miR-27b-3p Attenuates Muscle Atrophy by Targeting Cbl-b in Skeletal Muscles
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Ethics Statement
2.2. Cell Culture and Treatment
2.3. miRNA and mRNA Screen
2.4. Cell Transfection
2.5. Dual-Luciferase Reporter Assay
2.6. Experimental Animals
2.7. RNA Extraction, cDNA Synthesis, and Quantitative Real-Time PCR (qRT-PCR)
2.8. Western Blotting Assay
2.9. Hematoxylin and Eosin Staining (HE)
2.10. Statistical Analysis
3. Results
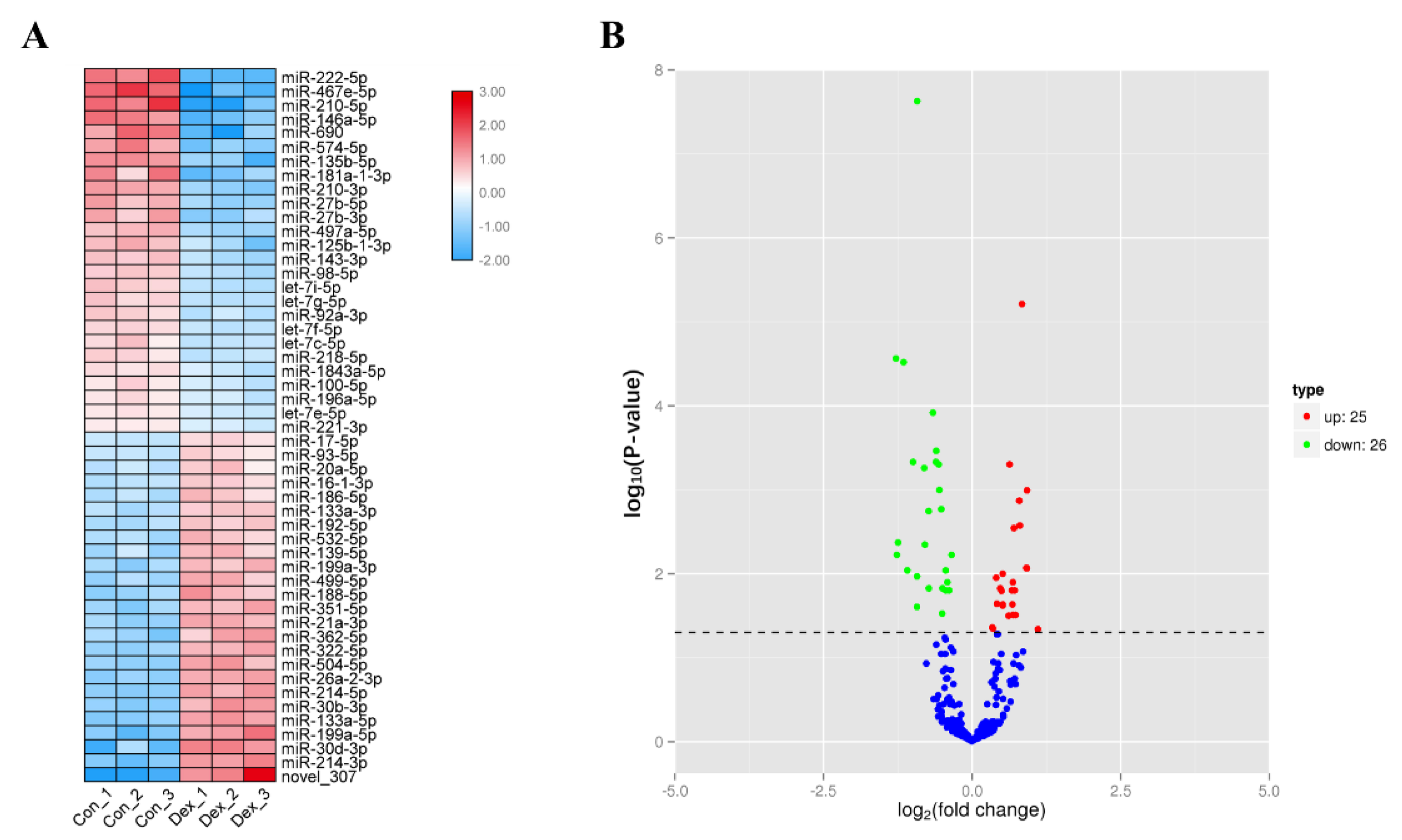
3.1. Screening of Differentially Expressed miRNAs in Atrophic Myotubes
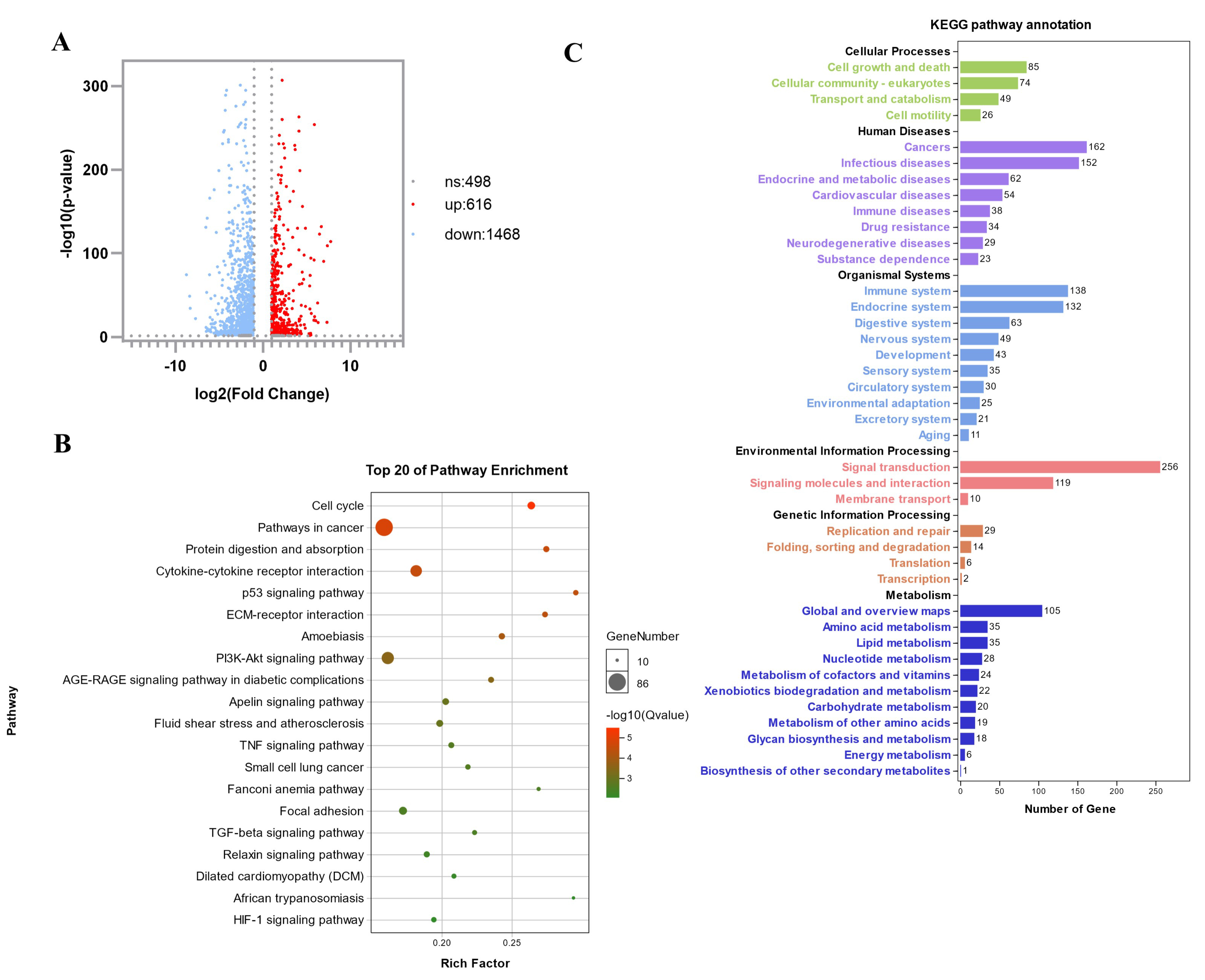
3.2. Transcriptome Sequencing Analysis to Identify and Screen Differentially Expressed Genes in Atrophic Myotubes
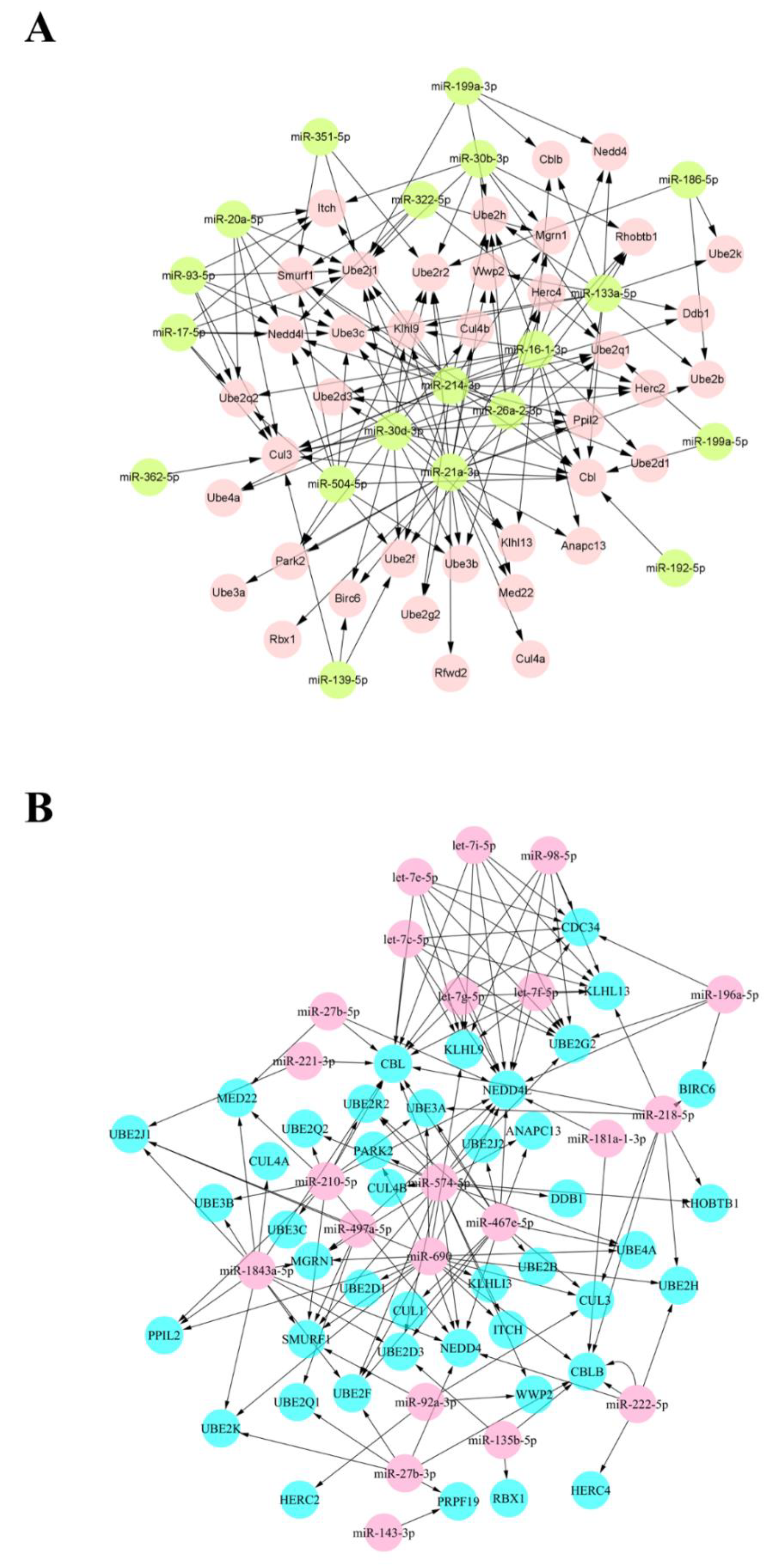
3.3. Joint Analysis and Interactive Network of Differentially Expressed miRNAs and Genes
3.4. miR-27b-3p Can Affect Muscle Atrophy
3.5. Cbl-b Is the Target Gene of miR-27b-3p
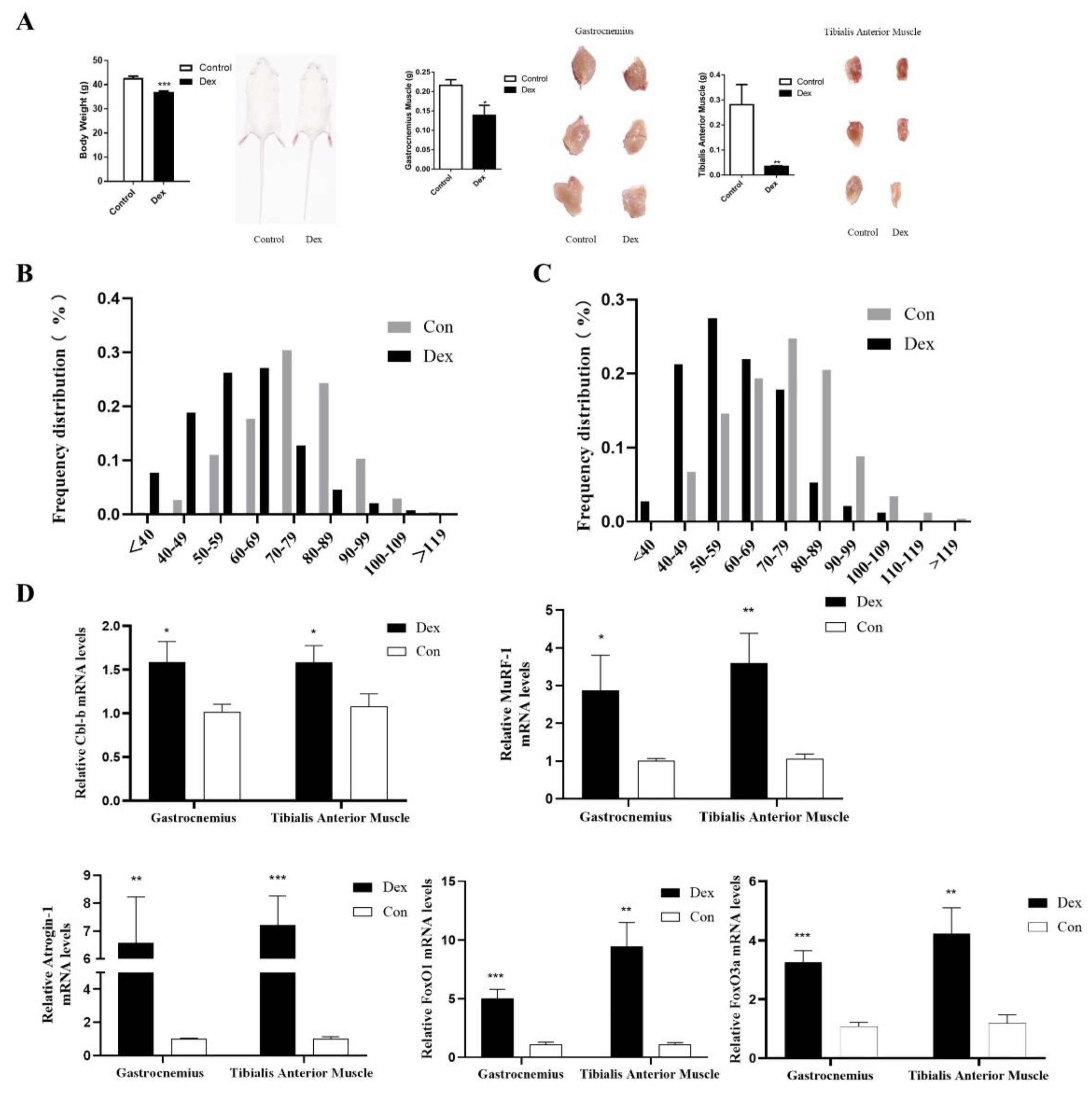
3.6. UpRegulation of Cbl-b Expression in Mice with Muscle Atrophy
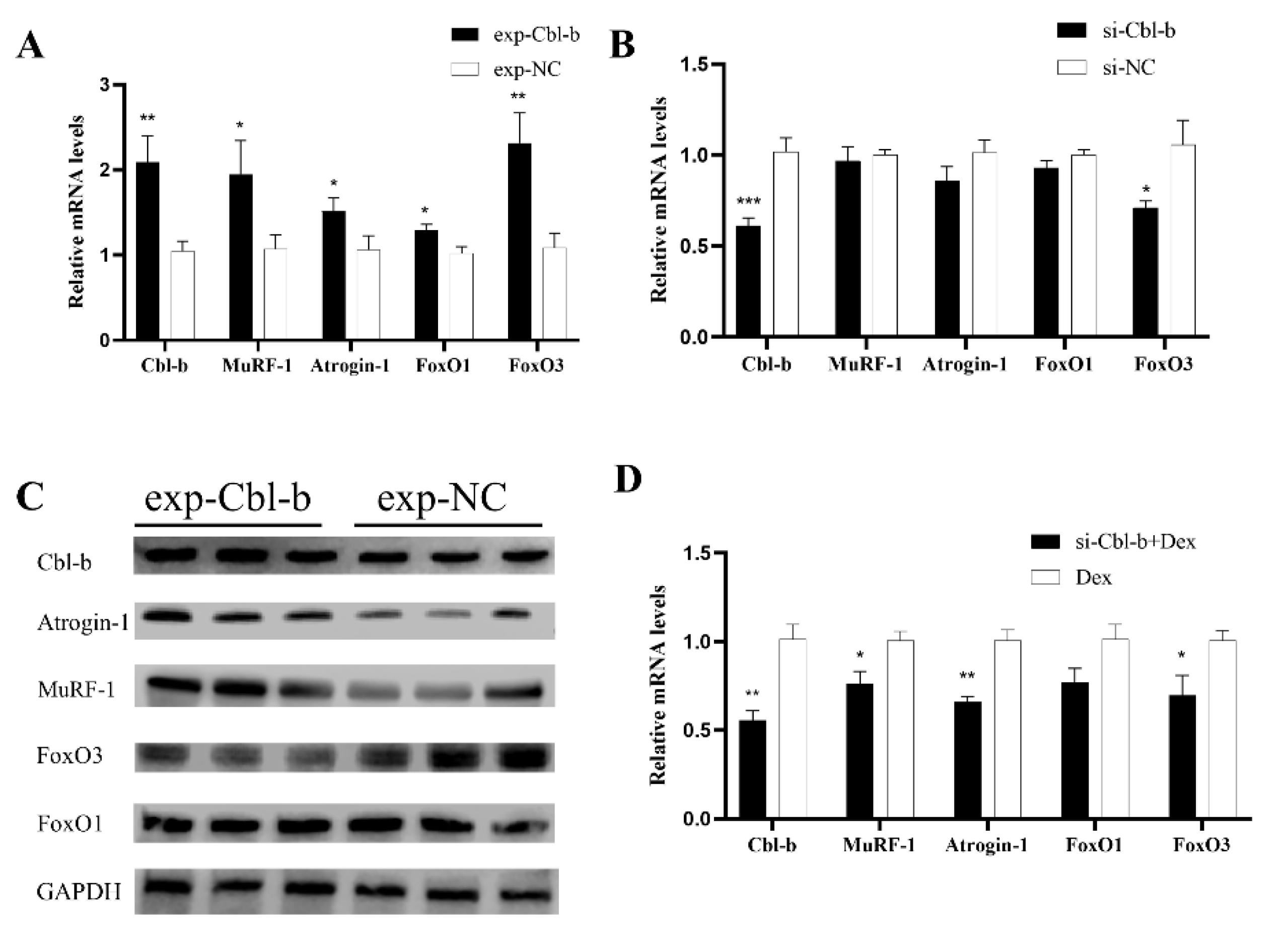
3.7. Cbl-b Gene Can Promote Muscle Atrophy
3.8. Inhibition of Cbl-b Gene Expression Can Alleviate Muscle Atrophy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Deane, C.S.; Willis, C.R.G.; Phillips, B.E.; Atherton, P.J.; Harries, L.W.; Ames, R.M.; Szewczyk, N.J.; Etheridge, T. Transcriptomic meta-analysis of disuse muscle atrophy vs. resistance exercise-induced hypertrophy in young and older humans. J. Cachexia Sarcopenia Muscle 2021, 12, 629–645. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liang, J.; Chen, N. Do not neglect the role of circadian rhythm in muscle atrophy. Ageing Res. Rev. 2020, 63, 101155. [Google Scholar] [CrossRef] [PubMed]
- Hiensch, A.E.; Bolam, K.A.; Mijwel, S.; Jeneson, J.A.L.; Huitema, A.D.R.; Kranenburg, O.; van der Wall, E.; Rundqvist, H.; Wengstrom, Y.; May, A.M. Doxorubicin-induced skeletal muscle atrophy: Elucidating the underlying molecular pathways. Acta Physiol. 2020, 229, e13400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wijaya, Y.T.; Setiawan, T.; Sari, I.N.; Nah, S.Y.; Kwon, H.Y. Amelioration of muscle wasting by gintonin in cancer cachexia. Neoplasia 2021, 23, 1307–1317. [Google Scholar] [CrossRef]
- Melstrom, L.G.; Melstrom, K.A., Jr.; Ding, X.Z.; Adrian, T.E. Mechanisms of skeletal muscle degradation and its therapy in cancer cachexia. Histol. Histopathol. 2007, 22, 805–814. [Google Scholar]
- Dutt, V.; Gupta, S.; Dabur, R.; Injeti, E.; Mittal, A. Skeletal muscle atrophy: Potential therapeutic agents and their mechanisms of action. Pharmacol. Res. 2015, 99, 86–100. [Google Scholar] [CrossRef]
- Cohen, S.; Nathan, J.A.; Goldberg, A.L. Muscle wasting in disease: Molecular mechanisms and promising therapies. Nat. Rev. Drug Discov. 2015, 14, 58–74. [Google Scholar] [CrossRef]
- Kumar, A.; Bhatnagar, S.; Paul, P.K. TWEAK and TRAF6 regulate skeletal muscle atrophy. Curr. Opin. Clin. Nutr. Metab. Care 2012, 15, 233–239. [Google Scholar] [CrossRef]
- Horak, M.; Novak, J.; Bienertova-Vasku, J. Muscle-specific microRNAs in skeletal muscle development. Dev. Biol. 2016, 410, 1–13. [Google Scholar] [CrossRef]
- Li, J.; Chan, M.C.; Yu, Y.; Bei, Y.; Chen, P.; Zhou, Q.; Cheng, L.; Chen, L.; Ziegler, O.; Rowe, G.C.; et al. miR-29b contributes to multiple types of muscle atrophy. Nat. Commun. 2017, 8, 15201. [Google Scholar] [CrossRef] [Green Version]
- Silva, W.J.; Graca, F.A.; Cruz, A.; Silvestre, J.G.; Labeit, S.; Miyabara, E.H.; Yan, C.Y.I.; Wang, D.Z.; Moriscot, A.S. miR-29c improves skeletal muscle mass and function throughout myocyte proliferation and differentiation and by repressing atrophy-related genes. Acta Physiol. 2019, 226, e13278. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Li, J.; Zhou, L.; Han, J.; Liu, R.; Zhang, H.; Ning, T.; Gao, Z.; Liu, B.; Chen, X.; et al. The c-Myc/miR-27b-3p/ATG10 regulatory axis regulates chemoresistance in colorectal cancer. Theranostics 2020, 10, 1981–1996. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhou, H.; Langdon, W.Y.; Zhang, J. E3 ubiquitin ligase Cbl-b in innate and adaptive immunity. Cell Cycle 2014, 13, 1875–1884. [Google Scholar] [PubMed]
- Lin, A.; Mak, T. The role of E3 ligases in autoimmunity and the regulation of autoreactive T cells. Curr. Opin. Immunol. 2007, 19, 665–673. [Google Scholar] [CrossRef]
- Moynagh, P. The roles of Pellino E3 ubiquitin ligases in immunity. Nat. Reviews. Immunol. 2014, 14, 122–131. [Google Scholar] [CrossRef]
- Nikawa, T.; Ishidoh, K. Ubiquitin ligase Cbl-b and inhibitory Cblin peptides. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2020, 1868, 140495. [Google Scholar] [CrossRef]
- Uchida, T.; Sakashita, Y.; Kitahata, K.; Yamashita, Y.; Tomida, C.; Kimori, Y.; Komatsu, A.; Hirasaka, K.; Ohno, A.; Nakao, R.; et al. Reactive oxygen species upregulate expression of muscle atrophy-associated ubiquitin ligase Cbl-b in rat L6 skeletal muscle cells. Am. J. Physiol. Cell Physiol. 2018, 314, C721–C731. [Google Scholar] [CrossRef]
- Peris-Moreno, D.; Cussonneau, L.; Combaret, L.; Polge, C.; Taillandier, D. Ubiquitin Ligases at the Heart of Skeletal Muscle Atrophy Control. Molecules 2021, 26, 407. [Google Scholar] [CrossRef]
- Nakao, R.; Hirasaka, K.; Goto, J.; Ishidoh, K.; Yamada, C.; Ohno, A.; Okumura, Y.; Nonaka, I.; Yasutomo, K.; Baldwin, K.M.; et al. Ubiquitin ligase Cbl-b is a negative regulator for insulin-like growth factor 1 signaling during muscle atrophy caused by unloading. Mol. Cell. Biol. 2009, 29, 4798–4811. [Google Scholar] [CrossRef] [Green Version]
- Hanwright, P.J.; Qiu, C.; Rath, J.; Zhou, Y.; von Guionneau, N.; Sarhane, K.A.; Harris, T.G.W.; Howard, G.P.; Malapati, H.; Lan, M.J.; et al. Sustained IGF-1 delivery ameliorates effects of chronic denervation and improves functional recovery after peripheral nerve injury and repair. Biomaterials 2021, 280, 121244. [Google Scholar] [CrossRef]
- Grunseich, C.; Miller, R.; Swan, T.; Glass, D.J.; El Mouelhi, M.; Fornaro, M.; Petricoul, O.; Vostiar, I.; Roubenoff, R.; Meriggioli, M.N.; et al. Safety, tolerability, and preliminary efficacy of an IGF-1 mimetic in patients with spinal and bulbar muscular atrophy: A randomised, placebo-controlled trial. Lancet Neurol. 2018, 17, 1043–1052. [Google Scholar] [CrossRef]
- Dalmay, T. Mechanism of miRNA-mediated repression of mRNA translation. Essays Biochem. 2013, 54, 29–38. [Google Scholar] [PubMed] [Green Version]
- Correia de Sousa, M.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs’ Action through miRNA Editing. Int. J. Mol. Sci. 2019, 20, 6249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, H.J.; Lee, K.P.; Milholland, B.; Shin, Y.J.; Kang, J.S.; Kwon, K.S.; Suh, Y. Comprehensive miRNA Profiling of Skeletal Muscle and Serum in Induced and Normal Mouse Muscle Atrophy During Aging. J. Gerontol. A Biol. Sci. Med. Sci. 2017, 72, 1483–1491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, R.; Shen, X.; Wang, F.; Wang, X.; DesJarlais, A.; Syed, A.; Saba, R.; Tan, Z.; Yu, F.; Ji, X.; et al. H19X-encoded miR-322(424)/miR-503 regulates muscle mass by targeting translation initiation factors. J. Cachexia Sarcopenia Muscle 2021, 12, 2174–2186. [Google Scholar] [CrossRef]
- Li, J.; Wang, L.; Hua, X.; Tang, H.; Chen, R.; Yang, T.; Das, S.; Xiao, J. CRISPR/Cas9-Mediated miR-29b Editing as a Treatment of Different Types of Muscle Atrophy in Mice. Mol. Ther. 2020, 28, 1359–1372. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Chen, L.; Liang, X.; Cao, Y.; Zhu, X.; Wang, S.; Li, J.; Gao, J.; Xiao, J. Exercise attenuates angiotensin-induced muscle atrophy by targeting PPARgamma/miR-29b. J. Sport Health Sci. 2021, 21, S2095. [Google Scholar]
- Li, J.; Yang, T.; Sha, Z.; Tang, H.; Hua, X.; Wang, L.; Wang, Z.; Gao, Z.; Sluijter, J.P.G.; Rowe, G.C.; et al. Angiotensin II-induced muscle atrophy via PPARgamma suppression is mediated by miR-29b. Mol. Ther. Nucleic Acids 2021, 23, 743–756. [Google Scholar] [CrossRef]
- Okamura, T.; Hashimoto, Y.; Osaka, T.; Senmaru, T.; Fukuda, T.; Hamaguchi, M.; Fukui, M. miR-23b-3p acts as a counter-response against skeletal muscle atrophy. J. Endocrinol. 2020, 244, 535–547. [Google Scholar] [CrossRef]
- Wang, B.; Wang, J.; He, W.; Zhao, Y.; Zhang, A.; Liu, Y.; Hassounah, F.; Ma, F.; Klein, J.D.; Wang, X.H.; et al. Exogenous miR-29a Attenuates Muscle Atrophy and Kidney Fibrosis in Unilateral Ureteral Obstruction Mice. Human Gene Ther. 2020, 31, 367–375. [Google Scholar] [CrossRef]
- Aoi, W.; Naito, Y.; Mizushima, K.; Takanami, Y.; Kawai, Y.; Ichikawa, H.; Yoshikawa, T. The microRNA miR-696 regulates PGC-1{alpha} in mouse skeletal muscle in response to physical activity. Am. J. Physiol.-Endocrinol. Metab. 2010, 298, E799–E806. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhang, C.; Zhang, A.; Cai, H.; Price, S.; Wang, X. MicroRNA-23a and MicroRNA-27a Mimic Exercise by Ameliorating CKD-Induced Muscle Atrophy. J. Am. Soc. Nephrol. JASN 2017, 28, 2631–2640. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lu, Y.; Zhu, L.; Zhang, H.; Feng, L. Inhibition of miR-27b Regulates Lipid Metabolism in Skeletal Muscle of Obese Rats During Hypoxic Exercise by Increasing PPARγ Expression. Front. Physiol. 2020, 11, 1090. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Qi, C.; Korenberg, J.R.; Chen, X.N.; Noya, D.; Rao, M.S.; Reddy, J.K. Structural organization of mouse peroxisome proliferator-activated receptor gamma (mPPAR gamma) gene: Alternative promoter use and different splicing yield two mPPAR gamma isoforms. Proc. Natl. Acad. Sci. USA 1995, 92, 7921–7925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manickam, R.; Duszka, K.; Wahli, W. PPARs and Microbiota in Skeletal Muscle Health and Wasting. Int. J. Mol. Sci. 2020, 21, 8056. [Google Scholar] [CrossRef]
- Gong, L.; Jin, H.; Li, Y.; Quan, Y.; Yang, J.; Tang, Q.; Zou, Z. Rosiglitazone ameliorates skeletal muscle insulin resistance by decreasing free fatty acids release from adipocytes. Biochem. Biophys. Res. Commun. 2020, 533, 1122–1128. [Google Scholar] [CrossRef]
- Das, S.K.; Eder, S.; Schauer, S.; Diwoky, C.; Temmel, H.; Guertl, B.; Gorkiewicz, G.; Tamilarasan, K.P.; Kumari, P.; Trauner, M.; et al. Adipose triglyceride lipase contributes to cancer-associated cachexia. Science 2011, 333, 233–238. [Google Scholar] [CrossRef] [Green Version]
- Kimura, K.; Morisasa, M.; Mizushige, T.; Karasawa, R.; Kanamaru, C.; Kabuyama, Y.; Hayasaka, T.; Mori, T.; Goto-Inoue, N. Lipid Dynamics due to Muscle Atrophy Induced by Immobilization. J. Oleo Sci. 2021, 70, 937–946. [Google Scholar] [CrossRef]
- Zhang, G.; He, M.; Wu, P.; Zhang, X.; Zhou, K.; Li, T.; Zhang, T.; Xie, K.; Dai, G.; Wang, J. MicroRNA-27b-3p Targets the Myostatin Gene to Regulate Myoblast Proliferation and Is Involved in Myoblast Differentiation. Cells 2021, 10, 423. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Tang, Y.; Liu, B.; Cong, W.; Liu, C.; Xiao, J. Retinoid acid-induced microRNA-27b-3p impairs C2C12 myoblast proliferation and differentiation by suppressing α-dystrobrevin. Exp. Cell Res. 2017, 350, 301–311. [Google Scholar] [CrossRef]
- Chen, D.; Si, W.; Shen, J.; Du, C.; Lou, W.; Bao, C.; Zheng, H.; Pan, J.; Zhong, G.; Xu, L.; et al. miR-27b-3p inhibits proliferation and potentially reverses multi-chemoresistance by targeting CBLB/GRB2 in breast cancer cells. Cell Death Dis. 2018, 9, 188. [Google Scholar] [CrossRef] [PubMed]
- Sandri, M. Signaling in muscle atrophy and hypertrophy. Physiology 2008, 23, 160–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latres, E.; Amini, A.R.; Amini, A.A.; Griffiths, J.; Martin, F.J.; Wei, Y.; Lin, H.C.; Yancopoulos, G.D.; Glass, D.J. Insulin-like growth factor-1 (IGF-1) inversely regulates atrophy-induced genes via the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR) pathway. J. Biol. Chem. 2005, 280, 2737–2744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stitt, T.; Drujan, D.; Clarke, B.; Panaro, F.; Timofeyva, Y.; Kline, W.; Gonzalez, M.; Yancopoulos, G.; Glass, D. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell 2004, 14, 395–403. [Google Scholar] [CrossRef]
- Zampieri, S.; Mosole, S.; Löfler, S.; Fruhmann, H.; Burggraf, S.; Cvečka, J.; Hamar, D.; Sedliak, M.; Tirptakova, V.; Šarabon, N.; et al. Physical Exercise in Aging: Nine Weeks of Leg Press or Electrical Stimulation Training in 70 Years Old Sedentary Elderly People. Eur. J. Transl. Myol. 2015, 25, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Carraro, U.; Kern, H.; Gava, P.; Hofer, C.; Loefler, S.; Gargiulo, P.; Edmunds, K.; Árnadóttir, Í.D.; Zampieri, S.; Ravara, B.; et al. Recovery from muscle weakness by exercise and FES: Lessons from Masters, active or sedentary seniors and SCI patients. Aging Clin. Exp. Res. 2017, 29, 579–590. [Google Scholar] [CrossRef]
- Mitchell, C.J.; D’Souza, R.F.; Mitchell, S.M.; Figueiredo, V.C.; Miller, B.F.; Hamilton, K.L.; Peelor, F.F., 3rd; Coronet, M.; Pileggi, C.A.; Durainayagam, B.; et al. Impact of dairy protein during limb immobilization and recovery on muscle size and protein synthesis; a randomized controlled trial. J. Appl. Physiol. 2018, 124, 717–728. [Google Scholar] [CrossRef]
- Arentson-Lantz, E.J.; Galvan, E.; Ellison, J.; Wacher, A.; Paddon-Jones, D. Improving Dietary Protein Quality Reduces the Negative Effects of Physical Inactivity on Body Composition and Muscle Function. J. Gerontol. A Biol. Sci. Med. Sci. 2019, 74, 1605–1611. [Google Scholar] [CrossRef]
- Howard, E.E.; Pasiakos, S.M.; Fussell, M.A.; Rodriguez, N.R. Skeletal Muscle Disuse Atrophy and the Rehabilitative Role of Protein in Recovery from Musculoskeletal Injury. Adv. Nutr. 2020, 11, 989–1001. [Google Scholar] [CrossRef]
- Ucci, S.; Renzini, A.; Russi, V.; Mangialardo, C.; Cammarata, I.; Cavioli, G.; Santaguida, M.G.; Virili, C.; Centanni, M.; Adamo, S.; et al. Thyroid Hormone Protects from Fasting-Induced Skeletal Muscle Atrophy by Promoting Metabolic Adaptation. Int. J. Mol. Sci. 2019, 20, 5754. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, X.; Li, Z.; Wang, Z.; Yu, J.; Ma, M.; Nie, Q. miR-27b-3p Attenuates Muscle Atrophy by Targeting Cbl-b in Skeletal Muscles. Biomolecules 2022, 12, 191. https://doi.org/10.3390/biom12020191
Yang X, Li Z, Wang Z, Yu J, Ma M, Nie Q. miR-27b-3p Attenuates Muscle Atrophy by Targeting Cbl-b in Skeletal Muscles. Biomolecules. 2022; 12(2):191. https://doi.org/10.3390/biom12020191
Chicago/Turabian StyleYang, Xin, Zhenhui Li, Zhijun Wang, Jiaao Yu, Manting Ma, and Qinghua Nie. 2022. "miR-27b-3p Attenuates Muscle Atrophy by Targeting Cbl-b in Skeletal Muscles" Biomolecules 12, no. 2: 191. https://doi.org/10.3390/biom12020191
APA StyleYang, X., Li, Z., Wang, Z., Yu, J., Ma, M., & Nie, Q. (2022). miR-27b-3p Attenuates Muscle Atrophy by Targeting Cbl-b in Skeletal Muscles. Biomolecules, 12(2), 191. https://doi.org/10.3390/biom12020191







