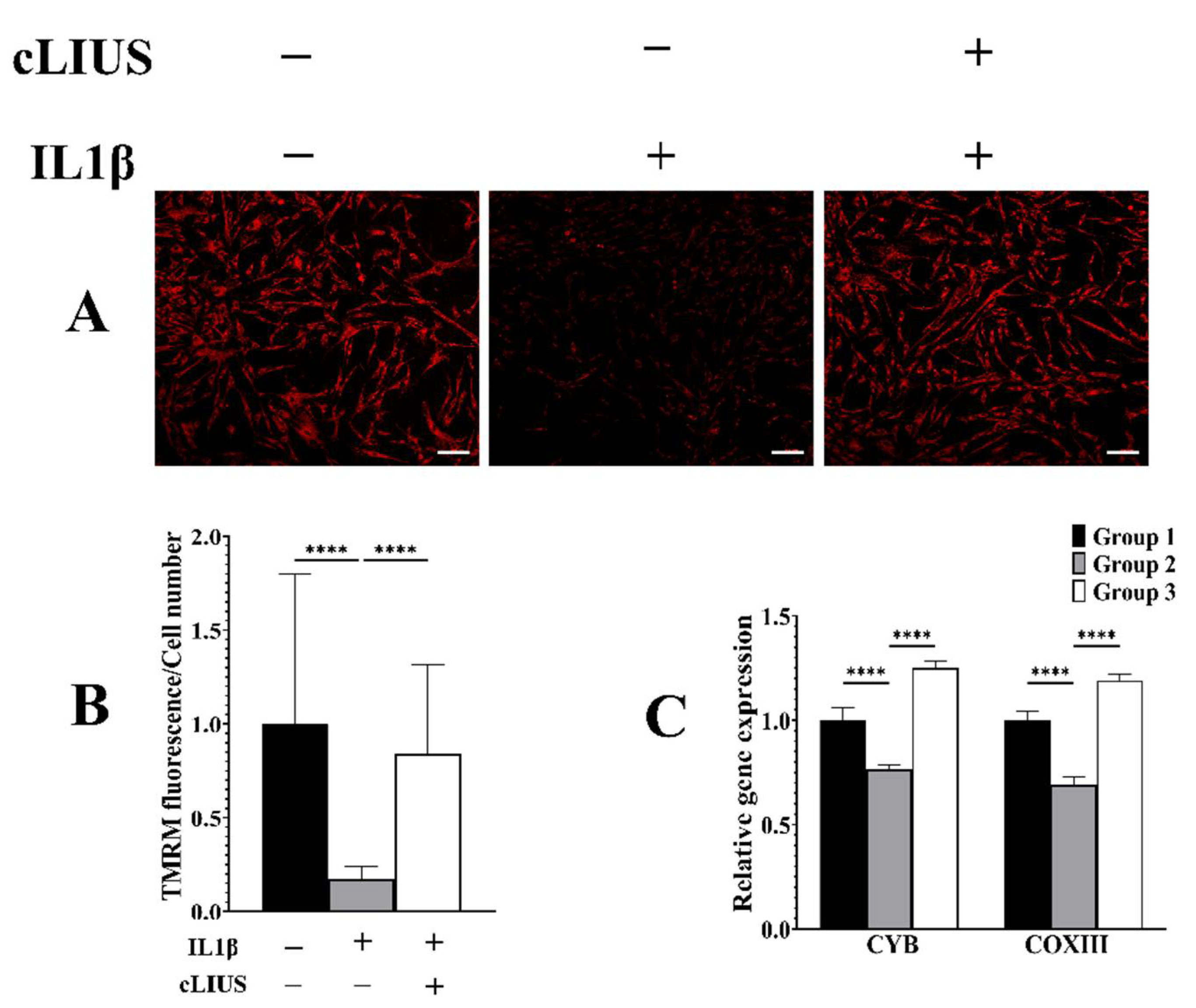
3.4. cLIUS Downregulates Catabolic and Upregulates Anabolic Gene Expression in the Presence of Proinflammatory Cytokines
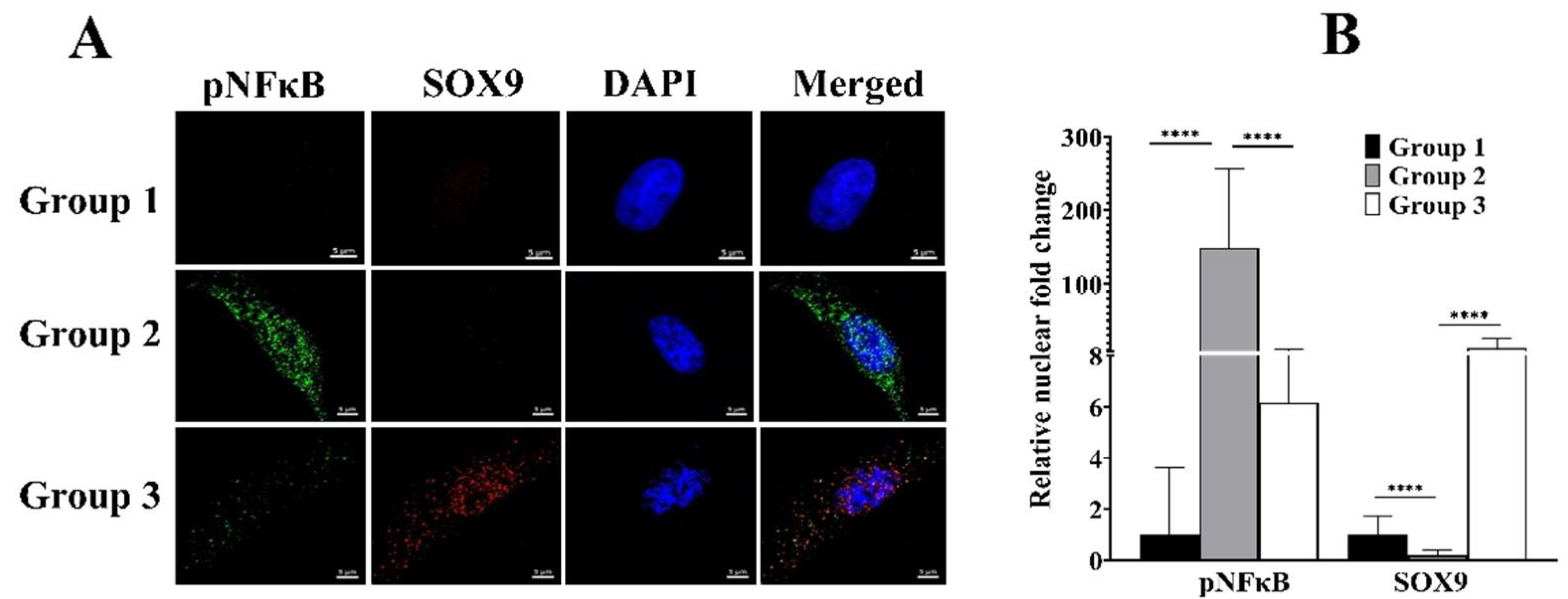
3.5. cLIUS Rescinds pNFκB and Promotes SOX9 Localization to the Nucleus in the Presence of IL1β
3.6. cLIUS Minimizes IL1β Induced pNFκB Expression and Persuades Total IκBα Expression
3.7. cLIUS Acts as a Mitochondrial Protective Agent in the Presence of IL1β
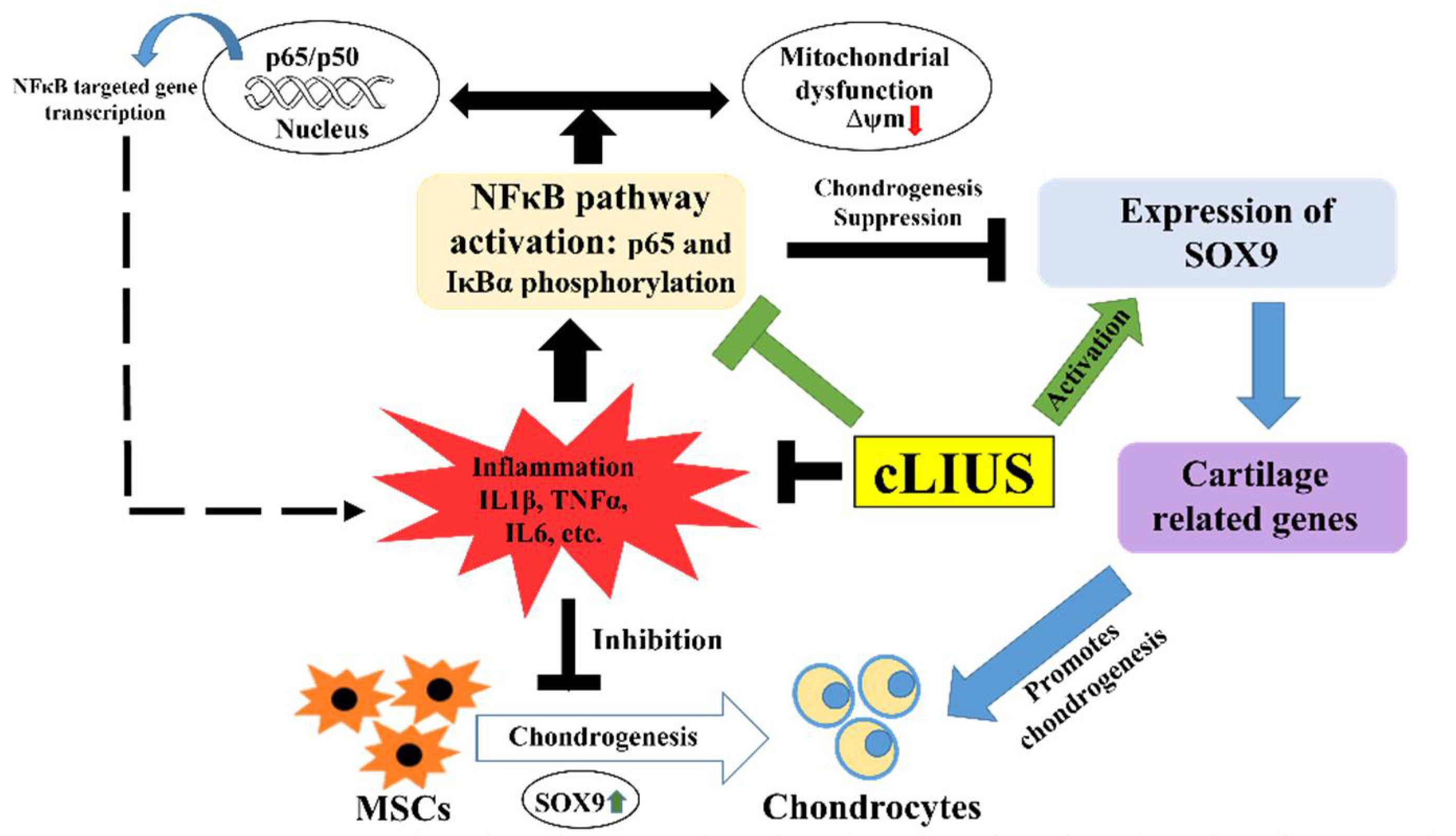
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cucchiarini, M.; Orth, P.; Rey-Rico, A.; Venkatesan, J.K.; Madry, H. Current perspectives in stem cell research for knee cartilage repair. Stem Cells Cloning Adv. Appl. 2014, 7, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Yao, J.Q.; Caplan, A. Stem cells for tissue engineering of articular cartilage. Proc. Inst. Mech. Eng. Part H J. Eng. Med. 2007, 221, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Martín, A.R.; Patel, J.; Zlotnick, H.M.; Carey, J.L.; Mauck, R.L. Emerging therapies for cartilage regeneration in currently excluded ‘red knee’ populations. NPJ Regen. Med. 2019, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Steinert, A.F.; Rackwitz, L.; Gilbert, F.; Nöth, U.; Tuan, R.S. Concise Review: The Clinical Application of Mesenchymal Stem Cells for Musculoskeletal Regeneration: Current Status and Perspectives. Stem Cells Transl. Med. 2012, 1, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Filardo, G.; Vannini, F.; Marcacci, M.; Andriolo, L.; Ferruzzi, A.; Giannini, S.; Kon, E. Matrix-assisted autologous chondrocyte transplantation for cartilage regeneration in osteoarthritic knees: Results and failures at midterm follow-up. Am. J. Sports Med. 2012, 41, 95–100. [Google Scholar] [CrossRef]
- Kondo, M.; Yamaoka, K.; Tanaka, Y. Acquiring Chondrocyte Phenotype from Human Mesenchymal Stem Cells under Inflammatory Conditions. Int. J. Mol. Sci. 2014, 15, 21270–21285. [Google Scholar] [CrossRef]
- Lolli, A.; Colella, F.; De Bari, C.; Van Osch, G.J.V.M. Targeting anti-chondrogenic factors for the stimulation of chondrogenesis: A new paradigm in cartilage repair. J. Orthop. Res. 2018, 37, 12–22. [Google Scholar] [CrossRef]
- Tsuchida, A.I.; Beekhuizen, M.; Ct Hart, M.; Radstake, T.R.; Dhert, W.J.; Saris, D.B.; van Osch, G.J.; Creemers, L.B. Cytokine profiles in the joint depend on pathology, but are different between synovial fluid, cartilage tissue and cultured chondrocytes. Arthritis Res. Ther. 2014, 16, 441. [Google Scholar] [CrossRef]
- Wojdasiewicz, P.; Poniatowski, L.A.; Szukiewicz, D. The Role of Inflammatory and Anti-Inflammatory Cytokines in the Pathogenesis of Osteoarthritis. Mediat. Inflamm. 2014, 2014, 561459. [Google Scholar] [CrossRef]
- Sitcheran, R.; Cogswell, P.C.; Baldwin, A.S. NF-κB mediates inhibition of mesenchymal cell differentiation through a posttranscriptional gene silencing mechanism. Genes Dev. 2003, 17, 2368–2373. [Google Scholar] [CrossRef]
- Wehling, N.; Palmer, G.D.; Pilapil, C.; Liu, F.; Wells, J.W.; Müller, P.E.; Evans, C.H.; Porter, R. Interleukin-1β and tumor necrosis factor α inhibit chondrogenesis by human mesenchymal stem cells through NF-κB-dependent pathways. Arthritis Care Res. 2009, 60, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Zayed, M.; Schumacher, J.; Misk, N.; Dhar, M. Effects of pro-inflammatory cytokines on chondrogenesis of equine mesenchymal stromal cells derived from bone marrow or synovial fluid. Vet. J. 2016, 217, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Jagielski, M.; Wolf, J.; Marzahn, U.; Völker, A.; Lemke, M.; Meier, C.; Ertel, W.; Godkin, O.; Arens, S.; Schulze-Tanzil, G. The Influence of IL-10 and TNFα on Chondrogenesis of Human Mesenchymal Stromal Cells in Three-Dimensional Cultures. Int. J. Mol. Sci. 2014, 15, 15821–15844. [Google Scholar] [CrossRef] [PubMed]
- Kondo, M.; Yamaoka, K.; Sonomoto, K.; Fukuyo, S.; Oshita, K.; Okada, Y.; Tanaka, Y. IL-17 Inhibits Chondrogenic Differentiation of Human Mesenchymal Stem Cells. PLoS ONE 2013, 8, e79463. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Gao, W.; Wu, Z.; Zhou, T.; Qiu, X.; Wang, X.; Lian, C.; Peng, Y.; Liang, A.; Qiu, J.; et al. Melatonin rescued interleukin 1β-impaired chondrogenesis of human mesenchymal stem cells. Stem Cell Res. Ther. 2018, 9, 162. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Xu, H.; Wang, C.; Qin, H.; An, Z. Magnesium enhances the chondrogenic differentiation of mesenchymal stem cells by inhibiting activated macrophage-induced inflammation. Sci. Rep. 2018, 8, 3406. [Google Scholar] [CrossRef]
- Armbruster, N.; Krieg, J.; Weißenberger, M.; Scheller, C.; Steinert, A.F. Rescued Chondrogenesis of Mesenchymal Stem Cells under Interleukin 1 Challenge by Foamyviral Interleukin 1 Receptor Antagonist Gene Transfer. Front. Pharmacol. 2017, 8, 255. [Google Scholar] [CrossRef]
- Buhrmann, C.; Mobasheri, A.; Matis, U.; Shakibaei, M. Curcumin mediated suppression of nuclear factor-κB promotes chondrogenic differentiation of mesenchymal stem cells in a high-density co-culture microenvironment. Arthritis Res. Ther. 2010, 12, R127. [Google Scholar] [CrossRef]
- Thakurta, S.G.; Kraft, M.; Viljoen, H.J.; Subramanian, A. Enhanced depth-independent chondrocyte proliferation and phenotype maintenance in an ultrasound bioreactor and an assessment of ultrasound dampening in the scaffold. Acta Biomater. 2014, 10, 4798–4810. [Google Scholar] [CrossRef][Green Version]
- Schumann, D.; Kujat, R.; Zellner, J.; Angele, M.K.; Nerlich, M.; Mayr, E.; Angele, P. Treatment of human mesenchymal stem cells with pulsed low intensity ultrasound enhances the chondrogenic phenotype in vitro. Biorheology 2006, 43, 431–443. Available online: http://www.ncbi.nlm.nih.gov/pubmed/16912415 (accessed on 19 February 2022).
- Lee, H.J.; Choi, B.H.; Min, B.-H.; Son, Y.S.; Park, S.R. Low-intensity Ultrasound Stimulation Enhances Chondrogenic Differentiation in Alginate Culture of Mesenchymal Stem Cells. Artif. Organs 2006, 30, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.H.; Park, K.; Park, S.R.; Min, B.-H. Effects of Low-Intensity Ultrasound on Chondrogenic Differentiation of Mesenchymal Stem Cells Embedded in Polyglycolic Acid: An in Vivo Study. Tissue Eng. 2006, 12, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Turner, J.A.; Budhiraja, G.; Thakurta, S.G.; Whitney, N.P.; Nudurupati, S.S. Ultrasonic Bioreactor as a Platform for Studying Cellular Response. Tissue Eng. Part C Methods 2013, 19, 244–255. [Google Scholar] [CrossRef] [PubMed]
- Sahu, N.; Budhiraja, G.; Subramanian, A. Preconditioning of mesenchymal stromal cells with low-intensity ultrasound: Influence on chondrogenesis and directed SOX9 signaling pathways. Stem Cell Res. Ther. 2020, 11, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Thakurta, S.G.; Budhiraja, G.; Subramanian, A. Growth factor and ultrasound-assisted bioreactor synergism for human mesenchymal stem cell chondrogenesis. J. Tissue Eng. 2015, 6, 2041731414566529. [Google Scholar] [CrossRef]
- Thakurta, S.G.; Sahu, N.; Miller, A.; Budhiraja, G.; Akert, L.; Viljoen, H.; Subramanian, A. Long-term culture of human mesenchymal stem cell-seeded constructs under ultrasound stimulation: Evaluation of chondrogenesis. Biomed. Phys. Eng. Express 2016, 2, 055016. [Google Scholar] [CrossRef]
- Sahu, N.; Viljoen, H.J.; Subramanian, A. Continuous low-intensity ultrasound attenuates IL-6 and TNFα-induced catabolic effects and repairs chondral fissures in bovine osteochondral explants. BMC Musculoskelet. Disord. 2019, 20, 193. [Google Scholar] [CrossRef]
- Marrella, A.; Lagazzo, A.; Barberis, F.; Catelani, T.; Quarto, R.; Scaglione, S. Enhanced mechanical performances and bioactivity of cell laden-graphene oxide/alginate hydrogels open new scenario for articular tissue engineering applications. Carbon 2017, 115, 608–616. [Google Scholar] [CrossRef]
- Louw, T.M.; Jackson, T.C.; Subramanian, A.; Viljoen, H.J. Simulation of acoustic fields in fluid-, solid- and porous layers by the combined transfer matrix/angular spectrum approach with applications in bioacoustics. Wave Motion 2015, 56, 122–136. [Google Scholar] [CrossRef]
- Louw, T.M.; Subramanian, A.; Viljoen, H.J. Theoretical Evaluation of the Acoustic Field in an Ultrasonic Bioreactor. Ultrasound Med. Biol. 2015, 41, 1766–1778. [Google Scholar] [CrossRef]
- Hernandez, P.A.; Jacobsen, T.D.; Barati, Z.; Chahine, N.O. Confocal scanning of intervertebral disc cells in 3D: Inside alginate beads and in native microenvironment. JOR Spine 2020, 3, e1106. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.-C.; Jo, J.; Park, J.; Kang, H.K.; Park, Y. NF-B Signaling Pathways in Osteoarthritic Cartilage Destruction. Cells 2019, 8, 734. [Google Scholar] [CrossRef] [PubMed]
- Cogswell, P.C.; Kashatus, D.F.; Keifer, J.A.; Guttridge, D.C.; Reuther, J.Y.; Bristow, C.; Roy, S.; Nicholson, D.W.; Baldwin, A.S. NF-κB and IκBα Are Found in the Mitochondria. J. Biol. Chem. 2003, 278, 2963–2968. [Google Scholar] [CrossRef]
- Murakami, S.; Lefebvre, V.; de Crombrugghe, B. Potent Inhibition of the Master Chondrogenic FactorSox9 Gene by Interleukin-1 and Tumor Necrosis Factor-α. J. Biol. Chem. 2000, 275, 3687–3692. [Google Scholar] [CrossRef] [PubMed]
- Goldring, M.B.; Marcu, K.B. Cartilage homeostasis in health and rheumatic diseases. Arthritis Res. Ther. 2009, 11, 224. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; May, M.J.; Kopp, E.B. NF-κB AND REL PROTEINS: Evolutionarily Conserved Mediators of Immune Responses. Annu. Rev. Immunol. 1998, 16, 225–260. [Google Scholar] [CrossRef]
- Kanichai, M.; Ferguson, D.; Prendergast, P.J.; Campbell, V.A. Hypoxia promotes chondrogenesis in rat mesenchymal stem cells: A role for AKT and hypoxia-inducible factor (HIF)-1α. J. Cell. Physiol. 2008, 216, 708–715. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhogoju, S.; Khan, S.; Subramanian, A. Continuous Low-Intensity Ultrasound Preserves Chondrogenesis of Mesenchymal Stromal Cells in the Presence of Cytokines by Inhibiting NFκB Activation. Biomolecules 2022, 12, 434. https://doi.org/10.3390/biom12030434
Bhogoju S, Khan S, Subramanian A. Continuous Low-Intensity Ultrasound Preserves Chondrogenesis of Mesenchymal Stromal Cells in the Presence of Cytokines by Inhibiting NFκB Activation. Biomolecules. 2022; 12(3):434. https://doi.org/10.3390/biom12030434
Chicago/Turabian StyleBhogoju, Sarayu, Shahid Khan, and Anuradha Subramanian. 2022. "Continuous Low-Intensity Ultrasound Preserves Chondrogenesis of Mesenchymal Stromal Cells in the Presence of Cytokines by Inhibiting NFκB Activation" Biomolecules 12, no. 3: 434. https://doi.org/10.3390/biom12030434
APA StyleBhogoju, S., Khan, S., & Subramanian, A. (2022). Continuous Low-Intensity Ultrasound Preserves Chondrogenesis of Mesenchymal Stromal Cells in the Presence of Cytokines by Inhibiting NFκB Activation. Biomolecules, 12(3), 434. https://doi.org/10.3390/biom12030434






