Abstract
Maintenance of protein homeostasis is crucial for virtually every aspect of eukaryotic biology. The ubiquitin-proteasome system (UPS) represents a highly regulated quality control machinery that protects cells from a variety of stress conditions as well as toxic proteins. A large body of evidence has shown that UPS dysfunction contributes to the pathogenesis of cardiovascular diseases. This review highlights the latest findings regarding the physiological and pathological roles of cullin-RING ubiquitin ligases (CRLs), an essential player in the UPS, in the cardiovascular system. To inspire potential therapeutic invention, factors regulating CRL activities are also discussed.
1. Introduction
For many years, systematic cohort and mechanistic studies have been carried out to unveil risk factors and therapeutic targets for cardiovascular diseases. Consequently, proper management of identified risk factors and application of cognate therapeutic strategies have led to a remarkable decrease in the incidence and fatality of cardiovascular diseases. Despite these efforts, cardiovascular diseases remain a leading cause of mortality around the globe, with 18.6 million recorded deaths in 2019 [1]. Furthermore, as a result of the world population aging, the number of people dying from cardiovascular diseases has been increasing over the last decade, especially in low- and middle-income countries [2]. It is thus still urgent to explore new avenues for the prevention and treatment of cardiovascular diseases.
Under various stress conditions, cells need to properly maintain a functional proteome for fundamental biological processes [3]. The ubiquitin-proteasome system (UPS) represents a pivotal player in regulating protein homeostasis. It has been well-established that UPS-mediated proteolysis is capable of targeting individual proteins for proteasomal degradation [4]. The signal that triggers such degradation is ubiquitination, a post-translational modification where a chain of the small protein ubiquitin (Ub) is covalently linked to a lysine residue of the target protein. Ubiquitination is catalyzed by sequential enzymatic reactions involving the Ub activating enzyme (E1), the Ub conjugating enzyme (E2) and the Ub ligase (E3). The substrate specificity of the ubiquitination system is conferred by E3s, which mediate the transfer of Ub to the target protein. Hundreds of E3s are encoded by the human genome, of which about 40% are predicted to cooperate with the UPS [5]. Based on typical structural characterizations, E3 ligases are classified into three main classes: the really interesting new gene (RING) type, the homologous to E6-AP carboxyl terminus (HECT) type and the RING-between-RING (RBR) type. With more than two hundred members, the multi-unit cullin-RING ligases (CRLs) represent the largest and most studied subfamily of E3 ligases [6,7]. The modular CRL complexes are characterized by a common cullin (CUL) scaffold, a small RING protein (RBX1 or RBX2), and an interchangeable substrate receptor module. Structurally, the C-terminus of the elongated CUL scaffold binds the RBX protein that recruits the E2 conjugated Ub (E2~Ub), whereas the N-terminus of CUL binds the various substrate receptor modules [8]. Seven different types of CUL proteins have been identified in human cells (CUL1, CUL2, CUL3, CUL4A, CUL4B, CUL5, CUL7 and CUL9), each of which engages a family of substrate receptors to assemble active CRLs (Figure 1). In addition, CUL6 has been identified in C. elegans [6], and Cul8 has been found in S. cerevisiae [9,10], but neither have been reported in humans. Through bringing the substrate and E2~Ub in close proximity, CRLs facilitate substrate ubiquitination [11].
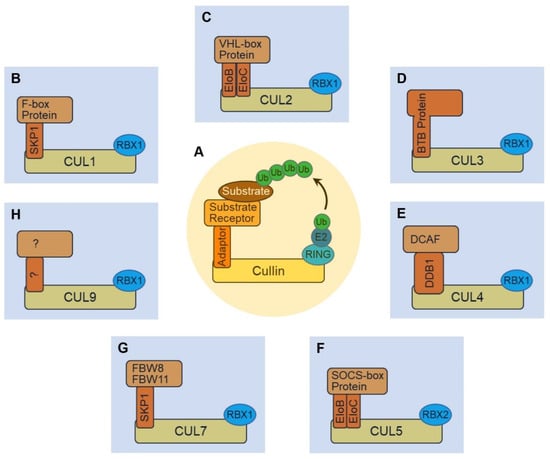
Figure 1.
Schematic illustrating the structure of cullin-RING ubiquitin ligases (CRLs). (A) CRL complexes are modular. They all contain a cullin protein that serves as the backbone for the complex. A RING finger protein binds the C-terminus of cullin and serves as an adaptor for E2~Ub. The N-terminus of cullin recruits various substrate receptors—usually through an adaptor protein—to recognize and specify target substrates for ubiquitination. (B) The CUL1•RBX1 enzymatic core of CRL1 uses SKP1 as an adaptor to recruit F-box proteins as substrate receptors. (C) CRL2 use EloB (elongin B) and EloC (elongin C) complex as the adaptor to recruit VHL-box proteins as substrate receptors. (D) CRL3 directly recruits BTB proteins without the need of an adaptor protein. (E) CRL4 (including CRL4A and CRL4B) uses DDB1 as the adaptor to recruit DCAF proteins as substrate receptors. (F) CRL5 is composed of CUL5 and RBX2, and it uses EloB and EloC complex as the adaptor to recruit SOCS-box proteins as substrate receptors. (G) CRL7 has been known to use the SKP1 adaptor protein to recruit FBW8 or FBW11 F-box protein substrate receptors. (H) CRL9 comprises CUL9 and RBX1, while the adaptor and substrate receptor complex remain unknown.
Growing evidence implicates that cardiovascular UPS plays a key role in regulating diverse cellular events such as the function of cardiac membrane channel and receptor, apoptosis of cardiomyocyte and vascular smooth muscle cells, as well as the quality control of sarcomere [12,13,14]. Given the importance of CRLs in the UPS, herein, we review the recent understanding on the role of CRLs in the context of cardiovascular diseases.
2. The CUL1-RING Ligase (CRL1)
The prototype of the CRL family is the CUL1-based CRL1 (also known as SCF) complex, which consists of the CUL1 core, the adaptor protein SKP1 (S-phase kinase-associated protein 1), and an F-box protein substrate receptor (Figure 1B) [8,15]. The RING protein RBX1 binds the C-terminal domain of CUL1 and serves as the docking site for E2~Ub. The N-terminal domain of CUL1 binds SKP1, and SKP1 recruits F-box proteins through their conserved F-box motif. Up to 69 different F-box proteins can be made in human cells [16], each of which recognizes specific protein targets for ubiquitination. Through engaging different F-box protein substrate receptors, CRL1s can change their substrate specificity, and therefore, various proteins can be targeted for ubiquitination and degradation [17,18]. While a multitude of studies have suggested F-box proteins as promising cancer drug targets [19], there is emerging evidence suggesting that therapeutic targeting of F-box proteins could be beneficial in other pathologies as well.
2.1. Implications for CRL1FBXO32 in Cardiac Function
An F-box protein well-known for its involvement in cardiovascular disease is FBXO32 (or Atrogin-1), a protein specifically expressed in skeletal and cardiac muscles [20]. FBXO32 was firstly identified as a key regulator for skeletal muscle atrophy [21,22,23,24], and it targets the initiation factor eIF3-f [25] and the myogenic regulatory factor MyoD [26] for ubiquitination and degradation. Besides a critical role in skeletal muscle, FBXO32 was also found to enhance ischemia/reperfusion-induced apoptosis in cardiomyocytes through promoting the proteosome-dependent degradation of MAPK phosphatase-1 [27]. In addition, studies using mouse models showed that Fbxo32 regulates the ubiquitination of Foxo1/3 [28], and it is involved in cardiac hypertrophy and cardiac aging [28,29,30,31]. More recently, human mutations in FBXO32 were identified to cause dilated cardiomyopathy [32,33,34], through dysregulating autophagy [32] and upregulating CHOP (C/EBP homologous protein)-mediated apoptosis [34]. Taken together, FBXO32 is a key player in cardiac physiology and the pathogenesis of heart disease.
2.2. CRL1FBXL2 and Ca2+ Homeostasis
A recent study highlighted the role of another F-box protein, FBXL2, in obesity-related cardiac dysfunction [35]. FBXL2 is widely expressed in various human tissues [36,37], and it was first found to bind and ubiquitinate the unphosphorylated p85β subunit of PI(3)Ks (phosphatidylinositol-3-OH kinases) [38]. Defects in the FBXL2-mediated degradation of p85β could promote autophagy [38]. Further searches for the substrate of CRL1FBXL2 identified IP3R3, a receptor for IP3 (inositol 1,4,5-trisphosphate), as a direct target of FBXL2 [39]. The FBXL2-dependent degradation of IP3R3 can limit Ca2+ influx into mitochondria and reduce apoptosis. In addition, the tumor suppressor PTEN (phosphatase and tensin homologue) was found to compete with FBXL2 for IP3R3 binding, and as a result, PTEN can limit tumor growth through stabilizing IP3R3 and enhancing Ca2+-dependent apoptosis [39]. The abovementioned study by Ren et al. [35] found that FBXL2 also interacts with FUNDC1 (FUN14 domain containing 1), an integral mitochondrial outer-membrane protein, in mouse heart. This interaction changes the stability of FBXL2 and, consequently, the stability of IP3R3. Suppression of FUNDC1 activity, which can be triggered by high-fat diet, reduces FBXL2-dependent degradation of IP3R3, leading to mitochondrial Ca2+ overload and cardiomyocyte dysfunction [35]. While the mechanism by which FUNDC1 regulates the stability of FBXL2 remains to be fully elucidated, these studies suggest that through altering the activity of CRL1FBXL2, it is possible to enhance or suppress Ca2+-dependent apoptosis. Therefore, FBXL2 or regulators of CRL1FBXL2 can be targets for therapeutic development.
2.3. A Role for CRL1FBXW7 in Oxidative Stress
FBXW7 is an F-box protein that has been heavily studied as a tumor suppressor. It is frequently mutated in human cancers, and targets multiple oncoproteins for proteasome-dependent degradation (for review, see [40]). Among these oncoproteins, MCL-1 (myeloid cell leukaemia-1) is an apoptosis regulator that regulates the apoptosis of not only cancer cells but also cardiac cells [40,41,42]. Given the important role of MCL-1 in mitochondria and cardiomyocytes [41], it is unsurprising that FBXW7 is involved in oxidative stress-induced cardiac cell injury [43]. Further, FBXW7 has been reported as a regulator of cardiac hypertrophy, likely through controlling the protein stability of EZH2 (enhancer of zeste homology 2) and the transcription of SIX1 (sine oculis homeobox homolog 1) [44]. Besides regulating the degradation of downstream targets, the expression of FBXW7, in turn, can be regulated by various microRNAs (for review, see [40]), a machinery also implicated in the pathogenesis of cardiovascular disease [45,46]. In fact, FBXW7 is not the only F-box protein known to be regulated by microRNAs. The microRNA MiR-184 was reported to target FBXO28, and inhibiting MiR-184 could reduce H2O2-induced cardiomyocyte injury [47]. This finding suggests that FBXO28 may play a role in handling oxidative stress and apoptosis in cardiomyocytes.
2.4. CRL1SKP2 in Proliferation and Senescence
Another recent study suggests that the F-box protein SKP2 (S-phase kinase-associated protein 2) plays a role in the aging of endothelial progenitor cells [48]. CRL1SKP2 has been extensively studied, and through targeting a variety of cell cycle proteins for ubiquitination, SKP2 plays key roles in cell proliferation and tumorigenesis (for review, see [49]). Consistent with its general cellular function, SKP2 regulates the proliferation of vascular smooth muscle cells [50]. Through analyzing the senescence of human endothelial progenitor cells, Wang et al. [48] found that decreased SKP2 levels induce early senescence while increasing the level of SKP2 can partially reverse senescence in aged endothelial progenitor cells. This finding suggests that aging-related vascular disease may be managed by altering the level or activity of CRL1SKP2.
3. The CUL2-RING Ligase (CRL2)
3.1. CRL2VHL-HIF1α Regulatory Axis and Cardiovascular Diseases
CUL2 is a type of cullin that only exists in in multi-cellular organisms [51]. CRL2 employs CUL2•RBX1 as the catalytic scaffold and the Elongin B and Elongin C (EloB/C) protein complex as the adaptor for substrate receptors (Figure 1C) [52,53]. Existing connections between CRL2 and cardiovascular disease are mostly attributed to HIF1α (hypoxia-inducible factor 1α), a well-studied oxygen-responsive substrate of CRL2 [54]. CRL2-dependent degradation of HIF1α requires post-translational hydroxylation of proline residues located in its ODD (oxygen-dependent degradation) domain [55]. Once hydroxylated, HIF1α binds to VHL (von Hippel-Lindau) tumor suppressor protein, a CRL2 substrate receptor, making it a target for CRL2-mediated proteasomal degradation. The activities of prolyl hydroxylase domain (PHD) enzymes that hydroxylate HIF1α are oxygen-dependent [56], and therefore, in hypoxia, HIF1α does not bind to VHL for degradation and is stabilized. The stabilized HIF1α then translocates into the nucleus and binds HIF1β [57]. The resulting HIF1α/β complex functions as a transcription factor that recognizes and binds specific hypoxia response elements (HREs) on the target genes [58]. It has been well-established that the HIF transcriptional complex mediates a broad spectrum of cellular and physiological pathways demanding adaption to hypoxia [59].
HIF1α is closely related to cardiovascular diseases in multiple dimensions. For example, in response to ischemia-induced hypoxia, the HIF transcription factor complex activates protective metabolic adaptions, including repression in oxygen-consuming processes and elevation in oxygen-sparing pathways [60]. Furthermore, HIF transcription factors are able to promote angiogenesis by regulating expression of angiogenic factors [61]. A recent review has summarized the role of HIF-1 in cardiovascular diseases in detail [62].
3.2. CRL2VHL and PROTACs
In addition to regulating the ubiquitination of natural substrates, VHL also plays a key role in drug discovery via the PROteolysis TArgeting Chimera (PROTAC) strategy [63,64,65,66]. PROTAC molecules contain a chemical linker that combines two types of ligands: one binds to VHL, and the other binds a protein that is not a natural substrate of VHL. Therefore, PROTAC can recruit a protein of interest to CRL2VHL and induce its ubiquitination and degradation. Over the past few years, a large number of VHL-based PROTACs have been reported [67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85] and new PROTACs are actively being developed. While PROTACs, as the majority of other therapeutic strategies directed towards the UPS, are mostly studied for cancer treatment [63], their effects—either positive or negative—on the cardiovascular system are worth further investigation.
4. The CUL3-RING Ligase (CRL3)
Unlike CUL1 or CUL2 that use adaptor protein(s) to engage substrate receptors, the N-terminus of CUL3 binds substrate receptors directly via their BTB (Bric-a-Brac/Tramtrack/Broad) domain (Figure 1D) [8]. The BTB domain also dimerizes, and as a result, CRL3 complexes exist as homodimers [86,87]. Approximately 180 BTB proteins are encoded in the human genome, and about 50 of them have been confirmed as substrate receptors that can potentially recruit protein substrates to CRL3 for ubiquitination [88].
4.1. CUL3 Mutations and Cardiovascular Diseases
CUL3 has been shown to play a significant role in the cardiovascular system, particularly in regulating blood pressure [89,90,91,92,93,94,95,96,97,98]. Tissue-specific deletion of the cul3 gene in mouse skeletal muscle cells or cardiomyocytes led to neonatal lethality, because of skeletal muscle dysfunction or severe cardiomyopathy [91]. Deleting cul3 specifically in mouse smooth muscle cells resulted in impaired vascular function and hypertension [92]. Knocking down CUL3, but not other cullin genes (CUL1, CUL2, CUL4a, CUL5), in human umbilical vein endothelial cells caused a significantly reduced level of vascular endothelial (VE)-cadherin protein, suggesting that CUL3 plays a critical role in VE-cadherin-mediated endothelial barrier function [93]. What highlights the role of CUL3 in cardiovascular disease is the identification of dominant-negative CUL3 mutations in patients with severe forms of familial hyperkalemic hypertension (FHHt) [94,95]. These mutations led to impaired splicing and skipping of exon 9 that encodes amino acid residues 403–459, resulting in a truncated form of CUL3 (CUL3∆403–459 or CUL3∆9). The CUL3∆9 mutant can still bind RBX1 and BTB substrate receptor to form a CRL3 complex. However, the RBX1•CUL3∆9•BTB protein complex can no longer ubiquitinate its substrate. Instead, CUL3∆9 led to increased autoubiquitination and increased degradation of the substrate receptor protein [96]. In mouse models, CUL3∆9 induced severe hypertension when expressed ubiquitously [97] or in vascular smooth muscle cells [97,98].
4.2. CRL3KLHL3-WNK Regulatory Axis
Studies of FHHt patients revealed mutations in not only CUL3, but also KLHL3 (Kelch-like 3), WNK1 (with-no-lysine kinase 1) and WNK4 [95,99,100,101]. WNK1 and WNK4 are kinases that regulate blood pressure through a signaling cascade that eventually phosphorylates and activates NCC (Na+-Cl− cotransporter) and NKCC2 (Na+-K+-2Cl− co-transporter) [102]. KLHL3 is a BTB-domain-containing protein that binds CUL3 and recruits the WNK proteins for ubiquitination and degradation [103]. Thus, through controlling the turnover rate of WNK proteins, CRL3KLHL3 regulates blood pressure [103]. The interaction between KLHL3 and WNK4 was found to depend on the phosphorylation status of KLHL3: phosphorylation at Serine 433 prevented KLHL3 from binding to WNK4 [104]. Mutations in KLHL3 or WNK4 discovered in FHHt patients disrupt the interaction between KLHL3 and WNK4, leading to insufficient degradation of WNK4 and, ultimately, hypertension [105,106,107,108]. In addition, KLHL2, a homologue of KLHL3, was also reported to form a CRL3 that targets WNK4 for ubiquitination and degradation, so KLHL2 may also play a role in the pathogenesis of FHHt [109].
4.3. CRL3KEAP1-NRF2 Regulatory Axis
KEAP1 is one of the best-studied substrate receptors for CUL3 [8], and the CRL3KEAP1 complex targets NRF2 (nuclear factor-E2-related factor 2) for ubiquitination and degradation. The KEAP1-NRF2 system provides an important mechanism for defense against oxidative and electrophilic stress (for review, see [110]). NRF2 is a transcription factor that activates the transcription of antioxidant genes. Under normal conditions, KEAP1 binds newly synthesized NRF2 in the cytoplasm and keeps it constitutively ubiquitinated and degraded. When exposed to oxidative stress, the cysteine residues of KEAP1 are modified by reactive oxygen species, releasing NRF2 from CRL3KEAP1. NRF2 is then stabilized and translocates into nuclei to activate antioxidant defense responses [110]. This mechanism for NRF2 activation is important for cellular redox homeostasis in general and has been observed as a mechanism for cardiovascular protection [111,112,113,114,115,116]. Particularly, NRF2 plays a protective role in cardiomyocytes after myocardial ischemia and reperfusion injury [112,117,118], and in endothelial injuries induced by oxidative stress [119]. Dysregulation of NRF2 may result in chronic heart failure [120] or hypertension [121].
4.4. Other BTB Proteins Involved in Cardiovascular Diseases
A few additional BTB proteins have also been reported to contribute to cardiovascular health. First, RhoBTB1 (Rho-related BTB domain-containing 1) is the substrate receptor mediating the CUL3-dependent ubiquitination of PDE5 (phosphodiesterase 5), which is a key regulator for smooth muscle relaxation [122,123]. Consequently, the RhoBTB–PDE5 system regulates vascular smooth muscle function and can protect against hypertension [122]. Second, LZTR1 (leucine zipper-like transcription regulator 1) can bind CHMP1B (charged multivesicular protein 1B) and control the ubiquitination of CHMP1B in a cullin-dependent manner [124]. Mutations of LZTR1 were identified in Noonan syndrome patients with bleeding disorders, and these mutations led to defects in LZTR1-mediated ubiquitination of CHMP1B and, ultimately, impaired vesicle trafficking, resulting in cardiovascular dysfunction [124].
Rho-GTPases are key regulators of cytoskeleton dynamics and cell adhesion [125], and this mechanism also controls the function and activity of endothelial cells [126]. RhoA, a member of the Rho-GTPase family, was found to be recruited to CUL3 for ubiquitination through BTB proteins named BACURDs [127], and the CRL3BACURD1-mediated RhoA ubiquitination was impaired by the dominant-negative mutant CUL3∆9 [128]. Another BTB protein, KCTD10 (potassium channel tetramerization domain containing 10), was reported to target RhoB for ubiquitination [126]. The ubiquitinated RhoB was subsequently degraded by lysosomes in endothelial cells, which maintained the integrity of the endothelial barrier [126]. Moreover, mutations in KLHL24 (Kelch like family member 24) were found to cause hypertrophic cardiomyopathy, a common inherited cardiovascular disorder, and silencing the klhl24a gene in zebrafish caused defects in cardiac function [129]. These findings revealed a crucial role of KLHL24 in heart development and function, and it will be important to identify the specific protein targets that KLHL24 recognizes for ubiquitination in cardiomyocytes. In summary, through targeting diverse protein substrates for ubiquitination, the CRL3 ubiquitin ligases play key regulatory roles in the cardiovascular system from different perspectives.
5. The CUL4-RING Ligase (CRL4)
Two members of the CUL4 subfamily exist in humans, CUL4A and CUL4B. They share highly similar amino acid sequences, except that CUL4B contains an elongated N-terminal domain of ~150 amino acid residues [130,131]. Both forms of CUL4 use a large protein DDB1 (DNA damage protein 1) as the adaptor to dock DCAF (DDB1–CUL4-associated factor) substrate receptor proteins (Figure 1E). Based on sequence analysis, about 100 DCAF proteins are predicted to associate with DDB1 in human cells [132,133].
5.1. CRL4DCAF8 and CRL4DDB1-GRK5
While roles of CRL4s in the cardiovascular system remain largely unexplored, Cul4a overexpression in H9c2, a cell line derived from rat heart tissue, was reported to reduce oxidative stress-induced apoptosis, whereas CUL4A knockdown had the reverse effect [134]. When cul4b was specifically knocked out in adipocytes, the mutant mice on a high-fat diet accumulated more body fat and were, thus, more likely to develop obesity [135]. In addition, in the mouse myoblast cell line C2C12, DCAF8 was found to bind TRIM63 (tripartite motif containing 63), a muscle-specific RING-finger ubiquitin ligase that facilitates the ubiquitination of MyHC (myosin heavy chain proteins). C2C12 cells lacking DCAF8 were defective in MyHC degradation and were resistant to atrophy [136]. The mechanism by which TRIM63 and CRL4DCAF8 controls the degradation of MyHC is still unclear, and if/how the TRIM63•CRL4DCAF8 complex contributes to cardiac function warrants further investigation. Lastly, dysregulation of GRKs (G-protein-coupled receptor kinases) can be associated with pathological conditions including cardiovascular disease [137], and GRK5 was found to form a complex with CUL4 through DDB1 and undergo CUL4•DDB1-dependent ubiquitination [138]. Whether the DDB1–GRK5 interaction requires a DCAF remains unclear and needs further investigation.
5.2. Non-Canonical CRL4s Involved in Cardiovascular Diseases
An interesting link between CRL4 and the cardiovascular system comes from discoveries of non-canonical CRL4 complexes. For example, Grk2, another member of the GRK family, is recognized by Gβ (G protein β subunit) protein and recruited to Cul4a through Ddb1 in mice [139]. This CRL4Gβ-dependent degradation of Grk2 plays a protective role in the mouse heart [139]. Another example is the CUL4B•DDB1•FBXO44-dependent ubiquitination of RGS2 (regulator of G protein signaling 2) [140], a protein that regulates vasoconstriction and the lack of which leads to hypertension in mice [141]. Several RGS2 mutations, causing reduced expression due to an increased rate of proteasomal degradation, have also been associated with hypertension in humans [142,143,144]. RGS2 was found to be recruited to CUL4B via an F-box protein, FBXO44, using DDB1 as the adaptor protein. Like the other 68 members of the F-box protein family, FBXO44 can also associate with CUL1 ligases. However, FBXO44 is only capable of targeting RGS2 for ubiquitination in the context of CUL4B but not CUL1 [140]. The RGS2–FBXO44 interaction can be regulated by phosphorylation, and the phosphorylation of Ser3 on RGS2 could protect RGS2 from degradation through reducing its binding with FBXO44 [145]. Since low RGS2 protein level is associated with disease such as hypertension and heart failure, drugs that interfere with the RGS2–FBXO44 interaction can be beneficial for preventing cardiovascular diseases [145].
5.3. CRL4 and PROTACs
A couple of CRL4s have been identified as drug targets for small-molecule-induced protein degradation. As a type of PROTACs (see Section 3), when these “molecular glue” drugs bind to the DCAF protein, the DCAF can then bind a disease-causing protein, leading to CRL4-dependent degradation of the target protein. One such example is the Immunomodulatory Drugs (IMiDs), which bind the Cereblon (CRBN) DCAF protein and trigger the degradation of a variety of cellular regulators, including Ikaros, Aiolos, ZFP91 zinc finger protein, etc. [146,147,148,149,150,151,152,153]. The other example is indisulam, which enables CRL4DCAF15 to recruit RBM39 (RNA binding motif protein 39) for ubiquitination and degradation [154,155]. While IMiDs represent an exciting new strategy for drug discovery and have been successfully used to treat patients with multiple myeloma, they also appear to increase the risk of cardiotoxicity in multiple myeloma patients [156]. Thus, the side effects of IMiDs and other molecular glue drugs on the cardiovascular system need to be carefully evaluated to guide and improve their clinical use.
6. The CUL5-RING Ligase (CRL5)
CRL5s recruit E2~Ub through RBX2 that associates with the C-terminus of CUL5. Although recombinant CUL5 could bind recombinant RBX1 in vitro, the formation of the CUL5•RBX1 complex in human cells has not been reported [157]. Like CUL2, the N-terminus of CUL5 employs the EloB/C adaptor complex to associate with different substrate receptors (Figure 1F). However, unlike CUL2 that binds a VHL-box in substrate receptor proteins, CUL5 specifically recognizes the SOCS (suppressor of cytokine signaling)-box at the C-terminus of its substrate receptor proteins [158]. In the human genome, 37 SOCS-box substrate receptors have been identified [157].
CRL5ASB2 in Cardiac Development
While the importance of CRL5-dependent protein ubiquitination in cancers has been well-documented [157], what role each CRL5 plays in the cardiovascular system has just started to come to light. ASB2 (ankyrin repeat and SOCS box containing 2) is a CRL5 substrate receptor worth highlighting here. The expression of ASB2 was primarily detected in human cardiac and skeletal muscles [37,159], and Asb2 knockout mice were embryonic lethal due to cardiovascular defects [160]. ASB2 was found to target Filamin A for ubiquitination and degradation [160,161], thereby controlling actin remodeling in immature cardiomyocytes, which plays an essential role in heart development [160]. Furthermore, SMAD9 (SMAD family member 9) was also identified as a substrate for CRL5ASB2 [162]. SMAD9 is one of the transcriptional modulators for BMP (bone morphogenetic protein) signaling, and excessive levels of SMAD9 could lead to abnormal cardiac differentiation [162]. Thus, controlling the stability of SMAD9 and its downstream BMP signaling is another pathway by which CRL5ASB2 contributes to normal heart development.
7. The CUL7-RING Ligase (CRL7) and Cardiac Signal Transduction
CUL7 belongs to one of the non-canonical cullins, and its molecular weight is at least twice as large as that of the canonical cullins (CUL1-5). Consistent with canonical cullins, CUL7 contains the conserved cullin domain that binds RBX1 to recruit E2~Ub [163]. Similar to CUL1, CUL7 engages substrate receptors via the adaptor protein SKP1, but only FBXW8 and FBXW11 have been reported to form active E3 ligase complexes with CUL7 (Figure 1G) [164,165,166].
The CUL7 protein is present in human muscles (cardiac, smooth, and skeletal) [36,37], and CRL7FBXW8 has been shown to regulate insulin signaling in human cells and mice through targeting IRS1 (insulin receptor substrate 1) for ubiquitination and degradation [167,168,169]. In addition, knocking down Cul7 but not Cul2, Cul3 or Cul5 in cultured neonatal rat ventricular cardiomyocytes (NRVCs) led to reduced ubiquitination and degradation of Mst1, a key component of the Hippo–YAP signaling pathway [170]. Thus, through controlling the stability of Mst1, CRL7 activities are important for heart development. Furthermore, when the Cul7 gene was deleted specifically in mouse cardiomyocytes, phosphoinositide 3-kinase (PI3K)/AKT signaling was activated in the heart, cardiomyocyte apoptosis was reduced, and cardiac fibrosis following transverse aortic constriction (an experimental model for pressure overload-induced cardiac hypertrophy and heart failure) was attenuated [171]. These findings suggest that CRL7 can be a target for developing antimyocardial fibrosis therapeutics.
8. The CUL9-RING Ligase (CRL9) and Cardiovascular Disease
CUL9 is the other known non-canonical cullin protein whose functions and properties have not been fully characterized yet. Its sequence is highly similar to CUL7, and with over 2500 amino acids, it is the largest member in the cullin family [172]. The C-terminus of CUL9 binds RBX1, and it also contains a RING between the RING domain. To date, no adapter protein for CUL9 has been identified (Figure 1H) [173].
CUL9 has been shown to bind and activate p53, and this interaction is important for cell proliferation and genome integrity [174]. A genome-wide association study (GWAS) revealed that CUL9 is a risk factor for cardiovascular diseases [175], but the mechanistic link between CUL9 and cardiovascular diseases remains to be further explored.
9. Regulators of CRLs
9.1. Neural Precursor Cell Expressed, Developmentally Downregulated 8 (NEDD8)
9.1.1. Neddylation Promotes the Activity of CRLs
The activity of CRLs is tightly regulated by neddylation, a post-translational modification where the ubiquitin-like protein NEDD8 is conjugated to a conserved lysine located in the C-terminal domain of cullins [176,177]. Similar to ubiquitination, the process of cullin neddylation requires consecutive enzymatic reactions involving NEDD8 E1 activating enzyme (NAE), NEDD8 E2 conjugating enzyme, and NEDD8 E3 ligase [18]. Neddylation of cullins efficiently promotes the activity of CRLs, leading to increased ubiquitination of substrates [178,179,180,181]. The mechanism by which neddylation activates CRLs has been revealed from both biochemical and structural perspectives. Biochemically, neddylation promotes ubiquitin chain initiation and elongation through enhancing E2 recruitment and E2 activity [182]. Structurally, neddylation induces substantial conformational changes on the C-terminal domain of the CUL•RBX1 core [183] and triggers multiple protein–protein interactions within the CRL complex, including interactions between NEDD8 and E2, and interactions between E2 and the substrate receptor [184]. As a result, neddylation converts the CRL from an open conformation, which benefits the engagement of E2~Ub and diverse substrates, to a closed and compact conformation, which promotes the transfer of the donor ubiquitin to the acceptor lysine and triggers efficient substrate ubiquitination [185]. Furthermore, cullin neddylation enables the ARIH family of RBR E3 ligases to bind CRLs—ARIH1 binds neddylated CUL•RBX1, whereas ARIH2 binds neddylated CUL5•RBX2—and the ARIH E3 ligase accelerates the transfer of Ub from an E2 to the CRL-bound protein substrate [186,187,188].
9.1.2. Impaired Neddylation Results in Cardiomyopathy
Given the large number of CRL substrates that function as key regulators in the cardiovascular system, it is not surprising that neddylation was reported to be crucial for cardiac development. Genetic studies showed that mice lacking NAE exhibit proliferation arrest in cardiomyocytes and ventricular non-compaction, leading to heart failure and, eventually, neonatal lethality [170]. Transient treatment of MLN4924 (or pevonedistat), a small-molecule inhibitor of neddylation [189], also resulted in obvious cardiac abnormalities in neonatal rats such as reduced cardiomyocyte proliferation and cardiac hypertrophy [190], indicating a role of neddylation in perinatal cardiac growth. Mechanistically, neddylation was shown to promote cardiac chamber maturation partially via the CUL7–MST1–YAP axis (see Section 7) [170]. It is noteworthy that dozens of neddylation substrates other than cullin proteins have been reported [191], and how these substrates contribute to the cardiovascular system remains largely unknown.
9.2. COP9 Signalosome (CSN)
9.2.1. CSN Is Required for Maintaining the CRL Activities in Cells
The reverse process of cullin neddylation, termed deneddylation, is mediated by the constitutive photomorphogenesis 9 (COP9) signalosome (CSN) [192], an evolutionarily conserved multi-protein complex. The mammalian CSN complex contains eight subunits in which only CSN5 renders the isopeptidase activity required for deneddylation. Early in vitro biochemical studies showed that CSN inhibits the activity of CRL1 [192], which is consistent with its deneddylase activity. However, later genetic studies demonstrated that CSN is required for CRL-mediated substrate degradation [193,194,195], indicating a positive effect on CRL activity. This functional paradox was partially explained by the role of CSN in preventing autoubiquitination of CRL substrate receptors [196,197,198]. More recently, with the functional characterization of CAND1 (cullin-associated NEDD8-dissociated protein 1; see also Section 9.3) [199,200,201], an alternative model was proposed that CSN promotes CRL activities by allowing CAND1 to bind cullins and subsequently exchange the substrate receptor module associated with the CUL•RBX1 core [202] (see Section 9.3).
9.2.2. Multifaceted Roles of CSN in Cardiac Physiology
CSN has been implicated in the pathogenesis of cardiovascular diseases by various studies [203,204,205,206,207,208,209,210,211,212]. First, cardiac-specific knockout of Csn8 in mice resulted in defective Csn complex assembly, leading to cardiac hypertrophy, heart failure, and eventually, postnatal lethality four to five weeks after birth [210]. This finding, together with phenotypes observed in NAE-deficient mice [170], highlights the importance of the neddylation–deneddylation cycle in heart development. Moreover, conditional deletion of Csn8 in adult mice hearts caused striking cardiomyocyte necrosis and heart failure [213], suggesting that CSN also plays an essential role in post-mitotic cardiac physiology. Second, it has been shown that through regulating the degradation of misfolded proteins, CSN protects cardiomyocytes from proteotoxic stress [212]. Third, CSN exhibited anti-atherogenic capacity in endothelial and myeloid cells through negatively regulating inflammatory processes [214,215]. In addition to mediating the deneddylation of cullin proteins, CSN also functions as a docking platform for kinases and deubiquitinases [216]. Therefore, besides being a key regulator for CRL-dependent protein ubiquitination and degradation, CSN may also play a CRL-independent role in the cardiovascular system.
9.3. Cullin-Associated NEDD8-Dissociated Protein 1/2 (CAND1/2)
CAND1 is a substrate receptor exchange factor for CRLs, and it binds unneddylated cullins in a manner mutually exclusive with substrate receptors [202]. When cullins are bound by CAND1, cullin neddylation is inhibited but the engagement of neddylation enzymes to the cullin is increased [201]. In the absence of CAND1, a CRL1 complex is very stable and displays extremely slow dissociation. CAND1 can dramatically increase the dissociation rate of the CRL1 and binds tightly to the CUL1 “recycled” from the pre-existing CRL1 complex (Figure 2, Step 1). Subsequently, a new substrate receptor module can destabilize the CUL1•CAND1 complex, remove CAND1, and form a new CRL1 (Figure 2, Step 2). This exchange process is controlled by neddylation: immediately after the removal of CAND1 by the substrate receptor module, CUL1 is neddylated and can no longer bind to CAND1 (Figure 2, Step 3); only when NEDD8 is cleaved by CSN, CUL1 is subject to CAND1-mediated exchange (Figure 2, Step 4). Substrate binding inhibits the NEDD8 deconjugation by CSN [217,218], allowing the formation of an active and stable CRL1 that can efficiently ubiquitinate the substrate (Figure 2, Step 5). With the collaborated efforts from CAND1, NEDD8, and CSN, CRL1 complexes constantly undergo cycles of assembly and disassembly, and substrate receptor modules in CRL1 complexes are rapidly exchanged [201,219]. This CAND1-mediated cycling of CUL1 and substrate receptor exchange has been primarily studied in the CRL1 system, but the same mechanism has been shown to apply to CRL3s [220] and CRL4s [221]. The rapid exchange cycles allow cells to quickly adjust the CRL repertoire in response to changing substrate demands and, therefore, newly emerged CRL substrates can be timely ubiquitinated.
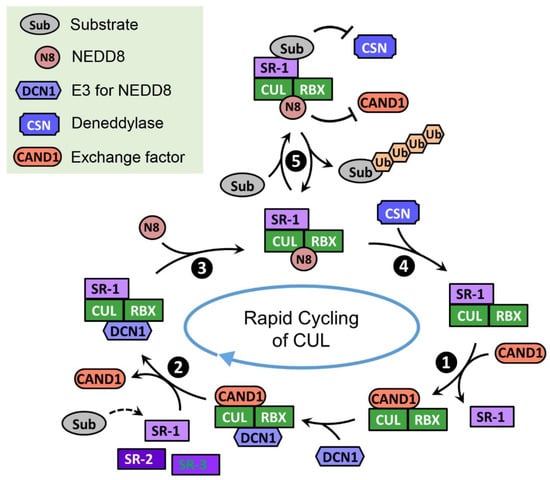
Figure 2.
Model for the rapid cycling of CUL. In the absence of CRL substrates, CUL quickly cycles through the CRL assembly (❷), neddylation (❸), deneddylation (❹), and exchange states (❶). This rapid cycling of CUL enables all kinds of substrate receptors to access the limited amount of CUL and assemble active CRLs that can potentially ubiquitinate their substrates. When the substrate is loaded on the CRL, it prevents CSN from binding and thus stabilizes CRL to allow substrate ubiquitination (❺). Key factors regulating the cycling of CUL are listed in the green box.
In mammals, CAND1 has a homologue, CAND2. The amino acid sequence of CAND2 is highly similar to CAND1, and CAND2 binds CUL1 in mammalian cells [201,222]. Unlike CAND1 that is ubiquitously expressed in all types of human cells, CAND2 protein is only detected in striated muscles (skeletal and cardiac) and testis [36,37,222]. While the role of CAND2 in regulating CRLs has not been well elucidated yet, population genetics and genome-wide association studies have identified CAND2 as a risk factor for multiple types of cardiovascular diseases (especially atrial fibrillation) [223,224,225,226,227,228,229]. A recent study aiming to determine the mechanism by which mTOR promotes pathological cardiac remodeling identified cand2 as a gene significantly upregulated by mTOR in mouse cardiomyocytes, and CAND2 depletion led to decreased protein level of Grk5 in a Cul1-dependent manner [230]. Furthermore, cand2 knockout mice exhibited pathological remodeling in the heart [230]. These results implied that an increased level of CAND2 would stabilize Grk5 and lead to adverse cardiac remodeling, providing one example for how CAND2 can be involved in cardiovascular diseases.
10. Concluding Remarks
Since the discovery of the F-box motif two and a half decades ago, the field of CRLs has been developing rapidly: components of the CRL family are defined, structures of diverse CRLs are solved, and substrates of individual CRL are discovered. It has now become clear that CRL-mediated ubiquitination modulates the activity and stability of proteins that play crucial roles in human cells, and thereby, CRLs regulate a broad spectrum of biological events. Furthermore, because of their capacity to selectively target cellular proteins for degradation, CRLs provide abundant opportunities for drug discovery, beyond the already clinically used proteasome inhibitors that have been proven successful for the treatment of cancer. The selectivity towards a limited number of substrates targeted by each CRL benefits drug discovery in other pathologies, where the tolerance for severe side effects is much lower, including hypertension and other cardiovascular diseases. In addition, the novel and promising PROTAC technique provides yet another avenue to harness the power of CRLs in the control of protein homeostasis.
With recent studies in cell biology, physiology, and disease genetics, CRL functions are implicated in various cardiovascular diseases. Continued efforts in understanding the mechanism and regulation of CRLs will undoubtedly expand our knowledge for the pathogenesis of cardiovascular diseases and uncover new avenues to develop therapeutics for disease treatment and prevention.
Author Contributions
Writing—original draft preparation, S.D.; writing—review and editing, K.W., B.S. and X.L.; supervision, X.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was aided by the National Institutes of Health grant R35GM138016 (to X.L.) and American Heart Association Career Development Award (to X.L.). The APC was funded by the AHA award.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Virani, S.S.; Alonso, A.; Aparicio, H.J.; Benjamin, E.J.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Cheng, S.; Delling, F.N.; et al. Heart Disease and Stroke Statistics-2021 Update: A Report From the American Heart Association. Circulation 2021, 143, e254–e743. [Google Scholar] [CrossRef]
- GBD 2016 Causes of Death Collaborators. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017, 390, 1151–1210. [Google Scholar] [CrossRef]
- Koga, H.; Kaushik, S.; Cuervo, A.M. Protein homeostasis and aging: The importance of exquisite quality control. Ageing Res. Rev. 2011, 10, 205–215. [Google Scholar] [CrossRef]
- Pohl, C.; Dikic, I. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science 2019, 366, 818–822. [Google Scholar] [CrossRef]
- Schapira, M.; Calabrese, M.F.; Bullock, A.N.; Crews, C.M. Targeted protein degradation: Expanding the toolbox. Nat. Rev. Drug Discov. 2019, 18, 949–963. [Google Scholar] [CrossRef]
- Sarikas, A.; Hartmann, T.; Pan, Z.Q. The cullin protein family. Genome Biol. 2011, 12, 220. [Google Scholar] [CrossRef]
- Harper, J.W.; Schulman, B.A. Cullin-RING Ubiquitin Ligase Regulatory Circuits: A Quarter Century Beyond the F-Box Hypothesis. Annu. Rev. Biochem. 2021, 90, 403–429. [Google Scholar] [CrossRef]
- Rusnac, D.V.; Zheng, N. Structural Biology of CRL Ubiquitin Ligases. Adv. Exp. Med. Biol. 2020, 1217, 9–31. [Google Scholar] [CrossRef]
- Michel, J.J.; McCarville, J.F.; Xiong, Y. A role for Saccharomyces cerevisiae Cul8 ubiquitin ligase in proper anaphase progression. J. Biol. Chem. 2003, 278, 22828–22837. [Google Scholar] [CrossRef]
- Mimura, S.; Yamaguchi, T.; Ishii, S.; Noro, E.; Katsura, T.; Obuse, C.; Kamura, T. Cul8/Rtt101 forms a variety of protein complexes that regulate DNA damage response and transcriptional silencing. J. Biol. Chem. 2010, 285, 9858–9867. [Google Scholar] [CrossRef]
- Petroski, M.D.; Deshaies, R.J. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005, 6, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Demasi, M.; Laurindo, F.R. Physiological and pathological role of the ubiquitin-proteasome system in the vascular smooth muscle cell. Cardiovasc. Res. 2012, 95, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Willis, M.S.; Patterson, C. Into the heart: The emerging role of the ubiquitin-proteasome system. J. Mol. Cell. Cardiol. 2006, 41, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Pagan, J.; Seto, T.; Pagano, M.; Cittadini, A. Role of the ubiquitin proteasome system in the heart. Circ. Res. 2013, 112, 1046–1058. [Google Scholar] [CrossRef] [PubMed]
- Deshaies, R.J.; Joazeiro, C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef]
- Gu, D.; Wang, S.; Kuiatse, I.; Wang, H.; He, J.; Dai, Y.; Jones, R.J.; Bjorklund, C.C.; Yang, J.; Grant, S.; et al. Inhibition of the MDM2 E3 Ligase induces apoptosis and autophagy in wild-type and mutant p53 models of multiple myeloma, and acts synergistically with ABT-737. PLoS ONE 2014, 9, e103015. [Google Scholar] [CrossRef]
- Hanna, J.; Guerra-Moreno, A.; Ang, J.; Micoogullari, Y. Protein Degradation and the Pathologic Basis of Disease. Am. J. Pathol. 2019, 189, 94–103. [Google Scholar] [CrossRef]
- Lydeard, J.R.; Schulman, B.A.; Harper, J.W. Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Rep. 2013, 14, 1050–1061. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, P.; Inuzuka, H.; Wei, W. Roles of F-box proteins in cancer. Nat. Rev. Cancer 2014, 14, 233–247. [Google Scholar] [CrossRef]
- Peris-Moreno, D.; Cussonneau, L.; Combaret, L.; Polge, C.; Taillandier, D. Ubiquitin Ligases at the Heart of Skeletal Muscle Atrophy Control. Molecules 2021, 26, 407. [Google Scholar] [CrossRef]
- Schisler, J.C.; Willis, M.S.; Patterson, C. You spin me round: MaFBx/Atrogin-1 feeds forward on FOXO transcription factors (like a record). Cell Cycle 2008, 7, 440–443. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.R.; Kim, H.J.; Lecker, S.H. Ubiquitin-protein ligases in muscle wasting. Int. J. Biochem. Cell Biol. 2005, 37, 2088–2097. [Google Scholar] [CrossRef] [PubMed]
- Gomes, M.D.; Lecker, S.H.; Jagoe, R.T.; Navon, A.; Goldberg, A.L. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. USA 2001, 98, 14440–14445. [Google Scholar] [CrossRef] [PubMed]
- Bodine, S.C.; Latres, E.; Baumhueter, S.; Lai, V.K.; Nunez, L.; Clarke, B.A.; Poueymirou, W.T.; Panaro, F.J.; Na, E.; Dharmarajan, K.; et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 2001, 294, 1704–1708. [Google Scholar] [CrossRef] [PubMed]
- Lagirand-Cantaloube, J.; Offner, N.; Csibi, A.; Leibovitch, M.P.; Batonnet-Pichon, S.; Tintignac, L.A.; Segura, C.T.; Leibovitch, S.A. The initiation factor eIF3-f is a major target for atrogin1/MAFbx function in skeletal muscle atrophy. EMBO J. 2008, 27, 1266–1276. [Google Scholar] [CrossRef] [PubMed]
- Tintignac, L.A.; Lagirand, J.; Batonnet, S.; Sirri, V.; Leibovitch, M.P.; Leibovitch, S.A. Degradation of MyoD mediated by the SCF (MAFbx) ubiquitin ligase. J. Biol. Chem. 2005, 280, 2847–2856. [Google Scholar] [CrossRef]
- Xie, P.; Guo, S.; Fan, Y.; Zhang, H.; Gu, D.; Li, H. Atrogin-1/MAFbx enhances simulated ischemia/reperfusion-induced apoptosis in cardiomyocytes through degradation of MAPK phosphatase-1 and sustained JNK activation. J. Biol. Chem. 2009, 284, 5488–5496. [Google Scholar] [CrossRef]
- Li, H.H.; Willis, M.S.; Lockyer, P.; Miller, N.; McDonough, H.; Glass, D.J.; Patterson, C. Atrogin-1 inhibits Akt-dependent cardiac hypertrophy in mice via ubiquitin-dependent coactivation of Forkhead proteins. J. Clin. Investig. 2007, 117, 3211–3223. [Google Scholar] [CrossRef]
- Li, H.H.; Kedar, V.; Zhang, C.; McDonough, H.; Arya, R.; Wang, D.Z.; Patterson, C. Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J. Clin. Investig. 2004, 114, 1058–1071. [Google Scholar] [CrossRef]
- Zaglia, T.; Milan, G.; Ruhs, A.; Franzoso, M.; Bertaggia, E.; Pianca, N.; Carpi, A.; Carullo, P.; Pesce, P.; Sacerdoti, D.; et al. Atrogin-1 deficiency promotes cardiomyopathy and premature death via impaired autophagy. J. Clin. Investig. 2014, 124, 2410–2424. [Google Scholar] [CrossRef]
- Mota, R.; Parry, T.L.; Yates, C.C.; Qiang, Z.; Eaton, S.C.; Mwiza, J.M.; Tulasi, D.; Schisler, J.C.; Patterson, C.; Zaglia, T.; et al. Increasing Cardiomyocyte Atrogin-1 Reduces Aging-Associated Fibrosis and Regulates Remodeling In Vivo. Am. J. Pathol. 2018, 188, 1676–1692. [Google Scholar] [CrossRef] [PubMed]
- Al-Yacoub, N.; Shaheen, R.; Awad, S.M.; Kunhi, M.; Dzimiri, N.; Nguyen, H.C.; Xiong, Y.; Al-Buraiki, J.; Al-Habeeb, W.; Alkuraya, F.S.; et al. FBXO32, encoding a member of the SCF complex, is mutated in dilated cardiomyopathy. Genome Biol. 2016, 17, 2. [Google Scholar] [CrossRef] [PubMed]
- Al-Hassnan, Z.N.; Shinwari, Z.M.; Wakil, S.M.; Tulbah, S.; Mohammed, S.; Rahbeeni, Z.; Alghamdi, M.; Rababh, M.; Colak, D.; Kaya, N.; et al. A substitution mutation in cardiac ubiquitin ligase, FBXO32, is associated with an autosomal recessive form of dilated cardiomyopathy. BMC Med. Genet. 2016, 17, 3. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Al-Yacoub, N.; Colak, D.; Mahmoud, S.A.; Hammonds, M.; Muhammed, K.; Al-Harazi, O.; Assiri, A.M.; Al-Buraiki, J.; Al-Habeeb, W.; Poizat, C. Mutation in FBXO32 causes dilated cardiomyopathy through up-regulation of ER-stress mediated apoptosis. Commun. Biol. 2021, 4, 884. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Sun, M.; Zhou, H.; Ajoolabady, A.; Zhou, Y.; Tao, J.; Sowers, J.R.; Zhang, Y. FUNDC1 interacts with FBXL2 to govern mitochondrial integrity and cardiac function through an IP3R3-dependent manner in obesity. Sci. Adv. 2020, 6, 943–953. [Google Scholar] [CrossRef]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Lee, J.E.; Sweredoski, M.J.; Graham, R.L.; Kolawa, N.J.; Smith, G.T.; Hess, S.; Deshaies, R.J. The steady-state repertoire of human SCF ubiquitin ligase complexes does not require ongoing Nedd8 conjugation. Mol. Cell. Proteom. MCP 2011, 10, M110006460. [Google Scholar] [CrossRef]
- Kuchay, S.; Duan, S.; Schenkein, E.; Peschiaroli, A.; Saraf, A.; Florens, L.; Washburn, M.P.; Pagano, M. FBXL2- and PTPL1-mediated degradation of p110-free p85beta regulatory subunit controls the PI(3)K signalling cascade. Nat. Cell Biol. 2013, 15, 472–480. [Google Scholar] [CrossRef]
- Kuchay, S.; Giorgi, C.; Simoneschi, D.; Pagan, J.; Missiroli, S.; Saraf, A.; Florens, L.; Washburn, M.P.; Collazo-Lorduy, A.; Castillo-Martin, M.; et al. PTEN counteracts FBXL2 to promote IP3R3- and Ca(2+)-mediated apoptosis limiting tumour growth. Nature 2017, 546, 554–558. [Google Scholar] [CrossRef]
- Yeh, C.H.; Bellon, M.; Nicot, C. FBXW7: A critical tumor suppressor of human cancers. Mol. Cancer 2018, 17, 115. [Google Scholar] [CrossRef]
- Wang, X.; Bathina, M.; Lynch, J.; Koss, B.; Calabrese, C.; Frase, S.; Schuetz, J.D.; Rehg, J.E.; Opferman, J.T. Deletion of MCL-1 causes lethal cardiac failure and mitochondrial dysfunction. Genes Dev. 2013, 27, 1351–1364. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.C.; Yeh, C.H. Inhibition of miR-302 Suppresses Hypoxia-Reoxygenation-Induced H9c2 Cardiomyocyte Death by Regulating Mcl-1 Expression. Oxid. Med. Cell. Longev. 2017, 2017, 7968905. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, N.; Zhang, Y.; Jia, P.; Guo, Y.; Tian, Y.; You, S.; Wu, S.; Sun, Y. E3 ligase Fbw7 participates in oxidative stressinduced myocardial cell injury via interacting with Mcl1. Mol. Med. Rep. 2019, 20, 1561–1568. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Guo, N.; Zhao, S.; Chen, Z.; Zhang, W.; Yan, F.; Liao, H.; Chi, K. FBXW7 promotes pathological cardiac hypertrophy by targeting EZH2-SIX1 signaling. Exp. Cell Res. 2020, 393, 112059. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Su, X.; Shen, Y.; Jin, Y.; Luo, T.; Kim, I.M.; Weintraub, N.L.; Tang, Y. MiR322 mediates cardioprotection against ischemia/reperfusion injury via FBXW7/notch pathway. J. Mol. Cell. Cardiol. 2019, 133, 67–74. [Google Scholar] [CrossRef]
- Wang, L.; Qin, D.; Shi, H.; Zhang, Y.; Li, H.; Han, Q. MiR-195-5p Promotes Cardiomyocyte Hypertrophy by Targeting MFN2 and FBXW7. Biomed. Res. Int. 2019, 2019, 1580982. [Google Scholar] [CrossRef]
- Zou, J.F.; Wu, X.N.; Shi, R.H.; Sun, Y.Q.; Qin, F.J.; Yang, Y.M. Inhibition of microRNA-184 reduces H2O2-mediated cardiomyocyte injury via targeting FBXO28. Eur. Rev. Med. Pharm. Sci. 2020, 24, 11251–11258. [Google Scholar] [CrossRef]
- Wang, H.H.; Lee, Y.N.; Su, C.H.; Shu, K.T.; Liu, W.T.; Hsieh, C.L.; Yeh, H.I.; Wu, Y.J. S-Phase Kinase-associated Protein-2 Rejuvenates Senescent Endothelial Progenitor Cells and Induces Angiogenesis In Vivo. Sci. Rep. 2020, 10, 6646. [Google Scholar] [CrossRef]
- Asmamaw, M.D.; Liu, Y.; Zheng, Y.C.; Shi, X.J.; Liu, H.M. Skp2 in the ubiquitin-proteasome system: A comprehensive review. Med. Res. Rev. 2020, 40, 1920–1949. [Google Scholar] [CrossRef]
- Wu, Y.J.; Sala-Newby, G.B.; Shu, K.T.; Yeh, H.I.; Nakayama, K.I.; Nakayama, K.; Newby, A.C.; Bond, M. S-phase kinase-associated protein-2 (Skp2) promotes vascular smooth muscle cell proliferation and neointima formation in vivo. J. Vasc. Surg. 2009, 50, 1135–1142. [Google Scholar] [CrossRef]
- Cai, W.; Yang, H. The structure and regulation of Cullin 2 based E3 ubiquitin ligases and their biological functions. Cell Div. 2016, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Pause, A.; Lee, S.; Worrell, R.A.; Chen, D.Y.; Burgess, W.H.; Linehan, W.M.; Klausner, R.D. The von Hippel-Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc. Natl. Acad. Sci. USA 1997, 94, 2156–2161. [Google Scholar] [CrossRef] [PubMed]
- Kamura, T.; Conrad, M.N.; Yan, Q.; Conaway, R.C.; Conaway, J.W. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 1999, 13, 2928–2933. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, P.H.; Wiesener, M.S.; Chang, G.W.; Clifford, S.C.; Vaux, E.C.; Cockman, M.E.; Wykoff, C.C.; Pugh, C.W.; Maher, E.R.; Ratcliffe, P.J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 1999, 399, 271–275. [Google Scholar] [CrossRef]
- Ivan, M.; Kondo, K.; Yang, H.; Kim, W.; Valiando, J.; Ohh, M.; Salic, A.; Asara, J.M.; Lane, W.S.; Kaelin, W.G., Jr. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science 2001, 292, 464–468. [Google Scholar] [CrossRef]
- Epstein, A.C.R.; Gleadle, J.M.; McNeill, L.A.; Hewitson, K.S.; O’Rourke, J.; Mole, D.R.; Mukherji, M.; Metzen, E.; Wilson, M.I.; Dhanda, A.; et al. C. elegans EGL-9 and Mammalian Homologs Define a Family of Dioxygenases that Regulate HIF by Prolyl Hydroxylation. Cell 2001, 107, 43–54. [Google Scholar] [CrossRef]
- Li, H.; Ko, H.P.; Whitlock, J.P. Induction of phosphoglycerate kinase 1 gene expression by hypoxia. Roles of Arnt and HIF1alpha. J. Biol. Chem. 1996, 271, 21262–21267. [Google Scholar] [CrossRef]
- Ema, M.; Hirota, K.; Mimura, J.; Abe, H.; Yodoi, J.; Sogawa, K.; Poellinger, L.; Fujii-Kuriyama, Y. Molecular mechanisms of transcription activation by HLF and HIF1alpha in response to hypoxia: Their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 1999, 18, 1905–1914. [Google Scholar] [CrossRef]
- Semenza, G.L. HIF-1: Mediator of physiological and pathophysiological responses to hypoxia. J. Appl. Physiol. 2000, 88, 1474–1480. [Google Scholar] [CrossRef]
- Semenza, G.L. Hypoxia-inducible factor 1: Regulator of mitochondrial metabolism and mediator of ischemic preconditioning. Biochim. Biophys. Acta 2011, 1813, 1263–1268. [Google Scholar] [CrossRef]
- Pugh, C.W.; Ratcliffe, P.J. Regulation of angiogenesis by hypoxia: Role of the HIF system. Nat. Med. 2003, 9, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Galli, G.; Wang, Y.; Fan, Q.; Wang, Z.; Wang, X.; Xiao, W. Novel Therapeutic Targets for Hypoxia-Related Cardiovascular Diseases: The Role of HIF-1. Front. Physiol. 2020, 11, 774. [Google Scholar] [CrossRef] [PubMed]
- Farnaby, W.; Koegl, M.; McConnell, D.B.; Ciulli, A. Transforming targeted cancer therapy with PROTACs: A forward-looking perspective. Curr. Opin. Pharm. 2021, 57, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Nalawansha, D.A.; Crews, C.M. PROTACs: An Emerging Therapeutic Modality in Precision Medicine. Cell Chem. Biol. 2020, 27, 998–1014. [Google Scholar] [CrossRef]
- Verma, R.; Mohl, D.; Deshaies, R.J. Harnessing the Power of Proteolysis for Targeted Protein Inactivation. Mol. Cell 2020, 77, 446–460. [Google Scholar] [CrossRef]
- Wu, T.; Yoon, H.; Xiong, Y.; Dixon-Clarke, S.E.; Nowak, R.P.; Fischer, E.S. Targeted protein degradation as a powerful research tool in basic biology and drug target discovery. Nat. Struct. Mol. Biol. 2020, 27, 605–614. [Google Scholar] [CrossRef]
- Buckley, D.L.; Gustafson, J.L.; Van Molle, I.; Roth, A.G.; Tae, H.S.; Gareiss, P.C.; Jorgensen, W.L.; Ciulli, A.; Crews, C.M. Small-molecule inhibitors of the interaction between the E3 ligase VHL and HIF1alpha. Angew. Chem. Int. Ed. Engl. 2012, 51, 11463–11467. [Google Scholar] [CrossRef]
- Buckley, D.L.; Van Molle, I.; Gareiss, P.C.; Tae, H.S.; Michel, J.; Noblin, D.J.; Jorgensen, W.L.; Ciulli, A.; Crews, C.M. Targeting the von Hippel-Lindau E3 ubiquitin ligase using small molecules to disrupt the VHL/HIF-1alpha interaction. J. Am. Chem. Soc. 2012, 134, 4465–4468. [Google Scholar] [CrossRef]
- Bond, M.J.; Chu, L.; Nalawansha, D.A.; Li, K.; Crews, C.M. Targeted Degradation of Oncogenic KRAS(G12C) by VHL-Recruiting PROTACs. ACS Cent. Sci. 2020, 6, 1367–1375. [Google Scholar] [CrossRef]
- Bondeson, D.P.; Mares, A.; Smith, I.E.; Ko, E.; Campos, S.; Miah, A.H.; Mulholland, K.E.; Routly, N.; Buckley, D.L.; Gustafson, J.L.; et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol. 2015, 11, 611–617. [Google Scholar] [CrossRef]
- Burslem, G.M.; Smith, B.E.; Lai, A.C.; Jaime-Figueroa, S.; McQuaid, D.C.; Bondeson, D.P.; Toure, M.; Dong, H.; Qian, Y.; Wang, J.; et al. The Advantages of Targeted Protein Degradation Over Inhibition: An RTK Case Study. Cell Chem. Biol. 2018, 25, 67–77.e63. [Google Scholar] [CrossRef] [PubMed]
- Cromm, P.M.; Samarasinghe, K.T.G.; Hines, J.; Crews, C.M. Addressing Kinase-Independent Functions of Fak via PROTAC-Mediated Degradation. J. Am. Chem. Soc. 2018, 140, 17019–17026. [Google Scholar] [CrossRef] [PubMed]
- Farnaby, W.; Koegl, M.; Roy, M.J.; Whitworth, C.; Diers, E.; Trainor, N.; Zollman, D.; Steurer, S.; Karolyi-Oezguer, J.; Riedmueller, C.; et al. BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design. Nat. Chem. Biol. 2019, 15, 672–680. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.E.; Wang, S.L.; Jaime-Figueroa, S.; Harbin, A.; Wang, J.; Hamman, B.D.; Crews, C.M. Differential PROTAC substrate specificity dictated by orientation of recruited E3 ligase. Nat. Commun. 2019, 10, 131. [Google Scholar] [CrossRef] [PubMed]
- Zoppi, V.; Hughes, S.J.; Maniaci, C.; Testa, A.; Gmaschitz, T.; Wieshofer, C.; Koegl, M.; Riching, K.M.; Daniels, D.L.; Spallarossa, A.; et al. Iterative Design and Optimization of Initially Inactive Proteolysis Targeting Chimeras (PROTACs) Identify VZ185 as a Potent, Fast, and Selective von Hippel-Lindau (VHL) Based Dual Degrader Probe of BRD9 and BRD7. J. Med. Chem. 2019, 62, 699–726. [Google Scholar] [CrossRef]
- Zengerle, M.; Chan, K.H.; Ciulli, A. Selective Small Molecule Induced Degradation of the BET Bromodomain Protein BRD4. ACS Chem. Biol. 2015, 10, 1770–1777. [Google Scholar] [CrossRef]
- Khan, S.; Zhang, X.; Lv, D.; Zhang, Q.; He, Y.; Zhang, P.; Liu, X.; Thummuri, D.; Yuan, Y.; Wiegand, J.S.; et al. A selective BCL-XL PROTAC degrader achieves safe and potent antitumor activity. Nat. Med. 2019, 25, 1938–1947. [Google Scholar] [CrossRef]
- Zhang, X.; Thummuri, D.; Liu, X.; Hu, W.; Zhang, P.; Khan, S.; Yuan, Y.; Zhou, D.; Zheng, G. Discovery of PROTAC BCL-XL degraders as potent anticancer agents with low on-target platelet toxicity. Eur. J. Med. Chem. 2020, 192, 112186. [Google Scholar] [CrossRef]
- Gadd, M.S.; Testa, A.; Lucas, X.; Chan, K.H.; Chen, W.; Lamont, D.J.; Zengerle, M.; Ciulli, A. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol. 2017, 13, 514–521. [Google Scholar] [CrossRef]
- Crew, A.P.; Raina, K.; Dong, H.; Qian, Y.; Wang, J.; Vigil, D.; Serebrenik, Y.V.; Hamman, B.D.; Morgan, A.; Ferraro, C.; et al. Identification and Characterization of Von Hippel-Lindau-Recruiting Proteolysis Targeting Chimeras (PROTACs) of TANK-Binding Kinase 1. J. Med. Chem. 2018, 61, 583–598. [Google Scholar] [CrossRef]
- Gechijian, L.N.; Buckley, D.L.; Lawlor, M.A.; Reyes, J.M.; Paulk, J.; Ott, C.J.; Winter, G.E.; Erb, M.A.; Scott, T.G.; Xu, M.; et al. Functional TRIM24 degrader via conjugation of ineffectual bromodomain and VHL ligands. Nat. Chem. Biol. 2018, 14, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Wang, C.; Qin, C.; Xiang, W.; Fernandez-Salas, E.; Yang, C.Y.; Wang, M.; Zhao, L.; Xu, T.; Chinnaswamy, K.; et al. Discovery of ARD-69 as a Highly Potent Proteolysis Targeting Chimera (PROTAC) Degrader of Androgen Receptor (AR) for the Treatment of Prostate Cancer. J. Med. Chem. 2019, 62, 941–964. [Google Scholar] [CrossRef] [PubMed]
- Nunes, J.; McGonagle, G.A.; Eden, J.; Kiritharan, G.; Touzet, M.; Lewell, X.; Emery, J.; Eidam, H.; Harling, J.D.; Anderson, N.A. Targeting IRAK4 for Degradation with PROTACs. ACS Med. Chem. Lett. 2019, 10, 1081–1085. [Google Scholar] [CrossRef] [PubMed]
- Maniaci, C.; Hughes, S.J.; Testa, A.; Chen, W.; Lamont, D.J.; Rocha, S.; Alessi, D.R.; Romeo, R.; Ciulli, A. Homo-PROTACs: Bivalent small-molecule dimerizers of the VHL E3 ubiquitin ligase to induce self-degradation. Nat. Commun. 2017, 8, 830. [Google Scholar] [CrossRef]
- Imaide, S.; Riching, K.M.; Makukhin, N.; Vetma, V.; Whitworth, C.; Hughes, S.J.; Trainor, N.; Mahan, S.D.; Murphy, N.; Cowan, A.D.; et al. Trivalent PROTACs enhance protein degradation via combined avidity and cooperativity. Nat. Chem. Biol. 2021, 17, 1157–1167. [Google Scholar] [CrossRef] [PubMed]
- Canning, P.; Cooper, C.D.O.; Krojer, T.; Murray, J.W.; Pike, A.C.W.; Chaikuad, A.; Keates, T.; Thangaratnarajah, C.; Hojzan, V.; Marsden, B.D.; et al. Structural basis for Cul3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases. J. Biol. Chem. 2013, 288, 7803–7814. [Google Scholar] [CrossRef] [PubMed]
- Ji, A.X.; Prive, G.G. Crystal structure of KLHL3 in complex with Cullin3. PLoS ONE 2013, 8, e60445. [Google Scholar] [CrossRef]
- Dubiel, W.; Dubiel, D.; Wolf, D.A.; Naumann, M. Cullin 3-Based Ubiquitin Ligases as Master Regulators of Mammalian Cell Differentiation. Trends Biochem. Sci. 2018, 43, 95–107. [Google Scholar] [CrossRef]
- Wu, J.; McCormick, J.A.; Sigmund, C.D. Cullin-3: Renal and Vascular Mechanisms Regulating Blood Pressure. Curr. Hypertens. Rep. 2020, 22, 61. [Google Scholar] [CrossRef]
- Sakaue, T.; Maekawa, M.; Nakayama, H.; Higashiyama, S. Prospect of divergent roles for the CUL3 system in vascular endothelial cell function and angiogenesis. J. Biochem. 2017, 162, 237–245. [Google Scholar] [CrossRef][Green Version]
- Papizan, J.B.; Vidal, A.H.; Bezprozvannaya, S.; Bassel-Duby, R.; Olson, E.N. Cullin-3-RING ubiquitin ligase activity is required for striated muscle function in mice. J. Biol. Chem. 2018, 293, 8802–8811. [Google Scholar] [CrossRef] [PubMed]
- Agbor, L.N.; Nair, A.R.; Wu, J.; Lu, K.T.; Davis, D.R.; Keen, H.L.; Quelle, F.W.; McCormick, J.A.; Singer, J.D.; Sigmund, C.D. Conditional deletion of smooth muscle Cullin-3 causes severe progressive hypertension. JCI Insight 2019, 5. [Google Scholar] [CrossRef] [PubMed]
- Sakaue, T.; Fujisaki, A.; Nakayama, H.; Maekawa, M.; Hiyoshi, H.; Kubota, E.; Joh, T.; Izutani, H.; Higashiyama, S. Neddylated Cullin 3 is required for vascular endothelial-cadherin-mediated endothelial barrier function. Cancer Sci. 2017, 108, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Boyden, L.M.; Choi, M.; Choate, K.A.; Nelson-Williams, C.J.; Farhi, A.; Toka, H.R.; Tikhonova, I.R.; Bjornson, R.; Mane, S.M.; Colussi, G.; et al. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature 2012, 482, 98–102. [Google Scholar] [CrossRef]
- Glover, M.; Ware, J.S.; Henry, A.; Wolley, M.; Walsh, R.; Wain, L.V.; Xu, S.; Van’t Hoff, W.G.; Tobin, M.D.; Hall, I.P.; et al. Detection of mutations in KLHL3 and CUL3 in families with FHHt (familial hyperkalaemic hypertension or Gordon’s syndrome). Clin. Sci. 2014, 126, 721–726. [Google Scholar] [CrossRef]
- Schumacher, F.R.; Siew, K.; Zhang, J.; Johnson, C.; Wood, N.; Cleary, S.E.; Al Maskari, R.S.; Ferryman, J.T.; Hardege, I.; Yasmin; et al. Characterisation of the Cullin-3 mutation that causes a severe form of familial hypertension and hyperkalaemia. EMBO Mol. Med. 2015, 7, 1285–1306. [Google Scholar] [CrossRef]
- Abdel Khalek, W.; Rafael, C.; Loisel-Ferreira, I.; Kouranti, I.; Clauser, E.; Hadchouel, J.; Jeunemaitre, X. Severe Arterial Hypertension from Cullin 3 Mutations Is Caused by Both Renal and Vascular Effects. J. Am. Soc. Nephrol. 2019, 30, 811–823. [Google Scholar] [CrossRef]
- Agbor, L.N.; Ibeawuchi, S.C.; Hu, C.; Wu, J.; Davis, D.R.; Keen, H.L.; Quelle, F.W.; Sigmund, C.D. Cullin-3 mutation causes arterial stiffness and hypertension through a vascular smooth muscle mechanism. JCI Insight 2016, 1, e91015. [Google Scholar] [CrossRef]
- Louis-Dit-Picard, H.; Barc, J.; Trujillano, D.; Miserey-Lenkei, S.; Bouatia-Naji, N.; Pylypenko, O.; Beaurain, G.; Bonnefond, A.; Sand, O.; Simian, C.; et al. KLHL3 mutations cause familial hyperkalemic hypertension by impairing ion transport in the distal nephron. Nat. Genet. 2012, 44, 456–460. [Google Scholar] [CrossRef]
- Hureaux, M.; Mazurkiewicz, S.; Boccio, V.; Vargas-Poussou, R.; Jeunemaitre, X. The variety of genetic defects explains the phenotypic heterogeneity of Familial Hyperkalemic Hypertension. Kidney Int. Rep. 2021, 6, 2639–2652. [Google Scholar] [CrossRef]
- Louis-Dit-Picard, H.; Kouranti, I.; Rafael, C.; Loisel-Ferreira, I.; Chavez-Canales, M.; Abdel-Khalek, W.; Argaiz, E.R.; Baron, S.; Vacle, S.; Migeon, T.; et al. Mutation affecting the conserved acidic WNK1 motif causes inherited hyperkalemic hyperchloremic acidosis. J. Clin. Investig. 2020, 130, 6379–6394. [Google Scholar] [CrossRef] [PubMed]
- McCormick, J.A.; Mutig, K.; Nelson, J.H.; Saritas, T.; Hoorn, E.J.; Yang, C.L.; Rogers, S.; Curry, J.; Delpire, E.; Bachmann, S.; et al. A SPAK isoform switch modulates renal salt transport and blood pressure. Cell Metab. 2011, 14, 352–364. [Google Scholar] [CrossRef] [PubMed]
- Shibata, S.; Zhang, J.; Puthumana, J.; Stone, K.L.; Lifton, R.P. Kelch-like 3 and Cullin 3 regulate electrolyte homeostasis via ubiquitination and degradation of WNK4. Proc. Natl. Acad. Sci. USA 2013, 110, 7838–7843. [Google Scholar] [CrossRef]
- Ishizawa, K.; Wang, Q.; Li, J.; Yamazaki, O.; Tamura, Y.; Fujigaki, Y.; Uchida, S.; Lifton, R.P.; Shibata, S. Calcineurin dephosphorylates Kelch-like 3, reversing phosphorylation by angiotensin II and regulating renal electrolyte handling. Proc. Natl. Acad. Sci. USA 2019, 116, 3155–3160. [Google Scholar] [CrossRef] [PubMed]
- Ohta, A.; Schumacher, F.R.; Mehellou, Y.; Johnson, C.; Knebel, A.; Macartney, T.J.; Wood, N.T.; Alessi, D.R.; Kurz, T. The CUL3-KLHL3 E3 ligase complex mutated in Gordon’s hypertension syndrome interacts with and ubiquitylates WNK isoforms: Disease-causing mutations in KLHL3 and WNK4 disrupt interaction. Biochem. J. 2013, 451, 111–122. [Google Scholar] [CrossRef]
- Schumacher, F.R.; Sorrell, F.J.; Alessi, D.R.; Bullock, A.N.; Kurz, T. Structural and biochemical characterization of the KLHL3-WNK kinase interaction important in blood pressure regulation. Biochem. J. 2014, 460, 237–246. [Google Scholar] [CrossRef]
- Wu, G.; Peng, J.B. Disease-causing mutations in KLHL3 impair its effect on WNK4 degradation. FEBS Lett. 2013, 587, 1717–1722. [Google Scholar] [CrossRef]
- Wakabayashi, M.; Mori, T.; Isobe, K.; Sohara, E.; Susa, K.; Araki, Y.; Chiga, M.; Kikuchi, E.; Nomura, N.; Mori, Y.; et al. Impaired KLHL3-mediated ubiquitination of WNK4 causes human hypertension. Cell Rep. 2013, 3, 858–868. [Google Scholar] [CrossRef]
- Zhang, C.; Meermeier, N.P.; Terker, A.S.; Blankenstein, K.I.; Singer, J.D.; Hadchouel, J.; Ellison, D.H.; Yang, C.L. Degradation by Cullin 3 and effect on WNK kinases suggest a role of KLHL2 in the pathogenesis of Familial Hyperkalemic Hypertension. Biochem. Biophys. Res. Commun. 2016, 469, 44–48. [Google Scholar] [CrossRef]
- Yamamoto, M.; Kensler, T.W.; Motohashi, H. The KEAP1-NRF2 System: A Thiol-Based Sensor-Effector Apparatus for Maintaining Redox Homeostasis. Physiol. Rev. 2018, 98, 1169–1203. [Google Scholar] [CrossRef]
- Bellezza, I.; Giambanco, I.; Minelli, A.; Donato, R. Nrf2-Keap1 signaling in oxidative and reductive stress. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Jakobs, P.; Serbulea, V.; Leitinger, N.; Eckers, A.; Haendeler, J. Nuclear Factor (Erythroid-Derived 2)-Like 2 and Thioredoxin-1 in Atherosclerosis and Ischemia/Reperfusion Injury in the Heart. Antioxid. Redox Signal. 2017, 26, 630–644. [Google Scholar] [CrossRef] [PubMed]
- Piantadosi, C.A.; Carraway, M.S.; Babiker, A.; Suliman, H.B. Heme oxygenase-1 regulates cardiac mitochondrial biogenesis via Nrf2-mediated transcriptional control of nuclear respiratory factor-1. Circ. Res. 2008, 103, 1232–1240. [Google Scholar] [CrossRef]
- Li, J.; Zhang, C.; Xing, Y.; Janicki, J.S.; Yamamoto, M.; Wang, X.L.; Tang, D.Q.; Cui, T. Up-regulation of p27(kip1) contributes to Nrf2-mediated protection against angiotensin II-induced cardiac hypertrophy. Cardiovasc. Res. 2011, 90, 315–324. [Google Scholar] [CrossRef]
- Strom, J.; Xu, B.; Tian, X.; Chen, Q.M. Nrf2 protects mitochondrial decay by oxidative stress. FASEB J. 2016, 30, 66–80. [Google Scholar] [CrossRef] [PubMed]
- Cominacini, L.; Mozzini, C.; Garbin, U.; Pasini, A.; Stranieri, C.; Solani, E.; Vallerio, P.; Tinelli, I.A.; Fratta Pasini, A. Endoplasmic reticulum stress and Nrf2 signaling in cardiovascular diseases. Free Radic. Biol. Med. 2015, 88, 233–242. [Google Scholar] [CrossRef]
- Shen, Y.; Liu, X.; Shi, J.; Wu, X. Involvement of Nrf2 in myocardial ischemia and reperfusion injury. Int. J. Biol. Macromol. 2019, 125, 496–502. [Google Scholar] [CrossRef]
- Cadenas, S. ROS and redox signaling in myocardial ischemia-reperfusion injury and cardioprotection. Free Radic. Biol. Med. 2018, 117, 76–89. [Google Scholar] [CrossRef]
- Chen, B.; Lu, Y.; Chen, Y.; Cheng, J. The role of Nrf2 in oxidative stress-induced endothelial injuries. J. Endocrinol. 2015, 225, R83–R99. [Google Scholar] [CrossRef]
- Tian, C.; Gao, L.; Zimmerman, M.C.; Zucker, I.H. Myocardial infarction-induced microRNA-enriched exosomes contribute to cardiac Nrf2 dysregulation in chronic heart failure. Am. J. Physiol. Heart Circ. Physiol. 2018, 314, H928–H939. [Google Scholar] [CrossRef]
- Gao, L.; Zimmerman, M.C.; Biswal, S.; Zucker, I.H. Selective Nrf2 Gene Deletion in the Rostral Ventrolateral Medulla Evokes Hypertension and Sympathoexcitation in Mice. Hypertension 2017, 69, 1198–1206. [Google Scholar] [CrossRef] [PubMed]
- Mukohda, M.; Fang, S.; Wu, J.; Agbor, L.N.; Nair, A.R.; Ibeawuchi, S.C.; Hu, C.; Liu, X.; Lu, K.T.; Guo, D.F.; et al. RhoBTB1 protects against hypertension and arterial stiffness by restraining phosphodiesterase 5 activity. J. Clin. Investig. 2019, 129, 2318–2332. [Google Scholar] [CrossRef] [PubMed]
- Rybalkin, S.D.; Yan, C.; Bornfeldt, K.E.; Beavo, J.A. Cyclic GMP phosphodiesterases and regulation of smooth muscle function. Circ. Res. 2003, 93, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Sewduth, R.N.; Pandolfi, S.; Steklov, M.; Sheryazdanova, A.; Zhao, P.; Criem, N.; Baietti, M.F.; Lechat, B.; Quarck, R.; Impens, F.; et al. The Noonan Syndrome Gene Lztr1 Controls Cardiovascular Function by Regulating Vesicular Trafficking. Circ. Res. 2020, 126, 1379–1393. [Google Scholar] [CrossRef]
- Schaefer, A.; Reinhard, N.R.; Hordijk, P.L. Toward understanding RhoGTPase specificity: Structure, function and local activation. Small GTPases 2014, 5, 6. [Google Scholar] [CrossRef]
- Kovacevic, I.; Sakaue, T.; Majolee, J.; Pronk, M.C.; Maekawa, M.; Geerts, D.; Fernandez-Borja, M.; Higashiyama, S.; Hordijk, P.L. The Cullin-3-Rbx1-KCTD10 complex controls endothelial barrier function via K63 ubiquitination of RhoB. J. Cell Biol. 2018, 217, 1015–1032. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, Z.; Meng, M.; Zhao, Y.; Dong, N.; Yan, H.; Liu, L.; Ding, M.; Peng, H.B.; Shao, F. Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol. Cell 2009, 35, 841–855. [Google Scholar] [CrossRef]
- Ibeawuchi, S.R.; Agbor, L.N.; Quelle, F.W.; Sigmund, C.D. Hypertension-causing Mutations in Cullin3 Protein Impair RhoA Protein Ubiquitination and Augment the Association with Substrate Adaptors. J. Biol. Chem. 2015, 290, 19208–19217. [Google Scholar] [CrossRef]
- Hedberg-Oldfors, C.; Abramsson, A.; Osborn, D.P.S.; Danielsson, O.; Fazlinezhad, A.; Nilipour, Y.; Hubbert, L.; Nennesmo, I.; Visuttijai, K.; Bharj, J.; et al. Cardiomyopathy with lethal arrhythmias associated with inactivation of KLHL24. Hum. Mol. Genet. 2019, 28, 1919–1929. [Google Scholar] [CrossRef]
- Sang, Y.; Yan, F.; Ren, X. The role and mechanism of CRL4 E3 ubiquitin ligase in cancer and its potential therapy implications. Oncotarget 2015, 6, 42590–42602. [Google Scholar] [CrossRef]
- Hannah, J.; Zhou, P. Distinct and overlapping functions of the cullin E3 ligase scaffolding proteins CUL4A and CUL4B. Gene 2015, 573, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Zhou, P. DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol. Cell 2007, 26, 775–780. [Google Scholar] [CrossRef] [PubMed]
- He, Y.J.; McCall, C.M.; Hu, J.; Zeng, Y.; Xiong, Y. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev. 2006, 20, 2949–2954. [Google Scholar] [CrossRef] [PubMed]
- Ye, N.; Zhang, N.; Zhang, Y.; Qian, H.; Wu, B.; Sun, Y. Cul4a as a New Interaction Protein of PARP1 Inhibits Oxidative Stress-Induced H9c2 Cell Apoptosis. Oxid. Med. Cell. Longev. 2019, 2019, 4273261. [Google Scholar] [CrossRef]
- Li, P.; Song, Y.; Zan, W.; Qin, L.; Han, S.; Jiang, B.; Dou, H.; Shao, C.; Gong, Y. Lack of CUL4B in Adipocytes Promotes PPARgamma-Mediated Adipose Tissue Expansion and Insulin Sensitivity. Diabetes 2017, 66, 300–313. [Google Scholar] [CrossRef]
- Nowak, M.; Suenkel, B.; Porras, P.; Migotti, R.; Schmidt, F.; Kny, M.; Zhu, X.; Wanker, E.E.; Dittmar, G.; Fielitz, J.; et al. DCAF8, a novel MuRF1 interaction partner, promotes muscle atrophy. J. Cell Sci. 2019, 132, jcs233395. [Google Scholar] [CrossRef]
- Penela, P. Chapter Three—Ubiquitination and Protein Turnover of G-Protein-Coupled Receptor Kinases in GPCR Signaling and Cellular Regulation. Prog. Mol. Biol. Transl. Sci. 2016, 141, 85–140. [Google Scholar] [CrossRef]
- Wu, Z.; Chen, Y.; Yang, T.; Gao, Q.; Yuan, M.; Ma, L. Targeted ubiquitination and degradation of G-protein-coupled receptor kinase 5 by the DDB1-CUL4 ubiquitin ligase complex. PLoS ONE 2012, 7, e43997. [Google Scholar] [CrossRef]
- Zha, Z.; Han, X.; Smith, M.D.; Liu, Y.; Giguere, P.M.; Kopanja, D.; Raychaudhuri, P.; Siderovski, D.P.; Guan, K.L.; Lei, Q.Y.; et al. A Non-Canonical Function of Gbeta as a Subunit of E3 Ligase in Targeting GRK2 Ubiquitylation. Mol. Cell 2015, 58, 794–803. [Google Scholar] [CrossRef]
- Sjogren, B.; Swaney, S.; Neubig, R.R. FBXO44-Mediated Degradation of RGS2 Protein Uniquely Depends on a Cullin 4B/DDB1 Complex. PLoS ONE 2015, 10, e0123581. [Google Scholar] [CrossRef]
- Heximer, S.P.; Knutsen, R.H.; Sun, X.; Kaltenbronn, K.M.; Rhee, M.H.; Peng, N.; Oliveira-dos-Santos, A.; Penninger, J.M.; Muslin, A.J.; Steinberg, T.H.; et al. Hypertension and prolonged vasoconstrictor signaling in RGS2-deficient mice. J. Clin. Investig. 2003, 111, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Bodenstein, J.; Sunahara, R.K.; Neubig, R.R. N-terminal residues control proteasomal degradation of RGS2, RGS4, and RGS5 in human embryonic kidney 293 cells. Mol. Pharm. 2007, 71, 1040–1050. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Kamide, K.; Kokubo, Y.; Takiuchi, S.; Tanaka, C.; Banno, M.; Miwa, Y.; Yoshii, M.; Horio, T.; Okayama, A.; et al. Genetic variations of regulator of G-protein signaling 2 in hypertensive patients and in the general population. J. Hypertens. 2005, 23, 1497–1505. [Google Scholar] [CrossRef] [PubMed]
- Phan, H.T.N.; Sjogren, B.; Neubig, R.R. Human Missense Mutations in Regulator of G Protein Signaling 2 Affect the Protein Function Through Multiple Mechanisms. Mol. Pharm. 2017, 92, 451–458. [Google Scholar] [CrossRef] [PubMed]
- McNabb, H.J.; Gonzalez, S.; Muli, C.S.; Sjogren, B. N-Terminal Targeting of Regulator of G Protein Signaling Protein 2 for F-Box Only Protein 44-Mediated Proteasomal Degradation. Mol. Pharm. 2020, 98, 677–685. [Google Scholar] [CrossRef]
- Chopra, R.; Sadok, A.; Collins, I. A critical evaluation of the approaches to targeted protein degradation for drug discovery. Drug Discov. Today Technol. 2019, 31, 5–13. [Google Scholar] [CrossRef]
- Matyskiela, M.E.; Lu, G.; Ito, T.; Pagarigan, B.; Lu, C.C.; Miller, K.; Fang, W.; Wang, N.Y.; Nguyen, D.; Houston, J.; et al. A novel cereblon modulator recruits GSPT1 to the CRL4(CRBN) ubiquitin ligase. Nature 2016, 535, 252–257. [Google Scholar] [CrossRef]
- Petzold, G.; Fischer, E.S.; Thoma, N.H. Structural basis of lenalidomide-induced CK1alpha degradation by the CRL4(CRBN) ubiquitin ligase. Nature 2016, 532, 127–130. [Google Scholar] [CrossRef]
- Sievers, Q.L.; Petzold, G.; Bunker, R.D.; Renneville, A.; Slabicki, M.; Liddicoat, B.J.; Abdulrahman, W.; Mikkelsen, T.; Ebert, B.L.; Thoma, N.H. Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science 2018, 362. [Google Scholar] [CrossRef]
- Hansen, J.D.; Condroski, K.; Correa, M.; Muller, G.; Man, H.W.; Ruchelman, A.; Zhang, W.; Vocanson, F.; Crea, T.; Liu, W.; et al. Protein Degradation via CRL4(CRBN) Ubiquitin Ligase: Discovery and Structure-Activity Relationships of Novel Glutarimide Analogs That Promote Degradation of Aiolos and/or GSPT1. J. Med. Chem. 2018, 61, 492–503. [Google Scholar] [CrossRef]
- Kronke, J.; Fink, E.C.; Hollenbach, P.W.; MacBeth, K.J.; Hurst, S.N.; Udeshi, N.D.; Chamberlain, P.P.; Mani, D.R.; Man, H.W.; Gandhi, A.K.; et al. Lenalidomide induces ubiquitination and degradation of CK1alpha in del(5q) MDS. Nature 2015, 523, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Kronke, J.; Udeshi, N.D.; Narla, A.; Grauman, P.; Hurst, S.N.; McConkey, M.; Svinkina, T.; Heckl, D.; Comer, E.; Li, X.; et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 2014, 343, 301–305. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Ponthier, C.M.; Sack, R.; Seebacher, J.; Stadler, M.B.; Donovan, K.A.; Fischer, E.S. pSILAC mass spectrometry reveals ZFP91 as IMiD-dependent substrate of the CRL4(CRBN) ubiquitin ligase. Nat. Commun. 2017, 8, 15398. [Google Scholar] [CrossRef] [PubMed]
- Uehara, T.; Minoshima, Y.; Sagane, K.; Sugi, N.H.; Mitsuhashi, K.O.; Yamamoto, N.; Kamiyama, H.; Takahashi, K.; Kotake, Y.; Uesugi, M.; et al. Selective degradation of splicing factor CAPERalpha by anticancer sulfonamides. Nat. Chem. Biol. 2017, 13, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Han, T.; Goralski, M.; Gaskill, N.; Capota, E.; Kim, J.; Ting, T.C.; Xie, Y.; Williams, N.S.; Nijhawan, D. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science 2017, 356, 6336. [Google Scholar] [CrossRef]
- Das, A.; Dasgupta, S.; Gong, Y.; Shah, U.A.; Fradley, M.G.; Cheng, R.K.; Roy, B.; Guha, A. Cardiotoxicity as an adverse effect of immunomodulatory drugs and proteasome inhibitors in multiple myeloma: A network meta-analysis of randomized clinical trials. Hematol. Oncol. 2021, 1–10. [Google Scholar] [CrossRef]
- Zhao, Y.; Xiong, X.; Sun, Y. Cullin-RING Ligase 5: Functional characterization and its role in human cancers. Semin. Cancer Biol. 2020, 67, 61–79. [Google Scholar] [CrossRef]
- Kamura, T.; Maenaka, K.; Kotoshiba, S.; Matsumoto, M.; Kohda, D.; Conaway, R.C.; Conaway, J.W.; Nakayama, K.I. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 2004, 18, 3055–3065. [Google Scholar] [CrossRef]
- Karlsson, M.; Zhang, C.; Mear, L.; Zhong, W.; Digre, A.; Katona, B.; Sjostedt, E.; Butler, L.; Odeberg, J.; Dusart, P.; et al. A single-cell type transcriptomics map of human tissues. Sci. Adv. 2021, 7, eabh2169. [Google Scholar] [CrossRef]
- Metais, A.; Lamsoul, I.; Melet, A.; Uttenweiler-Joseph, S.; Poincloux, R.; Stefanovic, S.; Valiere, A.; de Peredo, A.G.; Stella, A.; Burlet-Schiltz, O.; et al. Asb2 alpha-Filamin A Axis Is Essential for Actin Cytoskeleton Remodeling during Heart Development. Circ. Res. 2018, 122, e34–e48. [Google Scholar] [CrossRef]
- Heuze, M.L.; Lamsoul, I.; Baldassarre, M.; Lad, Y.; Leveque, S.; Razinia, Z.; Moog-Lutz, C.; Calderwood, D.A.; Lutz, P.G. ASB2 targets filamins A and B to proteasomal degradation. Blood 2008, 112, 5130–5140. [Google Scholar] [CrossRef] [PubMed]
- Min, K.D.; Asakura, M.; Shirai, M.; Yamazaki, S.; Ito, S.; Fu, H.Y.; Asanuma, H.; Asano, Y.; Minamino, T.; Takashima, S.; et al. ASB2 is a novel E3 ligase of SMAD9 required for cardiogenesis. Sci. Rep. 2021, 11, 23056. [Google Scholar] [CrossRef]
- Shi, L.; Du, D.; Peng, Y.; Liu, J.; Long, J. The functional analysis of Cullin 7 E3 ubiquitin ligases in cancer. Oncogenesis 2020, 9, 98. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Liu, Y.; Wu, L.; Ma, X.; Liu, Q.; Huang, F.; Zhang, X.; Zhang, Y.; Zhang, J.; Luo, H.; et al. CUL7 E3 Ubiquitin Ligase Mediates the Degradation of Activation-Induced Cytidine Deaminase and Regulates the Ig Class Switch Recombination in B Lymphocytes. J. Immunol. 2019, 203, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Tsutsumi, T.; Kuwabara, H.; Arai, T.; Xiao, Y.; Decaprio, J.A. Disruption of the Fbxw8 gene results in pre- and postnatal growth retardation in mice. Mol. Cell. Biol. 2008, 28, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Dias, D.C.; Dolios, G.; Wang, R.; Pan, Z.Q. CUL7: A DOC domain-containing cullin selectively binds Skp1.Fbx29 to form an SCF-like complex. Proc. Natl. Acad. Sci. USA 2002, 99, 16601–16606. [Google Scholar] [CrossRef]
- Scheufele, F.; Wolf, B.; Kruse, M.; Hartmann, T.; Lempart, J.; Muhlich, S.; Pfeiffer, A.F.H.; Field, L.J.; Charron, M.J.; Pan, Z.Q.; et al. Evidence for a regulatory role of Cullin-RING E3 ubiquitin ligase 7 in insulin signaling. Cell. Signal. 2014, 26, 233–239. [Google Scholar] [CrossRef]
- Xu, X.; Keshwani, M.; Meyer, K.; Sarikas, A.; Taylor, S.; Pan, Z.Q. Identification of the degradation determinants of insulin receptor substrate 1 for signaling cullin-RING E3 ubiquitin ligase 7-mediated ubiquitination. J. Biol. Chem. 2012, 287, 40758–40766. [Google Scholar] [CrossRef]
- Xu, X.; Sarikas, A.; Dias-Santagata, D.C.; Dolios, G.; Lafontant, P.J.; Tsai, S.C.; Zhu, W.; Nakajima, H.; Nakajima, H.O.; Field, L.J.; et al. The CUL7 E3 ubiquitin ligase targets insulin receptor substrate 1 for ubiquitin-dependent degradation. Mol. Cell 2008, 30, 403–414. [Google Scholar] [CrossRef]
- Zou, J.; Ma, W.; Li, J.; Littlejohn, R.; Zhou, H.; Kim, I.M.; Fulton, D.J.R.; Chen, W.; Weintraub, N.L.; Zhou, J.; et al. Neddylation mediates ventricular chamber maturation through repression of Hippo signaling. Proc. Natl. Acad. Sci. USA 2018, 115, E4101–E4110. [Google Scholar] [CrossRef]
- Anger, M.; Scheufele, F.; Ramanujam, D.; Meyer, K.; Nakajima, H.; Field, L.J.; Engelhardt, S.; Sarikas, A. Genetic ablation of Cullin-RING E3 ubiquitin ligase 7 restrains pressure overload-induced myocardial fibrosis. PLoS ONE 2020, 15, e0244096. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Pei, X.H.; Yan, J.; Yan, F.; Cappell, K.M.; Whitehurst, A.W.; Xiong, Y. CUL9 mediates the functions of the 3M complex and ubiquitylates survivin to maintain genome integrity. Mol. Cell 2014, 54, 805–819. [Google Scholar] [CrossRef] [PubMed]
- Ortolano, N.A.; Romero-Morales, A.I.; Rasmussen, M.L.; Bodnya, C.; Kline, L.A.; Joshi, P.; Connelly, J.P.; Rose, K.L.; Pruett-Miller, S.M.; Gama, V. A proteomics approach for the identification of cullin-9 (CUL9) related signaling pathways in induced pluripotent stem cell models. PLoS ONE 2021, 16, e0248000. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xiong, Y. Cytoplasmic E3 ubiquitin ligase CUL9 controls cell proliferation, senescence, apoptosis and genome integrity through p53. Oncogene 2017, 36, 5212–5218. [Google Scholar] [CrossRef]
- Andreassen, O.A.; Djurovic, S.; Thompson, W.K.; Schork, A.J.; Kendler, K.S.; O’Donovan, M.C.; Rujescu, D.; Werge, T.; van de Bunt, M.; Morris, A.P.; et al. Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. Am. J. Hum. Genet. 2013, 92, 197–209. [Google Scholar] [CrossRef]
- Osaka, F.; Kawasaki, H.; Aida, N.; Saeki, M.; Chiba, T.; Kawashima, S.; Tanaka, K.; Kato, S. A new NEDD8-ligating system for cullin-4A. Genes Dev. 1998, 12, 2263–2268. [Google Scholar] [CrossRef]
- Whitby, F.G.; Xia, G.; Pickart, C.M.; Hill, C.P. Crystal Structure of the Human Ubiquitin-like Protein NEDD8 and Interactions with Ubiquitin Pathway Enzymes. J. Biol. Chem. 1998, 273, 34983–34991. [Google Scholar] [CrossRef]
- Morimoto, M.; Nishida, T.; Honda, R.; Yasuda, H. Modification of cullin-1 by ubiquitin-like protein Nedd8 enhances the activity of SCF(skp2) toward p27(kip1). Biochem. Biophys. Res. Commun. 2000, 270, 1093–1096. [Google Scholar] [CrossRef]
- Kawakami, T.; Chiba, T.; Suzuki, T.; Iwai, K.; Yamanaka, K.; Minato, N.; Suzuki, H.; Shimbara, N.; Hidaka, Y.; Osaka, F.; et al. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 2001, 20, 4003–4012. [Google Scholar] [CrossRef]
- Read, M.A.; Brownell, J.E.; Gladysheva, T.B.; Hottelet, M.; Parent, L.A.; Coggins, M.B.; Pierce, J.W.; Podust, V.N.; Luo, R.S.; Chau, V.; et al. Nedd8 Modification of Cul-1 Activates SCFbeta TrCP-Dependent Ubiquitination of Ikappa Balpha. Mol. Cell. Biol. 2000, 20, 2326–2333. [Google Scholar] [CrossRef]
- Wang, K.; Reichermeier, K.M.; Liu, X. Quantitative analyses for effects of neddylation on CRL2(VHL) substrate ubiquitination and degradation. Protein Sci. 2021, 30, 2338–2345. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Deshaies, R.J. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell 2008, 32, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Duda, D.M.; Borg, L.A.; Scott, D.C.; Hunt, H.W.; Hammel, M.; Schulman, B.A. Structural insights into NEDD8 activation of cullin-RING ligases: Conformational control of conjugation. Cell 2008, 134, 995–1006. [Google Scholar] [CrossRef]
- Baek, K.; Krist, D.T.; Prabu, J.R.; Hill, S.; Klugel, M.; Neumaier, L.M.; von Gronau, S.; Kleiger, G.; Schulman, B.A. NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly. Nature 2020, 578, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Deshaies, R.J.; Pierce, N.W. Transfer of ubiquitin protein caught in the act. Nature 2020, 578, 372–373. [Google Scholar] [CrossRef]
- Kostrhon, S.; Prabu, J.R.; Baek, K.; Horn-Ghetko, D.; von Gronau, S.; Klugel, M.; Basquin, J.; Alpi, A.F.; Schulman, B.A. CUL5-ARIH2 E3-E3 ubiquitin ligase structure reveals cullin-specific NEDD8 activation. Nat. Chem. Biol. 2021, 17, 1075–1083. [Google Scholar] [CrossRef]
- Horn-Ghetko, D.; Krist, D.T.; Prabu, J.R.; Baek, K.; Mulder, M.P.C.; Klugel, M.; Scott, D.C.; Ovaa, H.; Kleiger, G.; Schulman, B.A. Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly. Nature 2021, 590, 671–676. [Google Scholar] [CrossRef]
- Scott, D.C.; Rhee, D.Y.; Duda, D.M.; Kelsall, I.R.; Olszewski, J.L.; Paulo, J.A.; de Jong, A.; Ovaa, H.; Alpi, A.F.; Harper, J.W.; et al. Two Distinct Types of E3 Ligases Work in Unison to Regulate Substrate Ubiquitylation. Cell 2016, 166, 1198–1214.e24. [Google Scholar] [CrossRef]
- Soucy, T.A.; Smith, P.G.; Milhollen, M.A.; Berger, A.J.; Gavin, J.M.; Adhikari, S.; Brownell, J.E.; Burke, K.E.; Cardin, D.P.; Critchley, S.; et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 2009, 458, 732–736. [Google Scholar] [CrossRef]
- Zou, J.; Ma, W.; Littlejohn, R.; Li, J.; Stansfield, B.K.; Kim, I.M.; Liu, J.; Zhou, J.; Weintraub, N.L.; Su, H. Transient inhibition of neddylation at neonatal stage evokes reversible cardiomyopathy and predisposes the heart to isoproterenol-induced heart failure. Am. J. Physiol. Heart Circ. Physiol. 2019, 316, H1406–H1416. [Google Scholar] [CrossRef]
- Enchev, R.I.; Schulman, B.A.; Peter, M. Protein neddylation: Beyond cullin-RING ligases. Nat. Rev. Mol. Cell Biol. 2015, 16, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Lyapina, S.; Cope, G.; Shevchenko, A.; Serino, G.; Tsuge, T.; Zhou, C.; Wolf, D.A.; Wei, N.; Shevchenko, A.; Deshaies, R.J. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 2001, 292, 1382–1385. [Google Scholar] [CrossRef] [PubMed]
- Schwechheimer, C.; Serino, G.; Callis, J.; Crosby, W.L.; Lyapina, S.; Deshaies, R.J.; Gray, W.M.; Estelle, M.; Deng, X.W. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science 2001, 292, 1379–1382. [Google Scholar] [CrossRef] [PubMed]
- Cope, G.A.; Suh, G.S.; Aravind, L.; Schwarz, S.E.; Zipursky, S.L.; Koonin, E.V.; Deshaies, R.J. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 2002, 298, 608–611. [Google Scholar] [CrossRef]
- Pintard, L.; Kurz, T.; Glaser, S.; Willis, J.H.; Peter, M.; Bowerman, B. Neddylation and Deneddylation of CUL-3 Is Required to Target MEI-1/Katanin for Degradation at the Meiosis-to-Mitosis Transition in C. elegans. Curr. Biol. 2003, 13, 911–921. [Google Scholar] [CrossRef]
- Zhou, C.; Wee, S.; Rhee, E.; Naumann, M.; Dubiel, W.; Wolf, D.A. Fission Yeast COP9/Signalosome Suppresses Cullin Activity through Recruitment of the Deubiquitylating Enzyme Ubp12p. Mol. Cell 2003, 11, 927–938. [Google Scholar] [CrossRef]
- Wee, S.; Geyer, R.K.; Toda, T.; Wolf, D.A. CSN facilitates Cullin-RING ubiquitin ligase function by counteracting autocatalytic adapter instability. Nat. Cell Biol. 2005, 7, 387–391. [Google Scholar] [CrossRef]
- He, Q.; Cheng, P.; He, Q.; Liu, Y. The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex. Genes Dev. 2005, 19, 1518–1531. [Google Scholar] [CrossRef]
- Pierce, N.W.; Lee, J.E.; Liu, X.; Sweredoski, M.J.; Graham, R.L.; Larimore, E.A.; Rome, M.; Zheng, N.; Clurman, B.E.; Hess, S.; et al. Cand1 Promotes Assembly of New SCF Complexes through Dynamic Exchange of F Box Proteins. Cell 2013, 153, 206–215. [Google Scholar] [CrossRef]
- Reitsma, J.M.; Liu, X.; Reichermeier, K.M.; Moradian, A.; Sweredoski, M.J.; Hess, S.; Deshaies, R.J. Composition and Regulation of the Cellular Repertoire of SCF Ubiquitin Ligases. Cell 2017, 171, 1326–1339.e14. [Google Scholar] [CrossRef]
- Liu, X.; Reitsma, J.M.; Mamrosh, J.L.; Zhang, Y.; Straube, R.; Deshaies, R.J. Cand1-Mediated Adaptive Exchange Mechanism Enables Variation in F-Box Protein Expression. Mol. Cell 2018, 69, 773–786.e6. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Deshaies, R.J.; Liu, X. Assembly and Regulation of CRL Ubiquitin Ligases. Adv. Exp. Med. Biol. 2020, 1217, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Milic, J.; Tian, Y.; Bernhagen, J. Role of the COP9 Signalosome (CSN) in Cardiovascular Diseases. Biomolecules 2019, 9, 217. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Martin, D.S. The COP9 signalosome and cullin-RING ligases in the heart. Am. J. Cardiovasc. Dis. 2015, 5, 1–18. [Google Scholar] [PubMed]
- Lykke-Andersen, K.; Schaefer, L.; Menon, S.; Deng, X.W.; Miller, J.B.; Wei, N. Disruption of the COP9 signalosome Csn2 subunit in mice causes deficient cell proliferation, accumulation of p53 and cyclin E, and early embryonic death. Mol. Cell. Biol. 2003, 23, 6790–6797. [Google Scholar] [CrossRef]
- Yan, J.; Walz, K.; Nakamura, H.; Carattini-Rivera, S.; Zhao, Q.; Vogel, H.; Wei, N.; Justice, M.J.; Bradley, A.; Lupski, J.R. COP9 signalosome subunit 3 is essential for maintenance of cell proliferation in the mouse embryonic epiblast. Mol. Cell. Biol. 2003, 23, 6798–6808. [Google Scholar] [CrossRef]
- Menon, S.; Chi, H.; Zhang, H.; Deng, X.W.; Flavell, R.A.; Wei, N. COP9 signalosome subunit 8 is essential for peripheral T cell homeostasis and antigen receptor-induced entry into the cell cycle from quiescence. Nat. Immunol. 2007, 8, 1236–1245. [Google Scholar] [CrossRef]
- Zhao, R.; Yeung, S.C.; Chen, J.; Iwakuma, T.; Su, C.H.; Chen, B.; Qu, C.; Zhang, F.; Chen, Y.T.; Lin, Y.L.; et al. Subunit 6 of the COP9 signalosome promotes tumorigenesis in mice through stabilization of MDM2 and is upregulated in human cancers. J. Clin. Investig. 2011, 121, 851–865. [Google Scholar] [CrossRef]
- Lei, D.; Li, F.; Su, H.; Liu, J.; Wei, N.; Wang, X. Hepatic deficiency of COP9 signalosome subunit 8 induces ubiquitin-proteasome system impairment and Bim-mediated apoptosis in murine livers. PLoS ONE 2013, 8, e67793. [Google Scholar] [CrossRef]
- Su, H.; Li, J.; Menon, S.; Liu, J.; Kumarapeli, A.R.; Wei, N.; Wang, X. Perturbation of cullin deneddylation via conditional Csn8 ablation impairs the ubiquitin-proteasome system and causes cardiomyocyte necrosis and dilated cardiomyopathy in mice. Circ. Res. 2011, 108, 40–50. [Google Scholar] [CrossRef]
- Su, H.; Li, F.; Ranek, M.J.; Wei, N.; Wang, X. COP9 signalosome regulates autophagosome maturation. Circulation 2011, 124, 2117–2128. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Li, J.; Zhang, H.; Ma, W.; Wei, N.; Liu, J.; Wang, X. COP9 signalosome controls the degradation of cytosolic misfolded proteins and protects against cardiac proteotoxicity. Circ. Res. 2015, 117, 956–966. [Google Scholar] [CrossRef] [PubMed]
- Su, H.B.; Li, J.; Osinska, H.; Li, F.Q.; Robbins, J.; Liu, J.B.; Wei, N.; Wang, X.J. The COP9 Signalosome Is Required for Autophagy, Proteasome-Mediated Proteolysis, and Cardiomyocyte Survival in Adult Mice. Circ. Heart Fail. 2013, 6, 1049–1057. [Google Scholar] [CrossRef] [PubMed]
- Asare, Y.; Ommer, M.; Azombo, F.A.; Alampour-Rajabi, S.; Sternkopf, M.; Sanati, M.; Gijbels, M.J.; Schmitz, C.; Sinitski, D.; Tilstam, P.V.; et al. Inhibition of atherogenesis by the COP9 signalosome subunit 5 in vivo. Proc. Natl. Acad. Sci. USA 2017, 114, E2766–E2775. [Google Scholar] [CrossRef] [PubMed]
- Asare, Y.; Shagdarsuren, E.; Schmid, J.A.; Tilstam, P.V.; Grommes, J.; El Bounkari, O.; Schutz, A.K.; Weber, C.; de Winther, M.P.; Noels, H.; et al. Endothelial CSN5 impairs NF-kappaB activation and monocyte adhesion to endothelial cells and is highly expressed in human atherosclerotic lesions. Thromb. Haemost. 2013, 110, 141–152. [Google Scholar] [CrossRef]
- Dubiel, W.; Chaithongyot, S.; Dubiel, D.; Naumann, M. The COP9 Signalosome: A Multi-DUB Complex. Biomolecules 2020, 10, 1082. [Google Scholar] [CrossRef]
- Cavadini, S.; Fischer, E.S.; Bunker, R.D.; Potenza, A.; Lingaraju, G.M.; Goldie, K.N.; Mohamed, W.I.; Faty, M.; Petzold, G.; Beckwith, R.E.; et al. Cullin-RING ubiquitin E3 ligase regulation by the COP9 signalosome. Nature 2016, 531, 598–603. [Google Scholar] [CrossRef]
- Emberley, E.D.; Mosadeghi, R.; Deshaies, R.J. Deconjugation of Nedd8 from Cul1 is directly regulated by Skp1-F-box and substrate, and the COP9 signalosome inhibits deneddylated SCF by a noncatalytic mechanism. J. Biol. Chem. 2012, 287, 29679–29689. [Google Scholar] [CrossRef]
- Reitsma, J.M.; Liu, X.; Reichermeier, K.M.; Moradian, A.; Sweredoski, M.J.; Hess, S.; Deshaies, R.J. Nedd8 conjugation and Cand1-mediated exchange sustain a non-equilibrium cellular landscape of SCF E3 ubiquitin ligases. Cell 2017. accepted in principle. [Google Scholar]
- Lo, S.C.; Hannink, M. CAND1-mediated substrate adaptor recycling is required for efficient repression of Nrf2 by Keap1. Mol. Cell. Biol. 2006, 26, 1235–1244. [Google Scholar] [CrossRef]
- Reichermeier, K.M.; Straube, R.; Reitsma, J.M.; Sweredoski, M.J.; Rose, C.M.; Moradian, A.; den Besten, W.; Hinkle, T.; Verschueren, E.; Petzold, G.; et al. PIKES Analysis Reveals Response to Degraders and Key Regulatory Mechanisms of the CRL4 Network. Mol. Cell 2020, 77, 1092–1106.e9. [Google Scholar] [CrossRef]
- Shiraishi, S.; Zhou, C.; Aoki, T.; Sato, N.; Chiba, T.; Tanaka, K.; Yoshida, S.; Nabeshima, Y.; Nabeshima, Y.; Tamura, T.A. TBP-interacting protein 120B (TIP120B)/cullin-associated and neddylation-dissociated 2 (CAND2) inhibits SCF-dependent ubiquitination of myogenin and accelerates myogenic differentiation. J. Biol. Chem. 2007, 282, 9017–9028. [Google Scholar] [CrossRef] [PubMed]
- Sinner, M.F.; Tucker, N.R.; Lunetta, K.L.; Ozaki, K.; Smith, J.G.; Trompet, S.; Bis, J.C.; Lin, H.; Chung, M.K.; Nielsen, J.B.; et al. Integrating genetic, transcriptional, and functional analyses to identify 5 novel genes for atrial fibrillation. Circulation 2014, 130, 1225–1235. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Song, J.; Xu, M.; Lv, L.; Liu, C.; Shen, J.; Huang, Y. NEURL rs6584555 and CAND2 rs4642101 contribute to postoperative atrial fibrillation: A prospective study among Chinese population. Oncotarget 2016, 7, 42617–42624. [Google Scholar] [CrossRef] [PubMed]
- Gregers, E.; Ahlberg, G.; Christensen, T.; Jabbari, J.; Larsen, K.O.; Herfelt, C.B.; Henningsen, K.M.; Andreasen, L.; Thiis, J.J.; Lund, J.; et al. Deep sequencing of atrial fibrillation patients with mitral valve regurgitation shows no evidence of mosaicism but reveals novel rare germline variants. Heart Rhythm 2017, 14, 1531–1538. [Google Scholar] [CrossRef] [PubMed]
- Koskimaki, J.; Zhang, D.; Li, Y.; Saadat, L.; Moore, T.; Lightle, R.; Polster, S.P.; Carrion-Penagos, J.; Lyne, S.B.; Zeineddine, H.A.; et al. Transcriptome clarifies mechanisms of lesion genesis versus progression in models of Ccm3 cerebral cavernous malformations. Acta Neuropathol. Commun. 2019, 7, 132. [Google Scholar] [CrossRef]
- Weng, L.C.; Hall, A.W.; Choi, S.H.; Jurgens, S.J.; Haessler, J.; Bihlmeyer, N.A.; Grarup, N.; Lin, H.; Teumer, A.; Li-Gao, R.; et al. Genetic Determinants of Electrocardiographic P-Wave Duration and Relation to Atrial Fibrillation. Circ. Genom. Precis. Med. 2020, 13, 387–395. [Google Scholar] [CrossRef]
- Moksnes, M.R.; Rosjo, H.; Richmond, A.; Lyngbakken, M.N.; Graham, S.E.; Hansen, A.F.; Wolford, B.N.; Gagliano Taliun, S.A.; LeFaive, J.; Rasheed, H.; et al. Genome-wide association study of cardiac troponin I in the general population. Hum. Mol. Genet. 2021, 30, 2027–2039. [Google Scholar] [CrossRef]
- Christophersen, I.E.; Rienstra, M.; Roselli, C.; Yin, X.; Geelhoed, B.; Barnard, J.; Lin, H.; Arking, D.E.; Smith, A.V.; Albert, C.M.; et al. Large-scale analyses of common and rare variants identify 12 new loci associated with atrial fibrillation. Nat. Genet. 2017, 49, 946–952. [Google Scholar] [CrossRef]
- Gorska, A.A.; Sandmann, C.; Riechert, E.; Hofmann, C.; Malovrh, E.; Varma, E.; Kmietczyk, V.; Olschlager, J.; Jurgensen, L.; Kamuf-Schenk, V.; et al. Muscle-specific Cand2 is translationally upregulated by mTORC1 and promotes adverse cardiac remodeling. EMBO Rep. 2021, 22, e52170. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).