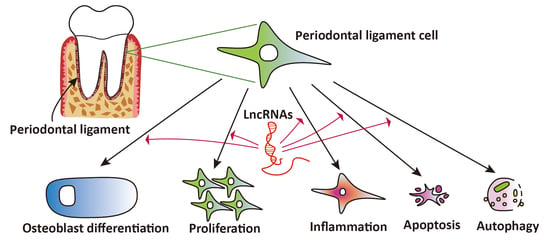
The Expression and Regulatory Roles of Long Non-Coding RNAs in Periodontal Ligament Cells: A Systematic Review
Abstract
:1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Selection Criteria
2.3. Selection of Studies
2.4. Quality Assessment
3. Results
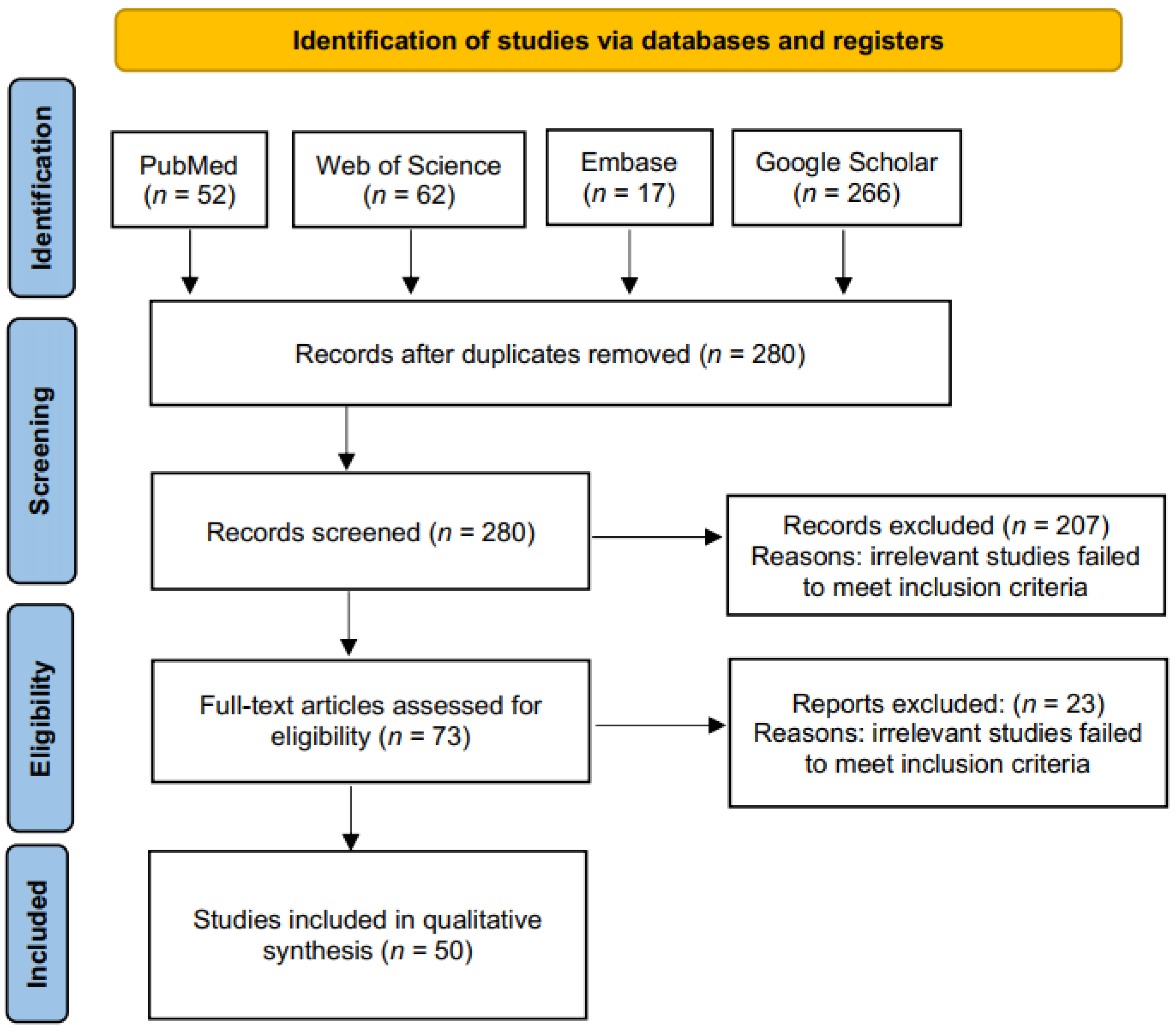
3.1. Literature Search and Screening of Studies
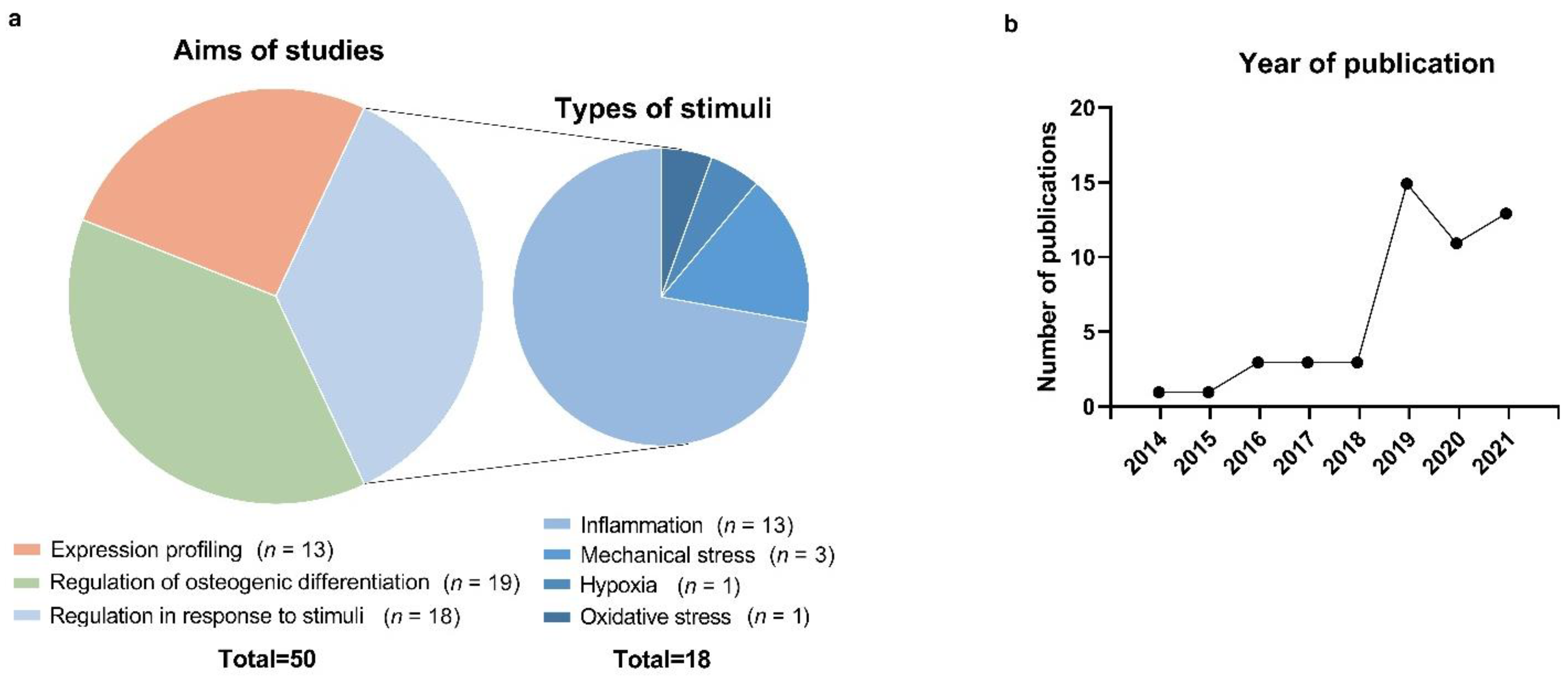
3.2. Studies on lncRNA Expression Profiling in PDL Cells
3.3. Studies on lncRNAs Involved in the Osteogenic Differentiation of PDL Cells
3.4. Studies on lncRNAs in PDL Cells Subjected to Inflammation, Mechanical Stress, and Other Stimuli
4. Discussion
4.1. Studies on lncRNA Expression Profiling in PDL Cells
4.2. Studies on lncRNAs Involved in the Osteogenic Differentiation of PDL Cells
4.3. Studies on lncRNAs in PDL Cells Subjected to Inflammation, Mechanical Stress, and Other Stimuli
4.4. Future Perspectives
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kinane, D.F.; Stathopoulou, P.G.; Papapanou, P.N. Periodontal diseases. Nat. Rev. Dis. Primers 2017, 3, 17038. [Google Scholar] [CrossRef] [PubMed]
- Ramseier, C.A.; Anerud, A.; Dulac, M.; Lulic, M.; Cullinan, M.P.; Seymour, G.J.; Faddy, M.J.; Bürgin, W.; Schätzle, M.; Lang, N.P. Natural history of periodontitis: Disease progression and tooth loss over 40 years. J. Clin. Periodontol. 2017, 44, 1182–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Ruan, J.; Weir, M.D.; Ren, K.; Schneider, A.; Wang, P.; Oates, T.W.; Chang, X.; Xu, H.H.K. Periodontal Bone-Ligament-Cementum, Regeneration via Scaffolds and Stem Cells. Cells 2019, 8, 537. [Google Scholar] [CrossRef] [Green Version]
- Nuñez, J.; Vignoletti, F.; Caffesse, R.G.; Sanz, M. Cellular therapy in periodontal regeneration. Periodontology 2000 2019, 79, 107–116. [Google Scholar] [CrossRef] [PubMed]
- McCulloch, C.A. Progenitor cell populations in the periodontal ligament of mice. Anat. Rec. 1985, 211, 258–262. [Google Scholar] [CrossRef] [PubMed]
- McCulloch, C.A.; Bordin, S. Role of fibroblast subpopulations in periodontal physiology and pathology. J. Periodontal Res. 1991, 26, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Pitaru, S.; McCulloch, C.A.; Narayanan, S.A. Cellular origins and differentiation control mechanisms during periodontal development and wound healing. J. Periodontal Res. 1994, 29, 81–94. [Google Scholar] [CrossRef]
- Lekic, P.; McCulloch, C.A. Periodontal ligament cell population: The central role of fibroblasts in creating a unique tissue. Anat. Rec. 1996, 245, 327–341. [Google Scholar] [CrossRef]
- Lekic, P.; Rojas, J.; Birek, C.; Tenenbaum, H.; McCulloch, C.A. Phenotypic comparison of periodontal ligament cells in vivo and in vitro. J. Periodontal Res. 2001, 36, 71–79. [Google Scholar] [CrossRef]
- Mattick, J.S.; Makunin, I.V. Non-coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef] [Green Version]
- Gay, I.C.; Chen, S.; MacDougall, M. Isolation and characterization of multipotent human periodontal ligament stem cells. Orthod. Craniofac. Res. 2007, 10, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Seo, B.M.; Miura, M.; Gronthos, S.; Bartold, P.M.; Batouli, S.; Brahim, J.; Young, M.; Robey, P.G.; Wang, C.Y.; Shi, S. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet 2004, 364, 149–155. [Google Scholar] [CrossRef]
- Shi, S.; Bartold, P.M.; Miura, M.; Seo, B.M.; Robey, P.G.; Gronthos, S. The efficacy of mesenchymal stem cells to regenerate and repair dental structures. Orthod. Craniofac. Res. 2005, 8, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Kawanabe, N.; Murata, S.; Murakami, K.; Ishihara, Y.; Hayano, S.; Kurosaka, H.; Kamioka, H.; Takano-Yamamoto, T.; Yamashiro, T. Isolation of multipotent stem cells in human periodontal ligament using stage-specific embryonic antigen-4. Differentiation 2010, 79, 74–83. [Google Scholar] [CrossRef]
- Ng, T.K.; Yung, J.S.; Choy, K.W.; Cao, D.; Leung, C.K.; Cheung, H.S.; Pang, C.P. Transdifferentiation of periodontal ligament-derived stem cells into retinal ganglion-like cells and its microRNA signature. Sci. Rep. 2015, 5, 16429. [Google Scholar] [CrossRef]
- Liu, Y.; Zheng, Y.; Ding, G.; Fang, D.; Zhang, C.; Bartold, P.M.; Gronthos, S.; Shi, S.; Wang, S. Periodontal ligament stem cell-mediated treatment for periodontitis in miniature swine. Stem Cells 2008, 26, 1065–1073. [Google Scholar] [CrossRef] [Green Version]
- Liu, O.; Xu, J.; Ding, G.; Liu, D.; Fan, Z.; Zhang, C.; Chen, W.; Ding, Y.; Tang, Z.; Wang, S. Periodontal ligament stem cells regulate B lymphocyte function via programmed cell death protein 1. Stem Cells 2013, 31, 1371–1382. [Google Scholar] [CrossRef]
- Iwasaki, K.; Akazawa, K.; Nagata, M.; Komaki, M.; Honda, I.; Morioka, C.; Yokoyama, N.; Ayame, H.; Yamaki, K.; Tanaka, Y.; et al. The Fate of Transplanted Periodontal Ligament Stem Cells in Surgically Created Periodontal Defects in Rats. Int. J. Mol. Sci. 2019, 20, 192. [Google Scholar] [CrossRef] [Green Version]
- Feng, F.; Akiyama, K.; Liu, Y.; Yamaza, T.; Wang, T.M.; Chen, J.H.; Wang, B.B.; Huang, G.T.; Wang, S.; Shi, S. Utility of PDL progenitors for in vivo tissue regeneration: A report of 3 cases. Oral Dis. 2010, 16, 20–28. [Google Scholar] [CrossRef]
- Romano, G.; Veneziano, D.; Acunzo, M.; Croce, C.M. Small non-coding RNA and cancer. Carcinogenesis 2017, 38, 485–491. [Google Scholar] [CrossRef] [Green Version]
- Ponjavic, J.; Ponting, C.P.; Lunter, G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007, 17, 556–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, J. The functional role of long non-coding RNAs and epigenetics. Biol. Proced. Online 2014, 16, 11. [Google Scholar] [CrossRef] [Green Version]
- Niland, C.N.; Merry, C.R.; Khalil, A.M. Emerging Roles for Long Non-Coding RNAs in Cancer and Neurological Disorders. Front. Genet. 2012, 3, 25. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Lu, X.; Yuan, L. LncRNA: A link between RNA and cancer. Biochim. Biophys. Acta 2014, 1839, 1097–1109. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.C.; Pan, H.F.; Leng, R.X.; Wang, D.G.; Li, X.P.; Li, X.M.; Ye, D.Q. Emerging role of long noncoding RNAs in autoimmune diseases. Autoimmun. Rev. 2015, 14, 798–805. [Google Scholar] [CrossRef] [PubMed]
- Lorenzen, J.M.; Thum, T. Long noncoding RNAs in kidney and cardiovascular diseases. Nat. Rev. Nephrol. 2016, 12, 360–373. [Google Scholar] [CrossRef]
- Dong, R.; Du, J.; Wang, L.; Wang, J.; Ding, G.; Wang, S.; Fan, Z. Comparison of long noncoding RNA and mRNA expression profiles in mesenchymal stem cells derived from human periodontal ligament and bone marrow. Biomed. Res. Int 2014, 2014, 317853. [Google Scholar] [CrossRef]
- Gu, X.; Li, M.; Jin, Y.; Liu, D.; Wei, F. Identification and integrated analysis of differentially expressed lncRNAs and circRNAs reveal the potential ceRNA networks during PDLSC osteogenic differentiation. BMC Genet. 2017, 18, 100. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Chen, L.; Cui, S.; Li, Y.; Zhao, Q.; Cao, W.; Lai, S.; Yin, S.; Zuo, Z.; Ren, J. Expression and regulation of long noncoding RNAs during the osteogenic differentiation of periodontal ligament stem cells in the inflammatory microenvironment. Sci. Rep. 2017, 7, 13991. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Li, X.; Huang, Y.; Jia, L.; Li, W. Time series clustering of mRNA and lncRNA expression during osteogenic differentiation of periodontal ligament stem cells. PeerJ 2018, 6, e5214. [Google Scholar] [CrossRef] [Green Version]
- Jia, Q.; Jiang, W.; Ni, L. Down-regulated non-coding RNA (lncRNA-ANCR) promotes osteogenic differentiation of periodontal ligament stem cells. Arch. Oral Biol. 2015, 60, 234–241. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Yang, S.; Gu, X.; Li, M.; Wang, C.; Wei, F. Long noncoding RNA TUG1 facilitates osteogenic differentiation of periodontal ligament stem cells via interacting with Lin28A. Cell Death Dis. 2018, 9, 455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Liu, C.; Zhang, A.; Yin, S.; Wang, T.; Wang, Y.; Wang, M.; Liu, Y.; Ying, Q.; Sun, J.; et al. Down-regulation of long non-coding RNA MEG3 suppresses osteogenic differentiation of periodontal ligament stem cells (PDLSCs) through miR-27a-3p/IGF1 axis in periodontitis. Aging 2019, 11, 5334–5350. [Google Scholar] [CrossRef] [PubMed]
- Wells, K.; Littell, J.H. Study quality assessment in systematic reviews of research on intervention effects. Res. Soc. Work Pr. 2008, 19, 52–62. [Google Scholar] [CrossRef]
- Qu, Q.; Fang, F.; Wu, B.; Hu, Y.; Chen, M.; Deng, Z.; Ma, D.; Chen, T.; Hao, Y.; Ge, Y. Potential Role of Long Non-Coding RNA in Osteogenic Differentiation of Human Periodontal Ligament Stem Cells. J. Periodontol. 2016, 87, e127–e137. [Google Scholar] [CrossRef]
- Xie, L.; Chen, J.; Ren, X.; Zhang, M.; Thuaksuban, N.; Nuntanaranont, T.; Guan, Z. Alteration of circRNA and lncRNA expression profile in exosomes derived from periodontal ligament stem cells undergoing osteogenic differentiation. Arch. Oral Biol. 2021, 121, 104984. [Google Scholar] [CrossRef]
- Guo, X.; Wang, J.; Chen, J. Identification of Aberrantly Expressed lncRNAs Involved in Orthodontic Force Using a Subpathway Strategy. Comput. Math. Methods Med. 2019, 2019, 9250129. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Zhang, Y.; Li, X.; Liu, H.; Yang, Q.; Jia, L.; Zheng, Y.; Li, W. The long non-coding RNA landscape of periodontal ligament stem cells subjected to compressive force. Eur. J. Orthod. 2019, 41, 333–342. [Google Scholar] [CrossRef]
- Lin, Y.; Cheng, T.; Zhu, S.; Gu, M.; Jin, L.; Yang, Y. mRNA and long non-coding RNA expression profiling of human periodontal ligament cells under tension loading. Eur. J. Orthod. 2021, 43, 698–707. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, Y.; Niu, Q.; Qiu, N.; Liu, S.; Li, C.; Li, C.; Miao, P.; Yan, L.; Li, Q.; et al. Long Noncoding RNA Expression Profiles of Periodontal Ligament Stem Cells from the Periodontitis, Microenvironment in Response to Static, Mechanical Strain. Stem Cells Int. 2021, 2021, 6655526. [Google Scholar] [CrossRef]
- Wang, H.; Feng, C.; Li, M.; Zhang, Z.; Liu, J.; Wei, F. Analysis of lncRNAs-miRNAs-mRNAs networks in periodontal ligament stem cells under mechanical force. Oral Dis. 2021, 27, 325–337. [Google Scholar] [CrossRef]
- Jia, L.; Zhang, Y.; Ji, Y.; Li, X.; Xing, Y.; Wen, Y.; Huang, H.; Xu, X. Comparative analysis of lncRNA and mRNA expression profiles between periodontal ligament stem cells and gingival mesenchymal stem cells. Gene 2019, 699, 155–164. [Google Scholar] [CrossRef]
- Wu, L.; Deng, L.; Hong, H.; Peng, C.; Zhang, X.; Chen, Z.; Ling, J. Comparison of long non-coding RNA expression profiles in human dental follicle cells and human periodontal ligament cells. Mol. Med. Rep. 2019, 20, 939–950. [Google Scholar] [CrossRef]
- Jia, Q.; Chen, X.; Jiang, W.; Wang, W.; Guo, B.; Ni, L. The Regulatory Effects of Long Noncoding RNA-ANCR on Dental Tissue-Derived Stem Cells. Stem Cells Int. 2016, 2016, 3146805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Wu, F.; Song, Y.; Li, X.; Wu, Q.; Duan, Y.; Jin, Z. Long noncoding RNA related to periodontitis interacts with miR-182 to upregulate osteogenic differentiation in periodontal mesenchymal stem cells of periodontitis patients. Cell Death Dis. 2016, 7, e2327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, D.; Wu, L.; Liu, L.; Gong, Q.; Zheng, J.; Peng, C.; Deng, J. Comparison of HIF1A-AS1 and HIF1A-AS2 in regulating HIF-1α and the osteogenic differentiation of PDLCs under hypoxia. Int. J. Mol. Med. 2017, 40, 1529–1536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, W.; Deng, W.; Zhang, J.; Pei, G.; Rong, Q.; Zhu, S. Long noncoding RNA ANCR suppresses bone formation of periodontal ligament stem cells via sponging miRNA-758. Biochem. Biophys. Res. Commun. 2018, 503, 815–821. [Google Scholar] [CrossRef]
- Jia, B.; Qiu, X.; Chen, J.; Sun, X.; Zheng, X.; Zhao, J.; Li, Q.; Wang, Z. A feed-forward regulatory network lncPCAT1/miR-106a-5p/E2F5 regulates the osteogenic differentiation of periodontal ligament stem cells. J. Cell. Physiol. 2019, 234, 19523–19538. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zeng, X.; Miao, J.; Liu, C.; Wei, F.; Liu, D.; Zheng, Z.; Ting, K.; Wang, C.; Guo, J. Upregulation of long noncoding RNA MEG3 inhibits the osteogenic differentiation of periodontal ligament cells. J. Cell. Physiol. 2019, 234, 4617–4626. [Google Scholar] [CrossRef]
- Xu, Y.; Qin, W.; Guo, D.; Liu, J.; Zhang, M.; Jin, Z. LncRNA-TWIST1 Promoted Osteogenic Differentiation Both in PPDLSCs and in HPDLSCs by Inhibiting TWIST1 Expression. Biomed. Res. Int. 2019, 2019, 8735952. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Wan, P.; Yin, L. Long Noncoding RNA X-Inactive Specific Transcript (XIST) Promotes Osteogenic Differentiation of Periodontal Ligament Stem Cells by Sponging MicroRNA-214-3p. Med. Sci. Monit. 2020, 26, e918932. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Han, Y.; Guo, R.; Liu, H.; Li, X.; Jia, L.; Zheng, Y.; Li, W. Long non-coding RNA FER1L4 promotes osteogenic differentiation of human periodontal ligament stromal cells via miR-874-3p and vascular endothelial growth factor A. Stem Cell Res. 2020, 11, 5. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Guo, X.; Wu, S. Epigenetic silencing of KLF2 by long non-coding RNA SNHG1 inhibits periodontal ligament stem cell osteogenesis differentiation. Stem Cell Res. 2020, 11, 435. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Huang, Y.; Tan, L. Downregulation of lncRNA DANCR promotes osteogenic differentiation of periodontal ligament stem cells. BMC Dev. Biol. 2020, 20, 2. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Yin, L.; Sun, D.; Wang, F.; Wu, Q.; Xu, Q.; Xin, B. Long noncoding RNA TUG1 promotes osteogenic differentiation of human periodontal ligament stem cell through sponging microRNA-222-3p to negatively regulate Smad2/7. Arch. Oral Biol. 2020, 117, 104814. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Han, Y.; Liu, P.; Huang, Y.; Li, X.; Jia, L.; Zheng, Y.; Li, W. Long Noncoding RNA GAS5 Promotes Osteogenic Differentiation of Human Periodontal Ligament Stem Cells by Regulating GDF5 and p38/JNK Signaling Pathway. Front. Pharm. 2020, 11, 701. [Google Scholar] [CrossRef] [PubMed]
- Assis, R.I.F.; Feltran, G.D.S.; Silva, M.E.S.; Palma, I.; Rovai, E.S.; Miranda, T.B.; Ferreira, M.R.; Zambuzzi, W.F.; Birbrair, A.; Andia, D.C.; et al. Non-coding RNAs repressive role in post-transcriptional processing of RUNX2 during the acquisition of the osteogenic phenotype of periodontal ligament mesenchymal stem cells. Dev. Biol. 2021, 470, 37–48. [Google Scholar] [CrossRef]
- Bian, M.; Yu, Y.; Li, Y.; Zhou, Z.; Wu, X.; Ye, X.; Yu, J. Upregulating the Expression of LncRNA ANRIL Promotes Osteogenesis via the miR-7-5p/IGF-1R Axis in the Inflamed Periodontal Ligament Stem Cells. Front. Cell Dev. Biol. 2021, 9, 604400. [Google Scholar] [CrossRef]
- Xiang, J.; Bian, Y. PWAR6 interacts with miR-106a-5p to regulate the osteogenic differentiation of human periodontal ligament stem cells. Mol. Med. Rep. 2021, 23, 268. [Google Scholar] [CrossRef]
- Chen, P.; Huang, Y.; Wang, Y.; Li, S.; Chu, H.; Rong, M. MALAT1 overexpression promotes the proliferation of human periodontal ligament stem cells by upregulating fibroblast growth factor 2. Exp. Med. 2019, 18, 1627–1632. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Zheng, Y.; Chen, B.; Ke, T.; Shi, Z. LncRNA papillary thyroid carcinoma susceptibility candidate 3 (PTCSC3) regulates the proliferation of human periodontal ligament stem cells and toll-like receptor 4 (TLR4) expression to improve periodontitis. BMC Oral Health 2019, 19, 108. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, Y.; Zheng, P.; Cai, C.; Jiang, Y.; Zhang, H.; Li, Z.; Cai, Q. Long non-coding RNAs mortal obligate RNA transcript regulates the proliferation of human periodontal ligament stem cells and affects the recurrence of periodontitis. Arch. Oral Biol. 2019, 105, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Wangzhou, K.; Gong, L.; Liu, C.; Tan, Y.; Chen, J.; Li, C.; Lai, Z.; Hao, C. LncRNA MAFG-AS1 regulates human periodontal ligament stem cell proliferation and Toll-like receptor 4 expression. Oral Dis. 2020, 26, 1302–1307. [Google Scholar] [CrossRef]
- Wu, X.; Cao, Z.; Chen, H.; Ou, Q.; Huang, X.; Wang, Y. Downregulation of Linc-RNA activator of myogenesis lncRNA participates in FGF2-mediated proliferation of human periodontal ligament stem cells. J. Periodontol. 2020, 91, 422–427. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y. LncRNA DCST1-AS1 inhibits PDLCs’ proliferation in periodontitis and may bind with miR-21 precursor to upregulate PLAP-1. J. Periodontal Res. 2021, 56, 256–264. [Google Scholar] [CrossRef]
- Chen, H.; Lan, Z.; Li, Q.; Li, Y. Abnormal expression of long noncoding RNA FGD5-AS1 affects the development of periodontitis through regulating miR-142-3p/SOCS6/NF-κB pathway. Artif. Cells Nanomed. Biotechnol. 2019, 47, 2098–2106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.; Wang, F.; Shao, L.; Huang, P.; Xu, Y. LncRNA TUG1 mediates lipopolysaccharide-induced proliferative inhibition and apoptosis of human periodontal ligament cells by sponging miR-132. Acta Biochim. Biophys. Sin. 2019, 51, 1208–1215. [Google Scholar] [CrossRef]
- Dong, Y.; Feng, S.; Dong, F. Maternally-Expressed, Gene 3 (MEG3)/miR-143-3p Regulates, Injury to Periodontal Ligament Cells by Mediating the AKT/Inhibitory κB Kinase (IKK) Pathway. Med. Sci. Monit. 2020, 26, e922486. [Google Scholar] [CrossRef]
- Zhu, Y.; Ai, R.; Ding, Z.; He, Q.; Zhang, X.; Dong, Y.; He, Y. LncRNA-01126 inhibits the migration of human periodontal ligament cells through MEK/ERK signaling pathway. J. Periodontal Res. 2020, 55, 631–641. [Google Scholar] [CrossRef]
- Chen, Q.; Cao, M.; Ge, H. Knockdown of MALAT1 Inhibits the Progression of Chronic, Periodontitis via Targeting miR-769-5p/HIF3A Axis. BioMed Res. Int. 2021, 2021, 8899863. [Google Scholar] [CrossRef]
- Huang, N.; Li, C.; Sun, W.; Wu, J.; Xiao, F. Long non-coding RNA TUG1 participates in LPS-induced periodontitis by regulating miR-498/RORA pathway. Oral Dis. 2021, 27, 600–610. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Huang, Y.; Liu, H.; Zheng, Y.; Jia, L.; Li, W. Long Non-Coding RNA H19 Participates in Periodontal Inflammation via Activation of Autophagy. J. Inflamm. Res. 2020, 13, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, Y.; Zhao, Z.; Han, X.; Chen, Y. Knockdown of DANCR reduces osteoclastogenesis and root resorption induced by compression force via Jagged1. Cell Cycle 2019, 18, 1759–1769. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Yang, Q.; Huang, Y.; Li, X.; Zhu, Y.; Jia, L.; Zheng, Y.; Li, W. Mechanical force inhibited hPDLSCs proliferation with the downregulation of MIR31HG via DNA methylation. Oral Dis. 2021, 27, 1268–1282. [Google Scholar] [CrossRef]
- Huang, Y.; Liu, H.; Guo, R.; Han, Y.; Yang, Y.; Zhao, Y.; Zheng, Y.; Jia, L.; Li, W. Long Non-coding RNA FER1L4 Mediates the Autophagy of Periodontal Ligament Stem Cells Under Orthodontic Compressive Force via AKT/FOXO3 Pathway. Front. Cell Dev. Biol. 2021, 9, 631181. [Google Scholar] [CrossRef]
- Zhou, M.; Hu, H.; Han, Y.; Li, J.; Zhang, Y.; Tang, S.; Yuan, Y.; Zhang, X. Long non-coding RNA 01126 promotes periodontitis pathogenesis of human periodontal ligament cells via miR-518a-5p/HIF-1α/MAPK pathway. Cell Prolif. 2021, 54, e12957. [Google Scholar] [CrossRef]
- Shi, B.; Shao, B.; Yang, C.; Guo, Y.; Fu, X.; Gan, N. Upregulation of JHDM1D-AS1 protects PDLSCs from H2O2-induced apoptosis by decreasing DNAJC10 via phosphorylation of eIF2α. Biochimie 2019, 165, 48–56. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Dykes, I.M.; Emanueli, C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genom. Proteom. Bioinform. 2017, 15, 177–186. [Google Scholar] [CrossRef]
- He, R.Z.; Luo, D.X.; Mo, Y.Y. Emerging roles of lncRNAs in the post-transcriptional regulation in cancer. Genes Dis. 2019, 6, 6–15. [Google Scholar] [CrossRef]
- Kim, C.; Kang, D.; Lee, E.K.; Lee, J.S. Long Noncoding RNAs and RNA-Binding Proteins in Oxidative Stress, Cellular Senescence, and Age-Related Diseases. Oxid. Med. Cell Longev. 2017, 2017, 2062384. [Google Scholar] [CrossRef] [Green Version]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Karreth, F.A.; Pandolfi, P.P. ceRNA cross-talk in cancer: When ce-bling rivalries go awry. Cancer Discov. 2013, 3, 1113–1121. [Google Scholar] [CrossRef] [Green Version]
- Sun, F.; Liang, W.; Tang, K.; Hong, M.; Qian, J. Profiling the lncRNA-miRNA-mRNA ceRNA network to reveal potential crosstalk between inflammatory bowel disease and colorectal cancer. PeerJ 2019, 7, e7451. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Jin, L.; Tong, W.M.; Leung, Y.Y.; Gu, M.; Yang, Y. Identification and integrated analysis of differentially expressed long non-coding RNAs associated with periodontitis in humans. J. Periodontal Res. 2021, 56, 679–689. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Zheng, W.; Wang, S.; Ma, D.; Tang, L.; Duan, Y.; Jin, Y. Loss of proliferation and differentiation capacity of aged human periodontal ligament stem cells and rejuvenation by exposure to the young extrinsic environment. Tissue Eng. Part A 2009, 15, 2363–2371. [Google Scholar] [CrossRef]
- Bhandi, S.; Alkahtani, A.; Reda, R.; Mashyakhy, M.; Boreak, N.; Maganur, P.C.; Vishwanathaiah, S.; Mehta, D.; Vyas, N.; Patil, V.; et al. Parathyroid Hormone Secretion and Receptor Expression Determine the Age-Related Degree of Osteogenic Differentiation in Dental Pulp Stem Cells. J. Pers. Med. 2021, 11, 349. [Google Scholar] [CrossRef]
- Young, T.L.; Matsuda, T.; Cepko, C.L. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr. Biol. 2005, 15, 501–512. [Google Scholar] [CrossRef] [Green Version]
- Ghaforui-Fard, S.; Vafaee, R.; Taheri, M. Taurine-upregulated gene 1: A functional long noncoding RNA in tumorigenesis. J. Cell. Physiol 2019, 234, 17100–17112. [Google Scholar] [CrossRef]
- Deng, L.; Hong, H.; Zhang, X.; Chen, D.; Chen, Z.; Ling, J.; Wu, L. Down-regulated lncRNA MEG3 promotes osteogenic differentiation of human dental follicle stem cells by epigenetically regulating Wnt pathway. Biochem. Biophys. Res. Commun. 2018, 503, 2061–2067. [Google Scholar] [CrossRef]
- Zhuang, W.; Ge, X.; Yang, S.; Huang, M.; Zhuang, W.; Chen, P.; Zhang, X.; Fu, J.; Qu, J.; Li, B. Upregulation of lncRNA MEG3 Promotes, Osteogenic Differentiation of Mesenchymal, Stem Cells from Multiple, Myeloma Patients by Targeting, BMP4 Transcription. Stem Cells 2015, 33, 1985–1997. [Google Scholar] [CrossRef]
- El-Awady, A.R.; Messer, R.L.; Gamal, A.Y.; Sharawy, M.M.; Wenger, K.H.; Lapp, C.A. Periodontal ligament fibroblasts sustain destructive immune modulators of chronic periodontitis. J. Periodontol. 2010, 81, 1324–1335. [Google Scholar] [CrossRef]
- Xia, Y.; Tang, H.N.; Wu, R.X.; Yu, Y.; Gao, L.N.; Chen, F.M. Cell Responses to Conditioned, Media Produced by Patient-Matched, Stem Cells, Derived from Healthy and Inflamed, Periodontal Ligament, Tissues. J. Periodontol. 2016, 87, e53–e63. [Google Scholar] [CrossRef]
- Tang, H.N.; Xia, Y.; Yu, Y.; Wu, R.X.; Gao, L.N.; Chen, F.M. Stem cells derived from “inflamed” and healthy periodontal ligament tissues and their sheet functionalities, a patient-matched comparison. J. Clin. Periodontol. 2016, 43, 72–84. [Google Scholar] [CrossRef]
- Liu, D.; Xu, J.; Liu, O.; Fan, Z.; Liu, Y.; Wang, F.; Ding, G.; Wei, F.; Zhang, C.; Wang, S. Mesenchymal stem cells derived from inflamed periodontal ligaments exhibit impaired immunomodulation. J. Clin. Periodontol. 2012, 39, 1174–1182. [Google Scholar] [CrossRef]
- Park, J.C.; Kim, J.M.; Jung, I.H.; Kim, J.C.; Choi, S.H.; Cho, K.S.; Kim, C.S. Isolation and characterization of human periodontal ligament (PDL) stem cells (PDLSCs) from the inflamed PDL tissue: In vitro and in vivo evaluations. J. Clin. Periodontol. 2011, 38, 721–731. [Google Scholar] [CrossRef]
| Study | Samples and Stimulation | Differential Expression of lncRNAs in PDL Cells | qPCR Validation |
|---|---|---|---|
| [27] | 3 PDLSC and 3 BMSC samples | 457↑ and 513↓ lncRNAs in PDLSCs | ↑: NR045555, NR027621, NR03365; ↓: NR037182, NR037595, XR111050 (in PDLSCs) |
| [28] | osteogenic-induced and non-induced PDLSC samples | 777↑ and ↓ lncRNAs in induced PDLSCs (|fold change| ≥ 2 and p < 0.05) | ↑: TCONS_00019601, TCONS_00227764, TCONS_00254538, TCONS_00198784, TCONS_00136898; ↓: TCONS_00085268, TCONS_00125934, TCONS_00115113 |
| [29] | 3 osteogenic-induced samples, 3 osteogenic- and TNFα-stimulated samples, and 3 non-induced/stimulated samples | 214↑ and 193↓ lncRNAs in osteogenic-induced PDLSCs; 149↑ and 169↓ lncRNAs in TNFα- and osteogenic-induced PDLSCs compared to non-induced PDLSCs (log2 fold-change ≥ 1 and adjusted p ≤ 0.05). | ↑: LINC-PDE10A-1, GK-AS-1; ↓: ZNF385D-AS-1, SGOL1-AS-1 |
| [30] | osteogenic-induced and non-induced PDLSC samples | 10, 36 and 69↑ and 44, 11 and 70↓ lncRNAs after 3 days, 7 days, and 14 days of osteogenic induction, respectively (fold-change ≥ 2 and adjusted p < 0.05). | ↑: MEG8, MIR22HG |
| [35] | 3 osteogenic-induced and 3 non-induced PDLSC samples from 15 individuals | 994↑ and 1177↓ lncRNAs in induced PDLSCs (|fold change| ≥ 2 and p < 0.05) | ↑: AC078851.1, RP11-45A16.4, XLOC_002932, RP4-613B23.1, RP11305L7.6 |
| [36] | osteogenic-induced and non-induced exosomes derived from PDLSCs | 118 (70↑ and 48↓) and 43 (24↑ and 19↓) lncRNAs after 5 or 7 days of osteogenic induction, respectively (p < 0.05 and log2 fold-change > 1). | SNHG5, LOC100130992, and ATP6V1B1-AS1: no significant difference. |
| [37] | 3 orthodontic force-induced and 3 non-induced PDL samples | DLEU2↑ and DNAJC3-AS1↓ in induced PDL samples (p ≤ 0.05) | / |
| [38] | compressive force-induced and non-induced PDLSCs | 72↑ and 18↓ lncRNAs in compression-induced PDLSCs (adjusted p < 0.05 and fold-change > 1.5) | ↑: FER1L4, HIF1A-AS2, MIAT, NEAT1, ADAMTS9-AS2, LUCAT1; ↓: MIR31HG and DHFRP1 |
| [39] | 5 tension-induced and 5 non-induced PDL cell samples | 107↑ and 88↓ lncRNAs in tension-induced-PDL cells (adjusted p < 0.05) | ↑: MIR22HG, CYTOR, SNHG3 |
| [40] | 3 H-PDLSC and 3 P-PDLSC samples | ENST00000411904 the most ↑ lncRNA in strained H-PDLSCs; lncRNA-XIST and ENST0000051750 the most ↑ and ↓ lncRNAs in strained P-PDLSCs, respectively. | ↓: TCONS_00008604, ENST00000428781, uc004arq.1, XIST |
| [41] | tensile force-induced and non-induced PDLSC | 799↑ and 540↓ lncRNAs in tension-induced PDLSC (p < 0.05, fold-change > 2) | ↑: TCONS_00103186, TCONS_00114231, TCONS_00015104, TCONS_00046925, TCONS_00022234; ↓: TCONS_00195572. |
| [42] | 3 PDLSC and 3 GMSC samples | 735↑ and 1427↓ lncRNAs in PDLSCs (fold-change ≥ 1.2). | ↑: NR_038849, TCONS_l2_00010766-XLOC_l2_005781, ENST00000450854; ↓: n341766, n337408, n385309 (in PDLSCs) |
| [43] | PDL cell and DFC samples from 4 individuals | 385↑ and 460↓ lncRNAs in PDL cells | ↑: NR_033917, NR_038367, NR_026861; ↓NR_102703, NR_110162, ENST00000430859 (in PDL cells) |
| Study | lncRNAs | Increased (↑) or Decreased (↓) Expression in PDL Cells upon Stimulation | Effect on Osteogenesis | Effect on the Associated Signaling Pathway |
|---|---|---|---|---|
| [31,44,47] | ANCR | ↓ upon osteogenic induction | ↓ | inhibition of miR-758, which upregulates Notch2- Wnt/β-catenin; inhibition of the Wnt/β-catenin signaling pathway |
| [54] | DANCR | ↓ upon osteogenic induction | ↓ | / |
| [52] | FER1L4 | ↑ upon osteogenic induction | ↑ | inhibition of miR-874-3p, which regulates the VEGFA axis |
| [56] | GAS5 | ↑ upon osteogenic induction | ↑ | upregulation of GDF5, which decreases the phosphorylation of p38/JNK |
| [46] | HIF1A-AS2 | ↑ upon hypoxia | ↓ | inhibition of HIF-1α |
| [58] | LncRNA ANRIL | ↓ in P-PDLSCs | ↑ | inhibition of miR-7-5p, which regulates the IGF-1R axis |
| [45] | LncRNA-POIR | ↓ in P-PDLSCs, ↑ upon osteogenic induction | ↑ | inhibition of miR-182, which downregulates the FoxO1/canonical Wnt pathway |
| [50] | LncRNA-TWIST1 | ↓ in P-PDLSCs, ↑ upon osteogenic induction | ↑ | activation of the Wnt/β-catenin signaling pathway |
| [33] | MEG3 | ↓ in P-PDLSCs, ↑ upon osteogenic induction in PDLSCs | ↑ | inhibition of miR-27a-3p, which regulates the IGF1 axis-regulated PI3K/AKT signaling pathway |
| [49] | MEG3 | ↓ upon osteogenic induction | ↓ | competes with BMP2 mRNA for RBP hnRNPI |
| [48] | PCAT1 | ↑ upon osteogenic induction | ↑ | inhibition of miR-106a-5p, which regulates the BMP2 and E2F5 feed-forward regulatory network |
| [59] | PWAR6 | ↑ upon osteogenic induction | ↑ | inhibition of miR-106a-5, which regulates the BMP2 axis |
| [53] | SNHG1 | ↓ upon osteogenic induction | ↓ | activation of H3K27 trimethylation of the KLF2 promoter |
| [32,55] | TUG1 | ↑ upon osteogenic induction | ↑ | inhibition of miR-222-3p, which downregulates the Smad2/7 ceRNA regulatory network; binding the RNA-binding protein (RBP) Lin28A |
| [51] | XIST | ↑ upon osteogenic induction | ↑ | inhibition of the miR-214-3p axis |
| [57] | XPO5, HOTAIR, HOTTIP | ↓ in PDLSCs with high osteogenic potentials | ↓ | / |
| Study | lncRNAs | Increased (↑) or Decreased (↓) Expression in PDLSCs upon Stimulation | Effect on PDLSCs upon Stimulation | Regulatory Mechanism | Associated Signaling Pathways or Biomarkers |
|---|---|---|---|---|---|
| [73] | DANCR | ↑ in H-PDL cells under compressive force | ↑ root resorption | miR-34a-5p/jagged1 | silences DANCR, downregulates number of TRAP-positive osteoclasts and the expression of RANKL. |
| [65] | DCST1-AS1 | ↓ in P-PDL cells | ↓ proliferation | miR-21/PLAP-1 | ↓ CDK4, CDK6, CCND1; ↑ PLAP-1 |
| [75] | FER1L4 | ↑ in H-PDLSC under compressive force | ↑ autophagy | AKT/FOXO3 signaling pathway | ↑ LC3 II/I, Beclin 1, autophagosomes, autolysosomes; ↓ p-FOXO3, p-AKT |
| [66] | FGD5-AS1 | ↓ in P-PDL cells and LPS-induced H-PDL cells | ↑ proliferation; ↓ apoptosis | miR-142-3p/SOCS6/NF-κB pathway | ↓ p/t-p65, BAX/Bcl-2, cleaved/pro-caspase-3, cleaved/pro-caspase-9, TNF-α, IL-6, IL-1β, and IL-8; ↑ p/t-IκBα |
| [72] | H19 | ↑ in TNF-α and LPS-induced H-PDL cells | ↑ autophagy | PI3K/AKT signaling pathway. | ↑ Beclin-1, LC3 II/I, TNF-α, and IL-6; ↓ p-AKT |
| [77] | JHDM1D-AS1 | ↓ in H2O2-induced H-PDLSC | ↓ apoptosis | DNAJC10/p-eIF2α/Bcl-2 regulatory axis | ↓ cleaved-caspase 3, cleaved-caspase 9, BAK, ROS, DNAJC10; ↑ p-PERK, p-eIF2α, Bcl-2/BAX |
| [69] | LINC01126 | ↑ in LPS-induced H-PDL cells | ↑ inflammation; ↓migration | MEK/ERK signaling pathway | ↓ p/t-MEK and p/t-ERK. |
| [76] | LINC01126 | ↑ in hypoxia-induced H-PDL cells | ↑ apoptosis, inflammation;↓ proliferation | miR-518a-5p/HIF-1α/MAPK pathway | ↑ p38, ERK1/2, JNK, IL-1β, IL-6, IL-8, TNF-α. |
| [64] | Linc-RAM | ↓ in P-PDLSC | ↑ proliferation | inhibits the effect of overexpression of FGF2 on proliferation | / |
| [63] | MAFG-AS1 | ↓ in P-PDLSC | ↑ inflammation; ↓ proliferation | miR-146a/TLR4 axis | ↑ TLR4 |
| [60] | MALAT1 | ↑ in P-PDLSC | ↑ proliferation | FGF2 axis | ↑ FGF2 |
| [70] | MALAT1 | ↑ in LPS-induced H-PDL cells | ↑ apoptosis, inflammation ↓ proliferation | miR-769-5p/HIF3A axis | ↑ IL-6, IL-1β, TNF-α, BAX, and caspase-3; ↑ Bcl-2. |
| [68] | MEG3 | ↓ in P-PDL cells and LPS-induced H-PDL cells | ↑ proliferation;↓ apoptosis, inflammation | miR-143-3p AKT/IKK pathway | ↓ p-AKT/AKT, p-IKK/IKK, p-p65, IL-6, IL-18, IL-1β, TNF-α. |
| [74] | MIR31HG | ↓ in H-PDLSC under compressive force | ↑ proliferation | DNMT1 and DNMT3B inhibited expression of MIR31HG | silences MIR31HG, inhibits cell viability. |
| [62] | MORT | ↓ in P-PDLSC | ↓ proliferation | inhibits cell viability | |
| [61] | PTCSC3 | ↓in P-PDL cells | ↓ proliferation | TLR4 | ↓ TLR4 |
| [67,71] | TUG1 | ↓ in P-PDL cells and LPS-induced H-PDL cells | ↑ proliferation;↓ apoptosis, inflammation | miR-498/RORA axis and Wnt/β-catenin signaling pathway; miR-132 axis | ↓ β-catenin, p/t-GSK-3β, p21, TNF-α, IL-1β, IL-6, and IL-8; ↑ CDK2 and cyclin D1. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, Y.; Tang, Z.; Jin, L.; Yang, Y. The Expression and Regulatory Roles of Long Non-Coding RNAs in Periodontal Ligament Cells: A Systematic Review. Biomolecules 2022, 12, 304. https://doi.org/10.3390/biom12020304
Lin Y, Tang Z, Jin L, Yang Y. The Expression and Regulatory Roles of Long Non-Coding RNAs in Periodontal Ligament Cells: A Systematic Review. Biomolecules. 2022; 12(2):304. https://doi.org/10.3390/biom12020304
Chicago/Turabian StyleLin, Yifan, Zhongyuan Tang, Lijian Jin, and Yanqi Yang. 2022. "The Expression and Regulatory Roles of Long Non-Coding RNAs in Periodontal Ligament Cells: A Systematic Review" Biomolecules 12, no. 2: 304. https://doi.org/10.3390/biom12020304
APA StyleLin, Y., Tang, Z., Jin, L., & Yang, Y. (2022). The Expression and Regulatory Roles of Long Non-Coding RNAs in Periodontal Ligament Cells: A Systematic Review. Biomolecules, 12(2), 304. https://doi.org/10.3390/biom12020304