Abstract
Ubiquitylation and ISGylation are protein post-translational modifications (PTMs) and two of the main events involved in the activation of pattern recognition receptor (PRRs) signals allowing the host defense response to viruses. As with similar viruses, SARS-CoV-2, the virus causing COVID-19, hijacks these pathways by removing ubiquitin and/or ISG15 from proteins using a protease called PLpro, but also by interacting with enzymes involved in ubiquitin/ISG15 machinery. These enable viral replication and avoidance of the host immune system. In this review, we highlight potential points of therapeutic intervention in ubiquitin/ISG15 pathways involved in key host–pathogen interactions, such as PLpro, USP18, TRIM25, CYLD, A20, and others that could be targeted for the treatment of COVID-19, and which may prove effective in combatting current and future vaccine-resistant variants of the disease.
1. Introduction
Coronaviruses (CoVs) are enveloped, single-stranded, positive-sense RNA viruses that cause the common cold in a broad range of mammals and avians [1]. Severe infection can lead to respiratory and multi-organ failure, as well as digestive and neurological insults [2]. This was previously evidenced with the emergence of the pathogens responsible for Severe Acute Respiratory Syndrome coronavirus (SARS-CoV) in 2002 [3], and Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012 [4]. Both diseases were linked to zoonotic origins and resulted in a highly contagious and sometimes lethal acute respiratory illness. In December 2019, the World Health Organization (WHO) was alerted to patient clusters of a pneumonia of unknown cause in the city of Wuhan, Hubei Province, China [2]. A novel coronavirus species, SARS-Cov-2, was subsequently isolated and identified, resulting in a highly contagious condition now commonly known as COVID-19 (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, 2020; [5]). The exact origins of SARS-Cov-2 are still unclear, but it is suspected to have ancestral origins in bats [1]. As of January 2022, it is estimated that ~300 million cases have been detected globally, which has resulted in ~6 million deaths (www.google/globalCOVIDcases; accessed on 28 January 2022).
COVID-19 has seen an unprecedented level of research and funding dedicated to fighting the disease. Novel viral vector and nucleic acid vaccines have been developed in record times, expedited by knowledge harbored from preceding outbreaks. Despite the success of global vaccine rollouts, several mutated variants of SARS-CoV-2 have evolved, resulting in subtle changes in disease indications and strengths of transmissibility [6]. Due to the likely emergence of future vaccine-resistant variants, it is imperative to develop novel strategies to combat the disease. The post-translational modification of proteins with ubiquitin (ubiquitylation) and/or ISG15 (ISGylation) plays a key role in mediating cellular host–pathogen interactions and antiviral signaling and defense by modulating key events of the innate immune activation signaling. In this review, we discuss how the machinery involved in ubiquitylation and ISGylation can be utilized for the development of novel therapeutics against SARS-CoV-2 infection.
2. SARS-CoV-2-General Biology and Mechanisms of Infection
2.1. SARS-CoV-2 Viral Genome Architecture
SARS-CoV-2, along with other CoVs, has a genome of approximately 30kB of single-stranded, positive-sense RNA [7], making them the largest RNA genomes described to date [8]. The virus has 79% sequence identity with SARS-CoV and 50% with MERS-CoV [9]. Its genome contains twelve functional open reading frames ordered from 5′–3′ that encode the viral replication transcription complex (ORF1a/1b), the four structural proteins, Spike (S), Envelope (E), Membrane (M), and Nucleocapsid (N), and the likelihood of highly variable accessory proteins encoded throughout [7] (Figure 1A). Once viral RNA is released into the cell, it translates ORF 1a/1b to generate the huge replicase polyproteins pp1a and pp1ab, which are responsible for viral transcription, replication, and higher-order RNA structure [7]. Sixteen non-structural proteins (nsps) are liberated by the proteolytic cleavage of pp1a (nsp1–11) and pp1ab (nsp1–10, nsp12–16). The enzymes responsible for this are two cysteine proteases; papain-like protease (PLpro) is positioned in the large nsp3 subunit, and the highly conserved main protease (Mpro)/chymotrypsin-like protease (3CLpro) is located within nsp5 (Figure 1B). Mpro cleaves the nsp4–nsp11 region of pp1a and nsp4–nsp16 of pp1ab, whereas PLpro cleaves the nsp1–nsp4 domain [10].
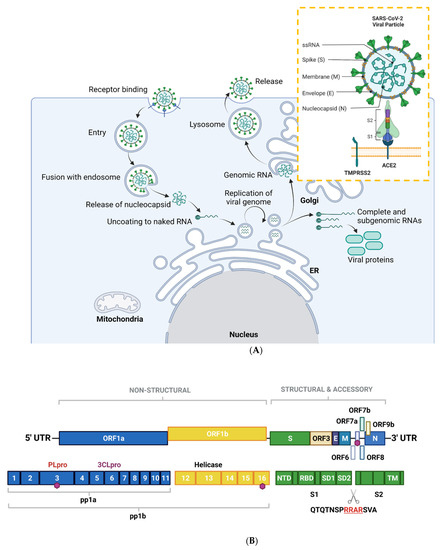
Figure 1.
Overview of the SARS-CoV-2 viral life cycle and structural organization. (A) SARS-CoV-2 comprises four structural proteins—the envelope (E) and membrane (M) proteins encase the single-stranded RNA and nucleocapsid (N). The spike (S) protein is made up of S1 and S2 subunits and facilitates receptor binding in the host cell. In humans, this is ACE2, in synergy with TMPRSS2 (orange box). SARS-CoV-2 binds to ACE2 on respiratory epithelial cells through the receptor binding domain on S. The SARS-CoV-2 viral particle is endocytosed following cleavage of S. SARS-CoV-2 nucleocapsid, a complex of viral RNA and N protein, is released into the cytosol. The N protein is removed from the capsid, leaving naked viral RNA, which is a capped positive-stranded RNA molecule that can immediately be translated to allow production of viral proteins (not shown). Viral replication centers are formed in the ER, using the positive-stranded RNA molecule as a template to form a double-stranded RNA intermediate, which in turns allows replication of the viral genome. Complete and sub-genomic RNAs are also formed during the replication process, which serve as the basis for production of a range of viral proteins. The replicated genomic RNA is then bound by N protein to form nucleocapsid, which is encapsulated in vesicles at the Golgi. The contents of the vesicles are then exocytosed, spreading viral particles from the infected cell. (B) Layout of SARS-CoV-2 genome. The virus encodes for ORF1a, ORF1b, ORF3, ORF6, ORF7a/b, ORF8, ORF9b, as well as S, E, M, N. Sixteen non-structural proteins (nsps1–16) of varying function are encoded by OF1a and ORF1b. nsp1 regulates viral mRNAs and interferes with host translation, nsp2–11 facilitate viral replication, nsp12 has RNA polymerase activity, whereas nsp14 is involved in RNA proofreading. A scissors indicates the site of furin and TMPRSS2 cleavage in S1/S2 and S2′, whereas sites of ubiquitin modification are identified by purple hexagons. Other regions of interest include PLpro and 3CLpro. S is composed of several sub-domains including the N-terminal domain (NTD), receptor-binding domain (RBD), subdomains 1 and 2 (SD1, SD2) and the transmembrane domain (TM). Panel B is adapted from Zhang et al., 2021. Figure created with BioRender.com in January 2022.
2.2. SARS-CoV-2 Mode of Entry and Proliferation
An overview of the SARS-CoV-2 viral life cycle is depicted in Figure 1A. As with the original SARS-CoV virus, human SARS-CoV-2 uses angiotensin-converting enzyme 2 (ACE2) as the main receptor for host cell invasion [11,12]. More recently, CD147 has also been identified as a novel route for infection [13]. The heavily glycosylated S protein mediates this attachment to host cell surface receptors [5]. Similar to other class I fusion glycoproteins, the viral S protein is cleaved and activated by host cell proteases to form an endosome [14]. The protease central to SARS-CoV-2 cell entry is Transmembrane Protease, Serine 2 (TMPSS2; Figure 1A) [15]. Once the endosome is formed, SARS-CoV-2 enters the cell either through acidification or the action of host protease cathepsin L.
The S protein is composed of two functional subunits: S1, which is responsible for binding to membrane-bound ACE2 [16], and S2, which contains the fusion domain. In the C-terminal to the S1 domain lies the receptor-binding domain (RBD), which is essential for viral entry [12,15,17]. One feature of SARS-CoV-2 that sets it apart from SARS-CoV and other beta-coronaviruses is a four-residue insertion of PRRA, forming a polybasic cleavage site RRAR at the junction of S1 and S2 (Figure 1B). This cleavage site can recruit furin and other host proteases, but it is unclear how this affects general virulence [17].
2.3. Clinical Manifestations of COVID-19
SARS-CoV-2 causes a wide spectrum of disease, from asymptomatic illness to severe acute respiratory failure. Findings on autopsy in terminal cases include diffuse alveolar damage, interstitial edema, and reactive type II pneumocytes [18]. Since SARS-CoV-2 enters cells via ACE2, there is a gradient of infection across the respiratory tract, with cells in the upper respiratory tract with high ACE2 and TMPRSS2 expression showing higher viral loads [19]. It is increasingly recognized that the host immune response is a major factor in the clinical manifestations of COVID-19. Indeed, aspects of the immune system are evaded, whereas others are magnified, leading to profound cytokine release, T cell activation, increased antibodies, and abnormalities of the granulocyte lineage [20]. In severe cases, T cells are both depleted and strongly activated [21], and peripheral blood has a “cytokine storm” profile, with large concentrations of inflammatory cytokines [20]. How SARS-CoV-2 achieves this in vivo has not been fully characterized, but work on airway epithelial cell lines has revealed a complex interaction of delayed, but powerful, type I interferon (IFN) response to SARS-CoV-2 infection, perhaps as a way of gaining time for viral replication [22]. Accumulating evidence has demonstrated that numerous IFN-stimulated genes (ISGs) and post-translational ubiquitylation play key roles in cellular antiviral signaling and defense by modulating key events of the innate immune activation signaling. Such mechanisms provide an attractive framework for the development of SARS-CoV-2 therapeutics.
4. Therapeutics-the Ub and ISG15 Machinery as a Target for COVID Treatment Regimes
Current pharmacological treatment regimens for COVID-19 can be broadly classified into antivirals, immunomodulators, and anticoagulant/antiplatelet therapies. Pharmacological regimens are primarily directed at mitigating complications in the later stages of the infection rather than early intervention during viral replication, and so far, only a handful have made it to clinical trials [96]. It is probable that many candidate compounds will have no therapeutic benefit in vivo; however, they can still provide invaluable insights into the druggability of the target, and act as molecular scaffolds in novel compound development.
Viral proteases have proven to be an attractive class of druggable enzymes and inhibitors directed against them are being investigated as possible therapeutic agents that may specifically target pathways within the SARS-CoV-2 lifecycle, or modulate the host immune response. The following section will explore the therapeutic potency of some of the most promising host (human) and viral proteases within the ubiquitin and ISG15 pathways.
4.1. Host Proteases
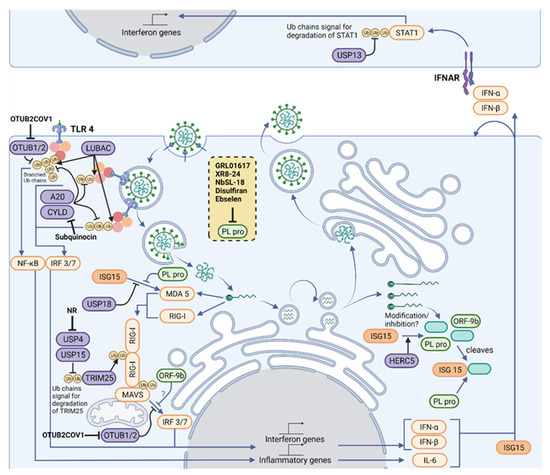
The IFN-I response is one of the first protective measures taken to encourage viral clearance during infection. It acts on downstream signaling pathways that result in the transcription of ISGs, leading to interactions between the innate and the adaptive immune systems [97]. As mentioned previously, there is an abnormal IFN-I response when cells are infected by SARS-CoV-2 [98,99]. This sometimes delayed but powerful IFN-I response has severe consequences as it hinders viral clearance and could contribute to the paradoxical hyperinflammatory state associated with cytokine storming. This regulation is tissue specific. For instance, a study has found that IFNα induces ACE2 expression as an ISG selectively in primary human upper airway basal cells [100], highlighting the importance of the host regulation of the IFN-I pathway in SARS-CoV-2 infection. PLpro and host proteases such as OTULIN, OTUB1/2, A20, CYLD, USP4, USP15, and USP18 are likely to be involved in such responses as key regulators of the innate immune response. Inhibitors of host proteases involved in antiviral response have already been described and could be explored in the context of COVID-19 infection. For example, Subquinocin, a CYLD inhibitor, was able to enhance activation of NF-κB signaling in cells [101]. Small molecule inhibitors for OTUB2 (OTUB2-COV-1) [102] and USP4 (NR) [103] have also been identified.
Small molecule inhibitors would be the traditional approach to downregulate the activity of an enzyme; however, as the case with some DUBs and most E3s, identifying suitable compounds that show high specificity whilst having limited off-target effects and good pharmacokinetic properties has been generally difficult [104,105]. Targeted degradation technologies such as degraders, and Proteolysis Targeting Chimeras (PROTACs) can be an alternative solution [106]. PROTACs enable a target protein to be ubiquitinated by an E3 ligase such that it undergoes proteasome-mediated degradation. This strategy has already been used for the FDA-approved drug ARV-110 which is used to treat metastatic castration-resistant prostate cancer [104]. The use of degraders or PROTACs could be the way to drug difficult-to-inhibit E3 ligases involved in antiviral responses such as TRIM25 and HERC5.
It may also be worth considering additional SARS-CoV-2-regulated host proteases, due to their likely involvement in inflammatory and immune response pathways. For instance, a study monitored lysine ubiquitylation on 8943 proteins from Vero E6 cells during SARS-CoV-2 infection by label-free LC-MS/MS [107]. Seventy-two hours post-infection, the authors observed a significant upregulation of 104 proteins and downregulation of a further 447. Several USP5 ubiquitin sites were downregulated during infection, whereas a single ubiquitylation (K558) site was upregulated. Similarly, in their global proteomic analysis, USP5 was upregulated by a factor of 1.2 [107]. This upregulation of USP5 provided insights into the deregulated IFN-I response during SARS-CoV-2 infection. The authors proposed a model whereby USP5 upregulation leads to the increased ubiquitylation of RIG-1, enabling the recruitment of STUB1 and inhibition of IFN-I production. Taken together, USP5 is an attractive target of pharmacological intervention that could be used in conjunction with other antivirals to induce a more adequate IFN response. Other targets of interest within the UPS include USP13, which has been reported to regulate IFN signaling via its ubiquitylation of STAT1 [108]. Furthermore, USP15 and USP25 are known negative regulators of IFN signaling [109,110]. Although most of the DUBs mentioned here have not been directly implicated in SARS-CoV-2 infection, they hold significant therapeutic potential.
4.2. Viral Proteases
SARS-CoV-2 encodes two proteases, the papain-like cysteine protease (PLpro) and the 3-chymotrypsin-like protease (3CLpro or Mpro) [111] and, unsurprisingly, both have garnered much attention in the exploration of therapeutic targets.
4.2.1. PLpro inhibitors
PLpro enables viral protein maturation through the cleavage of the viral polyprotein, and modulates host immune response via deubiquitylation and removal of ISG15 [72,85]. In an effort to find an effective pharmacological inhibitor against it, one study scanned a library of 3727 approved drugs and late-stage clinical drug candidates to repurpose, but failed to find any compounds which successfully inhibited the target [88]. Existing SARS PLpro inhibitors (rac3j, rac3k and rac5c) have been tested against SARS-CoV-2 PLpro. The most effective was rac5c, which was also shown to have acceptable cell toxicity, and decreased viral titers to similar levels as remdesivir and hydroxychloroquine regimes [88]. GRL0617 is a SARS PLpro inhibitor and has also been tested against SARS-COV-2 PLpro [91]. The study obtained an IC50 of 2.1 ± 0.2 μM for the compound, and also demonstrated that GRL0617 was capable of inhibiting the deubiquitylation and deISGylation activity of PLpro in HEK293T cells. GRL0617 is highly selective and does not alter the activity of USP18, the host-encoded deISGylating enzyme [91,112]. Furthermore, this drug was shown to have limited cytopathic effect, with good inhibition of viral replication in Vero E6 cells [91,113].
Molecular modelling approaches are also being used to find PLpro inhibitors. One group investigated 50 compounds with at least 65% similarity to GRL0617 through a molecular docking simulation [114]. They identified four potential inhibitors, which also had acceptable water solubility, toxicity, and gastrointestinal absorption [114]. Another study used similar methods to monitor the inhibitory potential of 67 approved compounds against PLpro. Of these, 26 had superior inhibitory potential compared with the GRL0617control [115]. Improving on the initial results, this latter study used the actual structure of PLpro rather than relying on homology modelling. It must be noted, however, that these drugs have not yet been approved for use in clinical trials.
Natural biflavones have also been reported as potential inhibitors of SARS-CoV-2 PLpro. Various phytochemicals such as polyphenols derived from plants such as Broussonetia papyrifera and Paulownia tomentosa have been shown to affect SARS-CoV-1 and MERS-CoV proteases [116,117]. Nine naturally occurring biflavones were identified from the national library of traditional Chinese medicine [118], and selected based on their inhibitory potential in molecular docking simulation against SARS-CoV-2 PLpro and subsequent validation by fluorogenic enzymatic assays. Of these, 4′-O-metyhlochnaflavone showed 60.7% inhibition even at concentrations as low as 2.5 μM, which is noteworthy as it is derived from L. japonica—a plant that has already shown to have antiviral effects against CoVs [118,119]. Other natural compounds such as ginkgolic acid and anacardic acid have been demonstrated to inhibit PLpro and have antiviral activity in vitro, with ginkgolic acid exhibiting higher antiviral potency amongst the two [120]. Tannic acid and methylcobalamin have also shown potent inhibitory effects on PLpro, independent of their other antiviral mechanisms [120,121,122].
Nanobodies (NbSL-17, -18 and -19) have been developed that targeted the S1 pocket of PLpro, a site that contributes to its deubiquitinating and deISGylating activity. NbSL-18 was shown to inhibit the cleavage of K48-linked triUB in a dose-dependent fashion, although there was less inhibition of ISG15-cleaving activity [123]. Interestingly, NbSL-18 inhibits Nsp3 mediated viral polyprotein processing, similar to rac5c [88,123]. It has been suggested that additional binding sites on PLpro distal from the active-site cysteine would allow development of compounds that exploit this binding cooperativity [124]. This included development of XR8-24, which could inhibit SARS-CoV-2 replication in human alveolar basal epithelial A549 cells. There was no inhibition of USP7 (the most similar human DUB to PLpro in structural terms), suggesting high target specificity. In addition, the authors argue that slower ligand dissociation and slow-off rates could be a possible consequence of positive binding cooperativity [124].
An attempt was made to repurpose 1971 FDA-approved drugs [123]; however, despite having identified five potential inhibitory compounds against SARS-CoV-2 PLpro and nsp3, these compounds demonstrated considerable off-target effects and low antiviral potency in cell-based models. More successful efforts have been made to repurpose approved, safe, cysteine modifiers that can inhibit the catalytic cysteines of PLpro, such as Disulfiran and Ebselen, two molecules that are currently part of the COVID-19 clinical trials (NCT04485130, NCT04483973 and NCT04484025) [125]. Another potential PLpro inhibitor undergoing clinical trials is Isotretinoin, a retinoic acid used for the treatment of severe acne (NCT04389580, NCT04382950, NCT04353180 and NCT04361422). This is encouraging and further supports targeting PLpro for the treatment of COVID-19.
4.2.2. CLpro/Mpro Inhibitors
Mpro is responsible for cleaving the majority of the viral polyprotein. Although it holds promise as a therapeutic target, we will only briefly summarize these findings as it does not play a known role in modulating host ubiquitin/ISG15 response. Different MPro inhibitors have been characterized [115,120,124,126,127,128,129,130,131,132,133,134]); however, the most successful has been Paxlovid, developed by Pfizer (PF-07321332) [126]. Paxlovid is an orally bioavailable Mpro inhibitor with in vitro and in vivo antiviral activities that has been tested in patients in combination with ritonavir (a protease inhibitor that extends the half-life of PF-07321332 in the body). As the results of a recent Phase 2/3 clinical trial show, paxlovid has been found to reduce the risk of hospitalization or death by 89% compared with placebo in non-hospitalized high-risk adults with COVID-19. Paxlovid has just been approved by the U.S. Food and Drug Administration issued with an emergency use authorization (EUA) and by the Medicines and Healthcare products Regulatory Agency (MHRA) in the UK (https://www.fda.gov/news-events/press-announcements/coronavirus-COVID-19-update-fda-authorizes-first-oral-antiviral-treatment-COVID-19; 28 January 2022). PF-07321332 is based on a previous compound (PF-835231) that was developed by Pfizer against SARS in 2003/4. Despite being based on an existing molecule, it is impressive that in only a matter of months, PF-07321332 was designed, synthesized, tested in vitro and in vivo, and subsequently approved for use in the clinic.
5. Conclusions and Future Perspectives
SARS-CoV-2 is a positive-sense single-stranded RNA virus, and it is the causative virus of COVID-19 in humans. Owing to increasing numbers of fatalities, the pandemic necessitates urgent therapeutic intervention. Genomic and proteomics analyses have been key in the design and implementation of specific and safe SARS-CoV-2 vaccines and other treatments such as monoclonal antibodies that have saved numerous lives (https://www.COVID19treatmentguidelines.nih.gov/therapies/anti-sars-cov-2-antibody-products; 28 January 2022). The severity of COVID-19 can widely vary between patients. Innate immune sensing and the subsequent activation of the adaptive immune response seem to play a critical role in this [31].
Machinery of the innate immune response is tightly regulated by selective expression of innate immune factors and PTMs of key enzymes in host tissues. For efficient infection, viruses need to evade the innate immune response. For this, they very often hijack the conjugation/deconjugation of ubiquitin and ISG15 on key viral and host proteins. Many virus proteases are able to revert such modifications encoding for Ub/ISG15 proteases. SARS-CoV-2 modulates and inhibits the host ubiquitin and ISG15 response through the expression of PLpro, which cleaves both, ubiquitin and ISG15 from proteins, disrupting antiviral functions. In addition, a number of host factors inside the ubiquitin system machinery interact with SARS-CoV-2 proteins [82,135] and several viral proteins are ubiquitylated [136]. Thus, it appears that SARS-CoV-2 attempts to take control of the host ubiquitin and ISG15 systems.
Viral proteases have been successfully used as targets for small-molecule therapies for the treatment of HIV and HCV [137,138]. The success of paxlovid supports the potential of using viral proteases in COVID-19. The ability of PLpro to inhibit immune responses mediated by ISG15 makes it a promising target for COVID-19 therapeutics. A number of PLpro inhibitors such as GRL0617 have shown antiviral efficacy against SARS-CoV-2 in vitro; however, it is too soon to know if they have the same effects in pre-clinical studies. Approved drugs with described and potential PLpro inhibition profiles as mechanism of action such Ebselen, Disulfiran and Isotretinoin are currently part of COVID-19 clinical trials. These drugs, despite not being highly selective against PLpro, suggest that targeting the deubiquitylating/deISGylating activities of PLpro can be a valid strategy for the treatment of COVID-19.
In addition to targeting viral proteins, it is worth considering host enzymes modulating ubiquitylation/ISGylation of innate immune regulators as targets to neutralize SARS-CoV-2 hijacking of cell functions. Not only can they prevent undesirable host–pathogen interactions, but as host proteins evolve at a much slower pace than ever-mutating viral proteins, they also provide a more stable form of therapy. For instance, USP18 is a potentially relevant target. Similar to PLpro, USP18 also cleaves ISG15 from proteins, acts as a key negative regulator of the interferon pathway, and has been linked to resistance to pathogens [104]. Therefore, USP18 could help to increase antiviral responses and to modulate cytokine storms. It would be also interesting to study whether there is a modulation of USP18 activity during COVID-19 infection as both PLpro and USP18 proteins share substrates. This could provide new avenues of therapeutic intervention. Interestingly, different ways could be used to target USP18: via catalytic activity inhibition with small molecules to prevent deISGylation; by blocking the inhibitory interaction with STAT2 with peptides [139]; or by inducing degradation of USP18, thus preventing both activities, either via degraders/PROTACs or by preventing stabilization by interaction with ISG15 [140].
Deubiquitylating enzymes are druggable enzymes and are currently being explored as pharmacological targets for other diseases [141,142]. Together with USP18, other enzymes with important roles in innate immune responses such as USP13, USP15, USP25, OTUB1, OTUB2, OTULIN, A20, and CYLD represent attractive antiviral targets, and inhibitors for some of them are already available (highlighted in Figure 2). Other DUBs, such as USP8, have been identified as interactors of the viral ORF3 protein and could have a role in viral infection [82]. In addition to DUBs, E3 ligases are equally interesting therapeutic targets. E3 ligases initiate ubiquitin and Ubl signaling during SARS-CoV-2 and other viral infections. Despite their central importance, E3 ligases remain challenging therapeutic targets as their catalytic activity is not easily inhibited by small molecules. However, small molecules promoting the protein degradation (PROTACs) of E3 ligases have already been used in the clinics and could be developed against SARS-CoV-2-relevant E3 enzymes.
It is important to mention that targeting components of the innate immune response could also lead to undesirable side effects, and this should always be considered in studies of a preclinical nature. For example, inherited inactivating mutations of USP18 have been linked to type I interferonopathies [143,144]. Immune-related toxicities are observed when targeting immunomodulatory proteins and can be severe. In the specific case of immune checkpoint inhibitors for cancer immunotherapy, these have been termed immune-related adverse events (irAEs) [145].
In summary, ubiquitylation and ISGylation are two important mechanisms underlying host–pathogen interactions and the defense response of humans infected with viruses such as SARS-CoV-2. The virus hijacks these pathways by either removing ubiquitin and/or ISG15 from proteins, but also by interacting with enzymes involved in ubiquitin/ISG15 machinery. We have highlighted likely points of therapeutic intervention for the treatment of COVID-19 that may prove helpful to combat future vaccine-resistant variants of SARS-CoV-2.
Author Contributions
“Conceptualization, B.M.K., D.P.O., and A.P.-F.; methodology, G.V., D.P.O., and A.P.-F.; software, G.V., R.K., D.P.O., and A.P.-F.; validation, not applicable; formal analysis, not applicable; investigation, G.V., M.R.A., S.F., D.P.O., and A.P.-F.; resources, B.M.K. and A.P.-F.; data curation, not applicable; writing—original draft preparation, G.V., M.R.A., S.F., D.P.O., and A.P.-F.; writing—review and editing, G.V., B.M.K., D.P.O., and A.P.-F.; visualization, G.V., R.K., D.P.O., and A.P.-F.; supervision, B.M.K., D.P.O., and A.P.-F.; project administration, B.M.K., D.P.O., and A.P.-F.; funding acquisition, B.M.K. and A.P.-F. All authors have read and agreed to the published version of the manuscript.”
Funding
B.M.K. and A.P.-F. were supported by the Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Science (CIFMS), China (grant number: 2018-I2M-2-002) and by Pfizer. D.P.O. was supported by a Bayer-Oxford Alliance in Women’s Healthcare award (B.M.K). G.V. is supported by the UKRI-funded E3-Exeter diabetes grant.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Zhong, N.S.; Zheng, B.J.; Li, Y.M.; Poon; Xie, Z.H.; Chan, K.H.; Li, P.H.; Tan, S.Y.; Chang, Q.; Xie, J.P.; et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet 2003, 362, 1353–1358. [Google Scholar] [CrossRef] [Green Version]
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.; Fouchier, R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 2012, 367, 1814–1820. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2020 19:3 2020, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Andreata-Santos, R.; Janini, L.M.R.; Duraes-Carvalho, R. From Alpha to Omicron SARS-CoV-2 variants: What their evolutionary signatures can tell us? J. Med. Virol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.; Kok, K.H.; Zhu, Z.; Chu, H.; To, K.K.; Yuan, S.; Yuen, K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorbalenya, A.E.; Enjuanes, L.; Ziebuhr, J.; Snijder, E.J. Nidovirales: Evolving the largest RNA virus genome. Virus Res. 2006, 117, 17–37. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Mielech, A.M.; Chen, Y.; Mesecar, A.D.; Baker, S.C. Nidovirus papain-like proteases: Multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Res. 2014, 194, 184–190. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.; Lian, J.Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.X.; Gong, L.; et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 2020 5:1 2020, 5, 1–10. [Google Scholar] [CrossRef]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 183, 1735. [Google Scholar] [CrossRef]
- Follis, K.E.; York, J.; Nunberg, J.H. Furin cleavage of the SARS coronavirus spike glycoprotein enhances cell–cell fusion but does not affect virion entry. Virology 2006, 350, 358–369. [Google Scholar] [CrossRef] [Green Version]
- Ackermann, M.; Verleden, S.E.; Kuehnel, M.; Haverich, A.; Welte, T.; Laenger, F.; Vanstapel, A.; Werlein, C.; Stark, H.; Tzankov, A.; et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19. N. Engl. J. Med. 2020, 383, 120–128. [Google Scholar] [CrossRef]
- Hou, Y.J.; Okuda, K.; Edwards, C.E.; Martinez, D.R.; Asakura, T.; Dinnon, K.H.; Kato, T.; Lee, R.E.; Yount, B.L.; Mascenik, T.M.; et al. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell 2020, 182, 429–446. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Liu, S.; Liu, J.; Zhang, Z.; Wan, X.; Huang, B.; Chen, Y.; Zhang, Y. COVID-19: Immunopathogenesis and Immunotherapeutics. Signal Transduct. Target. Ther. 2020, 5, 1–8. [Google Scholar] [CrossRef]
- Qin, C.; Zhou, L.; Hu, Z.; Zhang, S.; Yang, S.; Tao, Y.; Xie, C.; Ma, K.; Shang, K.; Wang, W.; et al. Dysregulation of Immune Response in Patients With Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020, 71, 762–768. [Google Scholar] [CrossRef]
- Lei, X.; Dong, X.; Ma, R.; Wang, W.; Xiao, X.; Tian, Z.; Wang, C.; Wang, Y.; Li, L.; Ren, L.; et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Jentsch, S.; Haendler, B. The Ubiquitin System in Health and Disease; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Kerscher, O.; Felberbaum, R.; Hochstrasser, M. Modification of Proteins by Ubiquitin and Ubiquitin-Like Proteins. Annu. Rev. Cell Dev. Biol. 2006, 22, 159–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clague, M.J.; Heride, C.; Urb, S. The demographics of the ubiquitin system. Trends Cell Biol. 2015, 25, 417–426. [Google Scholar] [CrossRef]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohtake, F.; Tsuchiya, H.; Tanaka, K.; Saeki, Y. Methods to measure ubiquitin chain length and linkage. Methods Enzymol. 2019, 618, 105–133. [Google Scholar] [CrossRef]
- Swatek, K.N.; Usher, J.L.; Kueck, A.F.; Gladkova, C.; Mevissen, T.E.T.; Pruneda, J.N.; Skern, T.; Komander, D. Insights into ubiquitin chain architecture using Ub-clipping. Nature 2019, 572, 533–537. [Google Scholar] [CrossRef]
- Heaton, S.M.; Borg, N.A.; Dixit, V.M. Ubiquitin in the activation and attenuation of innate antiviral immunity. J. Exp. Med. 2016, 213, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Ivashkiv, L.B.; Donlin, L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014, 14, 36–49. [Google Scholar] [CrossRef] [Green Version]
- Paludan, S.R.; Mogensen, T.H. Innate immunological pathways in COVID-19 pathogenesis. Sci. Immunol. 2022, 7, eabm5505. [Google Scholar] [CrossRef]
- Kasuga, Y.; Zhu, B.; Jang, J.-J.; Yoo, J.S. Innate immune sensing of coronavirus and viral evasion strategies. Exp. Mol. Med. 2021, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Pandolfi, L.; Fossali, T.; Frangipane, V.; Bozzini, S.; Morosini, M.; D’Amato, M.; Lettieri, S.; Urtis, M.A. Broncho-alveolar inflammation in COVID-19 patients: A correlation with clinical outcome. BMC BMC Pulm. Med. 2020, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Thwaites, R.S.; Sanchez Sevilla Uruchurtu, A.; Siggins, M.K.; Liew, F.; Russell, C.D.; Moore, S.C.; Fairfield, C.; Carter, E.; Abrams, S.; Short, C.E.; et al. Inflammatory profiles across the spectrum of disease reveal a distinct role for GM-CSF in severe COVID-19. Sci. Immunol. 2021, 6. [Google Scholar] [CrossRef] [PubMed]
- Lester, S.; Li, K. Toll-Like Receptors in Antiviral Innate Immunity. J. Mol. Biol. 2014, 426, 1246–1264. [Google Scholar] [CrossRef] [PubMed]
- Balka, K.R.a. Understanding early TLR signaling through the Myddosome. J. Leukoc. Biol. 2019, 105, 339–351. [Google Scholar] [CrossRef]
- Cohen, P.; Strickson, S. The role of hybrid ubiquitin chains in the MyD88 and other innate immune signalling pathways. Cell Death Differ. 2017, 24, 1153–1159. [Google Scholar] [CrossRef] [Green Version]
- Kurt-Jones, E.A.; Popova, L.; Kwinn, L.; Haynes, L.M.; Jones, L.P.; Tripp, R.A.; Walsh, E.E.; Freeman, M.W.; Golenbock, D.T.; Anderson, L.J.; et al. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat. Immunol. 2000, 1, 398–401. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Kuang, M.; Li, J.; Zhu, L.; Jia, Z.; Guo, X.; Hu, Y.; Kong, J.; Yin, H.; Wang, X.; et al. SARS-CoV-2 spike protein interacts with and activates TLR4. Cell Res. 2021, 31, 818–820. [Google Scholar] [CrossRef]
- Zheng, M.; Karki, R.; Williams, E.P.; Yang, D.; Fitzpatrick, E.; Vogel, P.; Jonsson, C.B.; Kanneganti, T.-d. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat. Immunol. 2021, 22, 829–838. [Google Scholar] [CrossRef]
- Moynagh, P.N. The roles of Pellino E3 ubiquitin ligases in immunity. Nat. Rev. Immunol. 2014, 14, 122–131. [Google Scholar] [CrossRef]
- Enesa, K.; Ordureau, A.; Smith, H.; Barford, D.; Cheung, P.C.F.; Patterson-Kane, J.; Arthur, J.S.C.; Cohen, P. Pellino1 is required for interferon production by viral double-stranded RNA. J. Biol. Chem. 2012, 287, 34825–34835. [Google Scholar] [CrossRef] [Green Version]
- Zinngrebe, J.; Rieser, E.; Taraborrelli, L.; Peltzer, N.; Hartwig, T.; Ren, H.; Kovcs, I.; Endres, C.; Draber, P.; Darding, M.a. LUBAC deficiency perturbs TLR3 signaling to cause immunodeficiency and autoinflammation. J. Exp. Med. 2016, 213, 2671–2689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sola, I.; Almazn, F.; Ziga, S.; Enjuanes, L. Continuous and Discontinuous RNA Synthesis in Coronaviruses. Annu. Rev. Virol. 2015, 2, 265–288. [Google Scholar] [CrossRef] [Green Version]
- van der Made, C.I.; Simons, A.; Schuurs-Hoeijmakers, J.; van den Heuvel, G.; Mantere, T.; Kersten, S.; van Deuren, R.C.; Steehouwer, M.; van Reijmersdal, S.V.; Jaeger, M.; et al. Presence of Genetic Variants Among Young Men With Severe COVID-19. JAMA 2020, 324, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Liu, Z.; Moncada-Velez, M.; Chen, J.; Ogishi, M.; Bigio, B.; Yang, R.; Arias, A.A.; Zhou, Q.; Han, J.E.; et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 2020, 370. [Google Scholar] [CrossRef] [PubMed]
- Totura, A.L.; Whitmore, A.; Agnihothram, S.; Schfer, A.; Katze, M.G.; Heise, M.T.; Baric, R.S. Toll-Like Receptor 3 Signaling via TRIF Contributes to a Protective Innate Immune Response to Severe Acute Respiratory Syndrome Coronavirus Infection. mBio 2015, 6, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Bortolotti, D.; Gentili, V.; Rizzo, S.; Schiuma, G.; Beltrami, S.; Strazzabosco, G.; Fernandez, M.; Caccuri, F.; Caruso, A.; Rizzo, R. TLR3 and TLR7 RNA Sensor Activation during SARS-CoV-2 Infection. Microorganisms 2021, 9, 1820. [Google Scholar] [CrossRef]
- Li, J.; Liu, Y.; Zhang, X. Murine Coronavirus Induces Type I Interferon in Oligodendrocytes through Recognition by RIG-I and MDA5. J. Virol. 2010, 84, 6472–6482. [Google Scholar] [CrossRef] [Green Version]
- Zalinger, Z.B.; Elliott, R.; Rose, K.M.; Weiss, S.R. MDA5 Is Critical to Host Defense during Infection with Murine Coronavirus. J. Virol. 2015, 89, 12330–12340. [Google Scholar] [CrossRef] [Green Version]
- Rehwinkel, J.; Gack, M.U. RIG-I-like receptors: Their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020, 20, 537–551. [Google Scholar] [CrossRef]
- Peisley, A.; Wu, B.; Xu, H.; Chen, Z.J.; Hur, S. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature 2014, 509, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Gack, M.U.; Shin, Y.C.; Joo, C.H.; Urano, T.; Liang, C.; Sun, L.; Takeuchi, O.; Akira, S.; Chen, Z.; Inoue, S.; et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 2007, 446, 916–920. [Google Scholar] [CrossRef]
- Yan, J.; Li, Q.; Mao, A.P.; Hu, M.M.; Shu, H.B. TRIM4 modulates type I interferon induction and cellular antiviral response by targeting RIG-I for K63-linked ubiquitination. J. Mol. Cell Biol. 2014, 6, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Song, Y.; Li, Y.; Zhu, Q.; Tan, P.; Qin, Y.; Wang, H.Y.; Wang, R.-f. USP3 inhibits type I interferon signaling by deubiquitinating RIG-I-like receptors. Nat. Publ. Group 2014, 24, 400–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedman, C.S.; O’Donnell, M.A.; Legarda-Addison, D.; Ng, A.; Crdenas, W.B.; Yount, J.S.; Moran, T.M.; Basler, C.F.; Komuro, A.; Horvath, C.M.; et al. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep. 2008, 9, 930–936. [Google Scholar] [CrossRef]
- Oshiumi, H.; Miyashita, M.; Inoue, N.; Okabe, M.; Matsumoto, M.; Seya, T. The ubiquitin ligase riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe 2010, 8, 496–509. [Google Scholar] [CrossRef] [Green Version]
- Pauli, E.K.; Chan, Y.K.; Davis, M.E.; Gableske, S.; Wang, M.K.; Feister, K.F.; Gack, M.U. The ubiquitin-specific protease USP15 promotes RIG-I-mediated antiviral signaling by deubiquitylating TRIM25. Sci. Signal. 2014, 7. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhao, W.; Zhang, M.; Wang, P.; Zhao, K.; Zhao, X.; Yang, S.; Gao, C. USP4 Positively Regulates RIG-I-Mediated Antiviral Response through Deubiquitination and Stabilization of RIG-I. J. Virol. 2013, 87, 4507–4515. [Google Scholar] [CrossRef] [Green Version]
- Lang, X.; Tang, T.; Jin, T.; Ding, C.; Zhou, R.; Jiang, W. TRIM65-catalized ubiquitination is essential for MDA5-mediated antiviral innate immunity. J. Exp. Med. 2017, 214, 459–473. [Google Scholar] [CrossRef]
- Liu, G.; Lee, J.-h.; Parker, Z.M.; Acharya, D.; Chiang, J.J.; Gent, M.V.; Riedl, W.; Davis-gardner, M.E.; Wies, E.; Chiang, C.; et al. ISG15-dependent activation of the sensor MDA5 is antagonized by the SARS-CoV-2 papain-like protease to evade host innate immunity. Nat. Microbiol. 2021, 6, 467–478. [Google Scholar] [CrossRef]
- Rebendenne, A.a. SARS-CoV-2 Triggers an MDA-5-Dependent Interferon Response Which Is Unable To Control Replication in Lung Epithelial Cells. J. Virol. 2021, 95, e02415-20. [Google Scholar] [CrossRef] [PubMed]
- Sampaio, N.G.; Chauveau, L.; Hertzog, J.; Bridgeman, A.; Fowler, G.; Moonen, J.P.; Dupont, M.; Russell, R.A.; Noerenberg, M.; Rehwinkel, J. The RNA sensor MDA5 detects SARS - CoV - 2 infection. Sci. Rep. 2021, 1–10. [Google Scholar] [CrossRef]
- Thorne, L.G.; Reuschl, A.-k.; Zuliani-alvarez, L.; Whelan, M.V.X.; Turner, J.; Noursadeghi, M.; Jolly, C.; Towers, G.J. SARS-CoV-2 sensing by RIG-I and MDA 5 links epithelial infection to macrophage inflammation. EMBO J. 2021, 40, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Riva, L.; Pu, Y.; Martin-Sancho, L.; Kanamune, J.; Yamamoto, Y.; Sakai, K.; Gotoh, S.; Miorin, L.a. MDA5 Governs the Innate Immune Response to SARS-CoV-2 in Lung Epithelial Cells. Cell Rep. 2021, 34. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Sato, S.; Sotoyama, Y.; Orba, Y.; Sawa, H.; Yamauchi, H.; Sasaki, M.; Takaoka, A. RIG-I triggers a signaling-abortive anti-SARS-CoV-2 defense in human lung cells. Nat. Immunol. 2021, 22, 820–828. [Google Scholar] [CrossRef]
- Castanier, C.; Zemirli, N.; Portier, A.; Garcin, D.; Bidre, N.; Vazquez, A.; Arnoult, D. MAVS ubiquitination by the E3 ligase TRIM25 and degradation by the proteasome is involved in type I interferon production after activation of the antiviral RIG-I-like receptors. BMC Biol. 2012, 10, 44. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Shi, Y.; Pan, X.; Wu, S.; Hou, R.; Zhang, Y.; Zhong, T.; Tang, H.; Du, W.; Wang, L.; et al. SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. Cell Rep. 2021, 34, 108761. [Google Scholar] [CrossRef] [PubMed]
- Daniel, J.; Liebau, E. The Ufm1 Cascade. Cells 2014, 3, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Hipp, M.S.; Kalveram, B.; Raasi, S.; Groettrup, M.; Schmidtke, G. FAT10, a Ubiquitin-Independent Signal for Proteasomal Degradation. Mol. Cell. Biol. 2005, 25, 3483–3491. [Google Scholar] [CrossRef] [Green Version]
- Hochstrasser, M. Origin and function of ubiquitin-like proteins. Nature 2009, 458, 422–429. [Google Scholar] [CrossRef] [Green Version]
- Perng, Y.C.; Lenschow, D.J. ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 2018, 16, 423–439. [Google Scholar] [CrossRef]
- Wong, J.J.; Pung, Y.F.; Sze, N.S.; Chin, K.C. HERC5 is an IFN-induced HECT-type E3 protein ligase that mediates type I IFN-induced ISGylation of protein targets. Proc. Natl. Acad. Sci. USA 2006, 103, 10735–10740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ketscher, L.; Basters, A.; Prinz, M.; Knobeloch, K.P. mHERC6 is the essential ISG15 E3 ligase in the murine system. Biochem. Biophys. Res. Commun. 2012, 417, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Durfee, L.A.; Lyon, N.; Seo, K.; Huibregtse, J.M. The ISG15 Conjugation System Broadly Targets Newly Synthesized Proteins: Implications for the Antiviral Function of ISG15. Mol. Cell 2010, 38, 722–732. [Google Scholar] [CrossRef] [PubMed]
- Okumura, F.; Okumura, A.J.; Uematsu, K.; Hatakeyama, S.; Zhang, D.E.; Kamura, T. Activation of double-stranded rna-activated protein kinase (PKR) by interferon-stimulated gene 15 (ISG15) modification down-regulates protein translation. J. Biol. Chem. 2013, 288, 2839–2847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Y.; Zhong, G.; Zhu, L.; Liu, X.; Shan, Y.; Feng, H.; Bu, Z.; Chen, H.; Wang, C. Herc5 Attenuates Influenza A Virus by Catalyzing ISGylation of Viral NS1 Protein. J. Immunol. 2010, 184, 5777–5790. [Google Scholar] [CrossRef] [Green Version]
- Skaug, B.; Chen, Z.J. Emerging Role of ISG15 in Antiviral Immunity. Cell 2010, 143, 187–190. [Google Scholar] [CrossRef] [Green Version]
- Munnur, D.; Teo, Q.; Eggermont, D.; Lee, H.H.Y.; Thery, F.; Ho, J.; Leur, S.W.V.; Ng, W.W.S.; Siu, L.Y.L.; Beling, A.; et al. Altered ISGylation drives aberrant macrophage-dependent immune responses during SARS-CoV-2 infection. Nat. Immunol. 2021. [Google Scholar] [CrossRef]
- Swaim, C.D.; Scott, A.F.; Canadeo, L.A.; Huibregtse, J.M. Extracellular ISG15 Signals Cytokine Secretion through the LFA-1 Integrin Receptor. Mol. Cell 2017, 68, 581–590.e5. [Google Scholar] [CrossRef] [Green Version]
- Kikkert, M. Innate Immune Evasion by Human Respiratory RNA Viruses. J. Innate. Immun. 2020, 12, 4–20. [Google Scholar] [CrossRef]
- Stukalov, A.; Girault, V.; Grass, V.; Karayel, O.; Bergant, V.; Urban, C.; Haas, D.A.; Huang, Y.; Oubraham, L.; Wang, A.; et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature 2021, 594, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Vere, G.; Kealy, R.; Kessler, B.M.; Pinto-Fernandez, A. Ubiquitomics: An overview and future. Biomolecules 2020, 10, 1453. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Bouch, R.J.; Blekhman, M.G.; He, Z. USP19 Suppresses Th17-Driven Pathogenesis in Autoimmunity. J. Immunol. 2021, 207, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Bez-Santos, Y.M.a. The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015, 115, 21–38. [Google Scholar] [CrossRef] [PubMed]
- Frias-Staheli, N.; Giannakopoulos, N.V.; Kikkert, M.; Taylor, S.L.; Bridgen, A.; Paragas, J.; Richt, J.A.; Rowland, R.R.; Schmaljohn, C.S.; Lenschow, D.J.; et al. Ovarian Tumor Domain-Containing Viral Proteases Evade Ubiquitin- and ISG15-Dependent Innate Immune Responses. Cell Host Microbe 2007, 2, 404–416. [Google Scholar] [CrossRef] [Green Version]
- Swatek, K.N.; Aumayr, M.; Pruneda, J.N.; Visser, L.J.; Berryman, S.; Kueck, A.F.; Geurink, P.P.; Ovaa, H.; van Kuppeveld, F.J.M.; Tuthill, T.J.; et al. Irreversible inactivation of ISG15 by a viral leader protease enables alternative infection detection strategies. Proc. Natl. Acad. Sci. USA 2018, 115, 2371–2376. [Google Scholar] [CrossRef] [Green Version]
- Klemm, T.; Ebert, G.; Calleja, D.J.; Allison, C.C.; Richardson, L.W.; Bernardini, J.P.; Lu, B.G.C.; Kuchel, N.W.; Grohmann, C.; Shibata, Y.; et al. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020, 39, 1–17. [Google Scholar] [CrossRef]
- Lindner, H.A.; Fotouhi-Ardakani, N.; Lytvyn, V.; Lachance, P.; Sulea, T.; Mnard, R. The Papain-Like Protease from the Severe Acute Respiratory Syndrome Coronavirus Is a Deubiquitinating Enzyme. J. Virol. 2005, 79, 15199–15208. [Google Scholar] [CrossRef] [Green Version]
- Shin, D.; Mukherjee, R.; Grewe, D.; Bojkova, D.; Baek, K.; Bhattacharya, A.; Schulz, L.; Widera, M.; Mehdipour, A.R.; Tascher, G.; et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 2020, 587, 657–662. [Google Scholar] [CrossRef]
- Fu, Z.; Huang, B.; Tang, J.; Liu, S.; Liu, M.; Ye, Y.; Liu, Z.; Xiong, Y.; Zhu, W.; Cao, D.; et al. The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery. Nat. Commun. 2021, 12, 488. [Google Scholar] [CrossRef]
- Leite, W.C.; Weiss, K.L.; Phillips, G.; Zhang, Q.; Qian, S.; Tsutakawa, S.E.; Coates, L.; O’Neill, H. Conformational Dynamics in the Interaction of SARS-CoV-2 Papain-like Protease with Human Interferon-Stimulated Gene 15 Protein. J. Phys. Chem. Lett. 2021, 12, 5608–5615. [Google Scholar] [CrossRef]
- Yan, S.; Ahmad, R.; Rosen, E.D.; Yan, S.; Kumari, M.; Xiao, H.; Jacobs, C.; Kochumon, S.; Jedrychowski, M.; Chouchani, E.; et al. IRF3 reduces adipose thermogenesis via ISG15- mediated reprogramming of glycolysis. J. Phys. Chem. Lett. 2021, 131. [Google Scholar] [CrossRef]
- Zhang, Y.; Thery, F.; Wu, N.C.; Luhmann, E.K.; Dussurget, O.; Foecke, M.; Bredow, C.; Jimnez-Fernndez, D.; Leandro, K.; Beling, A.; et al. The in vivo ISGylome links ISG15 to metabolic pathways and autophagy upon Listeria monocytogenes infection. Nat. Commun. 2019, 10, 5383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oxana, A.M.; Keun Il, K.; Jiann Kae, L.; Weiguo, Z.; Kumar, K.G.S.; Serge, Y.F.; Ke, S.; Dong Er, Z. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 2006, 25, 2358–2367. [Google Scholar] [CrossRef]
- Eleanor, Q.; Hasan, T.; Poornima, K.; Robert, H.; Rajeev, J. Treatment of COVID-19: A review of current and prospective pharmacotherapies. Br. J. Hosp. Med. 2021, 82, 50. [Google Scholar] [CrossRef]
- Yang, L.; Xie, X.; Tu, Z.; Fu, J.; Xu, D.; Zhou, Y. The signal pathways and treatment of cytokine storm in COVID-19. Signal Transduct. Target. 2021, 6, 255. [Google Scholar] [CrossRef] [PubMed]
- Daniel, B.-M.; Benjamin, E.N.-P.; Wen Chun, L.; Skyler, U.; Daisy, H.; Rasmus, M.; Tristan, X.J.; Kohei, O.; Maryline, P.; David, S.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045. [Google Scholar] [CrossRef]
- Jérôme, H.; Nader, Y.; Laura, B.; Aurélien, C.; Jeremy, B.; Nikaïa, S.; Hélène, P.; Bruno, C.; Vincent, B.; Camille, C.-G.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef]
- Ziegler, C.G.K.; Allon, S.J.; Nyquist, S.K.; Mbano, I.M.; Miao, V.N.; Tzouanas, C.N.; Cao, Y.; Yousif, A.S.; Bals, J.; Hauser, B.M.; et al. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell 2020, 181, 1016–1035.e19. [Google Scholar] [CrossRef]
- Yamanaka, S.; Sato, Y.; Oikawa, D.; Goto, E.; Fukai, S.; Tokunaga, F.; Takahashi, H.; Sawasaki, T. Subquinocin, a small molecule inhibitor of CYLD and USP-family deubiquitinating enzymes, promotes NF-kappaB signaling. Biochem. Biophys. Res. Commun. 2020, 524, 1–7. [Google Scholar] [CrossRef]
- Resnick, E.; Bradley, A.; Gan, J.; Douangamath, A.; Krojer, T.; Sethi, R.; Geurink, P.P.; Aimon, A.; Amitai, G.; Bellini, D.; et al. Rapid Covalent-Probe Discovery by Electrophile-Fragment Screening. J. Am. Chem. Soc. 2019, 141, 8951–8968. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, H.H.; Kim, T.; Nguyen, T.; Hahn, M.J.; Yun, S.I.; Kim, K.K. A Selective Inhibitor of Ubiquitin-Specific Protease 4 Suppresses Colorectal Cancer Progression by Regulating beta-Catenin Signaling. Cell Physiol. Biochem. 2019, 53, 157–171. [Google Scholar] [CrossRef] [PubMed]
- Daniel Jiménez, F.; Sandra, H.; Klaus Peter, K. Strategies to Target ISG15 and USP18 Toward Therapeutic Applications. Front. Chem. 2020, 7, 923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiaodong, H.; Vishva, M.D. Drugging the undruggables: Exploring the ubiquitin system for drug development. Cell Res. 2016, 26, 484–498. [Google Scholar] [CrossRef]
- Zhenyi, H.; Craig, M.C. Recent Developments in PROTAC-Mediated Protein Degradation: From Bench to Clinic. ChemBioChem 2021. [Google Scholar] [CrossRef]
- Zhang, H.; Zheng, H.; Zhu, J.; Dong, Q.; Wang, J.; Fan, H.; Chen, Y.; Zhang, X.; Han, X.; Li, Q.; et al. Ubiquitin-Modified Proteome of SARS-CoV-2-Infected Host Cells Reveals Insights into Virus-Host Interaction and Pathogenesis. J. Proteome Res. 2021, 20, 2224–2239. [Google Scholar] [CrossRef] [PubMed]
- Hom-Ming, Y.; Chia-Yi, Y.; Ho-Chun, Y.; Shih-Han, K.; Ching-Len, L.; Yi-Ling, L. Ubiquitin-Specific Protease 13 Regulates IFN Signaling by Stabilizing STAT1. J. Immunol. 2013, 191, 3328–3336. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, D.; Zhong, H.; Luo, R.; Shang, M.; Liu, D.; Chen, H.; Fang, L.; Xiao, S. Ubiquitin-specific Protease 15 Negatively Regulates Virus-induced Type I Interferon Signaling via Catalytically-dependent and -independent Mechanisms. Sci. Rep. 2015, 5, 11220. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; Wang, D.; Fang, L.; Zhang, H.; Luo, R.; Shang, M.; Ouyang, C.; Ouyang, H.; Chen, H.; Xiao, S. Ubiquitin-specific proteases 25 negatively regulates virus-induced type I interferon signaling. PLoS ONE 2013, 8, e80976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hylemariam Mihiretie, M.; Tebelay, D.; Tengchuan, J. Structural Basis of Potential Inhibitors Targeting SARS-CoV-2 Main Protease. Front. Chem. 2021, 9, 7. [Google Scholar] [CrossRef]
- Anja, B.; Paul, P.G.; Farid El, O.; Lars, K.; Marco, S.C.; Eberhard, K.; Huib, O.; Klaus Peter, K.; Günter, F. Molecular characterization of ubiquitin-specific protease 18 reveals substrate specificity for interferon-stimulated gene 15. FEBS J. 2014, 281, 1918–1928. [Google Scholar] [CrossRef]
- Brendan, T.F.; Ian, A.D.; Jackelyn, M.; Jaron, E.L.; Holden, C.M.; David, C.; Robert Jeff, H.; Ralph, A.T.; Scott, D.P. Characterization and Noncovalent Inhibition of the Deubiquitinase and deISGylase Activity of SARS-CoV-2 Papain-Like Protease. ACS Infect. Dis. 2020, 6, 2099–2109. [Google Scholar] [CrossRef]
- Mostafa, J.; Ebrahim, B.; Fathollah, G.-B. Structure-Based Screening to Discover New Inhibitors for Papain-like Proteinase of SARS-CoV-2: An in Silico Study. J. Proteome Res. 2021, 20, 1015–1026. [Google Scholar] [CrossRef]
- Shuvasish, C.; Debojyoti, M.; Anupom, B.; Purbajyoti, S.; Muhammed Khairujjaman, M. In search of drugs to alleviate suppression of the host’s innate immune responses against SARS-CoV-2 using a molecular modeling approach. Silico Pharmacol. 2021, 9. [Google Scholar] [CrossRef]
- Jung Keun, C.; Marcus, J.C.-L.; Kon Ho, L.; Dae Wook, K.; Hyung Won, R.; Heung Joo, Y.; Ki Hun, P. Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia tomentosa. Bioorganic Med. Chem. 2013, 21, 3051–3057. [Google Scholar] [CrossRef]
- Rajesh, G.; Ayon, C.; Ashis, B.; Snehasis, C. Identification of polyphenols from Broussonetia papyrifera as SARS CoV-2 main protease inhibitors using in silico docking and molecular dynamics simulation approaches. J. Biomol. Struct. Dyn. 2021, 39, 6747–6760. [Google Scholar] [CrossRef]
- Lingyu, L.; Liyan, M.; Yue, H.; Xiaoxue, L.; Meng, Y.; Hai, S.; Zhongmei, Z. Natural biflavones are potent inhibitors against SARS-CoV-2 papain-like protease. Phytochemistry 2021, 193, 112984. [Google Scholar] [CrossRef]
- Chung Yi, W.; Jia Tsrong, J.; Shiou Hwa, M.; Chin Jung, K.; Hsueh Fen, J.; Yih Shyun, E.C.; Hsien Hua, H.; Hsuan Cheng, H.; Douglass, W.; Ashraf, B.; et al. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. USA 2004, 101, 10012–10017. [Google Scholar] [CrossRef] [Green Version]
- Zinuo, C.; Qinghua, C.; Laura, C.; Pin, Z.; Hyun, L.; Zhaoyu, C.; Yanyan, W.; Xiaoyun, L.; Lijun, R.; Ruikun, D. Ginkgolic acid and anacardic acid are specific covalent inhibitors of SARS-CoV-2 cysteine proteases. Cell Biosci. 2021, 11. [Google Scholar] [CrossRef]
- Naveen, N.; Deepak, T.N. Vitamin B12 may inhibit RNA-dependent-RNA polymerase activity of nsp12 from the SARS-CoV-2 virus. IUBMB life 2020, 72, 2112–2120. [Google Scholar] [CrossRef]
- Sheng Teng, H.; Yeh, C.; Wei Chao, C.; Hsiao Fan, C.; Hsiang Chun, L.; Yu Chun, L.; Wei Jan, W.; Yu Chuan, W.; Chia Shin, Y.; Shao Chun, W.; et al. Scutellaria barbata d. Don inhibits the main proteases (mpro and tmprss2) of severe acute respiratory syndrome coronavirus 2 (sars-cov-2) infection. Viruses 2021, 13, 826. [Google Scholar] [CrossRef]
- Lee, A.A.; Sven, M.L.; Virginia Dee, C.; Stephen, P.M.; Raja Sekhar, N.; Isobel, C.; Anthony, H.; Fraser, C.; Rachel, T.; Rukmini, M.; et al. Biochemical characterization of protease activity of Nsp3 from SARS-CoV-2 and its inhibition by nanobodies. PLoS ONE 2021, 16, e0253364. [Google Scholar] [CrossRef]
- Zhengnan, S.; Kiira, R.; Laura, C.; Deyu, K.; Hyun, L.; Youngjin, K.; Yangfeng, L.; Saad, A.; Fei, H.; Oleksii, D.; et al. Design of SARS-CoV-2 PLpro Inhibitors for COVID-19 Antiviral Therapy Leveraging Binding Cooperativity. J. Med. Chem. 2021. [Google Scholar] [CrossRef]
- Narayanan, A.; Toner, S.A.; Jose, J. Structure-based inhibitor design and repurposing clinical drugs to target SARS-CoV-2 proteases. Biochem. Soc. Trans. 2022. [Google Scholar] [CrossRef]
- Dafydd, R.O.; Charlotte, M.N.A.; Annaliesa, S.A.; Lisa, A.; Melissa, A.; Simon, B.; Britton, B.; Rhonda, D.C.; Anthony, C.; Karen, J.C.; et al. An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19. Science 2021, 374, 1586–1593. [Google Scholar] [CrossRef]
- Robert, L.H.; Robert, S.K.; Mary, A.B.; Jay, F.D.; Rose, A.F.; Ketan, S.G.; Mingying, H.; Robert, J.H.; Kirk, K.; Lilian, Y.L.; et al. Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19. J. Med. Chem. 2020, 63, 12725–12747. [Google Scholar] [CrossRef]
- Linlin, Z.; Daizong, L.; Xinyuanyuan, S.; Ute, C.; Christian, D.; Lucie, S.; Stephan, B.; Katharina, R.; Rolf, H. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef] [Green Version]
- Britton, B.; Rhys, M.J.; Brandon, J.A.; Dan, A.; Lisa, A.; Malina, A.B.; Nathan, B.; Joseph, B.; Emily, C.; Heather, E.; et al. Preclinical characterization of an intravenous coronavirus 3CL protease inhibitor for the potential treatment of COVID19. Nat. Commun. 2021, 12, 1–17. [Google Scholar] [CrossRef]
- Temitope, I.A.; Misbaudeen, A.-H.; Emmanuel, M.O.; Qudus, K.O.; Ibrahim, D.B.; Ibrahim, O.A.; Olamide, T.O.; Ajayi, A.F.; Oladipo, E.K. Molecular docking assessment of clinically approved antiviral drugs against Mpro, spike glycoprotein and angiotensin converting enzyme-2 revealed probable anti-SARS-CoV-2 potential. Trop. J. Nat. Prod. Res. 2021, 5, 778–791. [Google Scholar] [CrossRef]
- Chunlong, M.; Michael Dominic, S.; Brett, H.; Julia Alma, T.; Yanmei, H.; Tommy, S.; Xiujun, Z.; Bart, T.; Michael Thomas, M.; Yu, C.; et al. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020, 30, 678–692. [Google Scholar] [CrossRef]
- Dale, L.B.; Valerie, D.H.; Jared, B.; Donald, F.S.; John, D.M.; Michael, J.O.; Robert, W.S. Inhibition of severe acute respiratory syndrome-associated coronavirus (SARSCoV) by calpain inhibitors and beta-D-N4-hydroxycytidine. Antivir. Chem. Chemother. 2004, 15, 15–22. [Google Scholar] [CrossRef] [Green Version]
- Bin, C.; Yeming, W.; Danning, W.; Wen, L.; Jingli, W.; Guohui, F.; Lianguo, R.; Bin, S.; Yanping, C.; Ming, W.; et al. A Trial of Lopinavir–Ritonavir in Adults Hospitalized with Severe Covid-19. N. Engl. J. Med. 2020, 382, 1787–1799. [Google Scholar] [CrossRef]
- Peter, W.H.; Marion, M.; Jennifer, L.B.; Louise, L.; Natalie, S.; Jonathan, E.; Adrian, P.; Jason, R.; Einas, E.; Benjamin, P.; et al. Lopinavir–ritonavir in patients admitted to hospital with COVID-19 (RECOVERY): A randomised, controlled, open-label, platform trial. Lancet 2020, 396, 1345–1352. [Google Scholar] [CrossRef]
- Terracciano, R.; Preiano, M.; Fregola, A.; Pelaia, C.; Montalcini, T.; Savino, R. Mapping the SARS-CoV-2-Host Protein-Protein Interactome by Affinity Purification Mass Spectrometry and Proximity-Dependent Biotin Labeling: A Rational and Straightforward Route to Discover Host-Directed Anti-SARS-CoV-2 Therapeutics. Int. J. Mol. Sci. 2021, 22, 532. [Google Scholar] [CrossRef]
- Schiller, H.B.; van Breugel, M.; Nawijn, M.C. SARS-CoV-2-specific hotspots in virus-host interaction networks. Nat. Immunol. 2021, 22, 806–808. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Osswald, H.L.; Prato, G. Recent Progress in the Development of HIV-1 Protease Inhibitors for the Treatment of HIV/AIDS. J. Med. Chem. 2016, 59, 5172–5208. [Google Scholar] [CrossRef] [Green Version]
- Cannalire, R.; Barreca, M.L.; Manfroni, G.; Cecchetti, V. A Journey around the Medicinal Chemistry of Hepatitis C Virus Inhibitors Targeting NS4B: From Target to Preclinical Drug Candidates. J. Med. Chem. 2016, 59, 16–41. [Google Scholar] [CrossRef]
- Arimoto, K.I.; Lochte, S.; Stoner, S.A.; Burkart, C.; Zhang, Y.; Miyauchi, S.; Wilmes, S.; Fan, J.B.; Heinisch, J.J.; Li, Z.; et al. STAT2 is an essential adaptor in USP18-mediated suppression of type I interferon signaling. Nat. Struct. Mol. Biol. 2017, 24, 279–289. [Google Scholar] [CrossRef] [Green Version]
- Tokarz, S.; Berset, C.; La Rue, J.; Friedman, K.; Nakayama, K.; Nakayama, K.; Zhang, D.E.; Lanker, S. The ISG15 isopeptidase UBP43 is regulated by proteolysis via the SCFSkp2 ubiquitin ligase. J. Biol. Chem. 2004, 279, 46424–46430. [Google Scholar] [CrossRef] [Green Version]
- Turnbull, A.P.; Ioannidis, S.; Krajewski, W.W.; Pinto-Fernandez, A.; Heride, C.; Martin, A.C.L.; Tonkin, L.M.; Townsend, E.C.; Buker, S.M.; Lancia, D.R.; et al. Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature 2017, 550, 481–486. [Google Scholar] [CrossRef]
- Pinto-Fernandez, A.; Kessler, B.M. DUBbing Cancer: Deubiquitylating Enzymes Involved in Epigenetics, DNA Damage and the Cell Cycle As Therapeutic Targets. Front. Genet. 2016, 7, 133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meuwissen, M.E.; Schot, R.; Buta, S.; Oudesluijs, G.; Tinschert, S.; Speer, S.D.; Li, Z.; van Unen, L.; Heijsman, D.; Goldmann, T.; et al. Human USP18 deficiency underlies type 1 interferonopathy leading to severe pseudo-TORCH syndrome. J. Exp. Med. 2016, 213, 1163–1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gruber, C.; Martin-Fernandez, M.; Ailal, F.; Qiu, X.; Taft, J.; Altman, J.; Rosain, J.; Buta, S.; Bousfiha, A.; Casanova, J.L.; et al. Homozygous STAT2 gain-of-function mutation by loss of USP18 activity in a patient with type I interferonopathy. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef] [PubMed]
- Conroy, M.; Naidoo, J. Immune-related adverse events and the balancing act of immunotherapy. Nat. Commun. 2022, 13, 392. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).