Comparative Transcriptomic Analysis of mRNAs, miRNAs and lncRNAs in the Longissimus dorsi Muscles between Fat-Type and Lean-Type Pigs
Abstract
1. Introduction
2. Materials and Methods
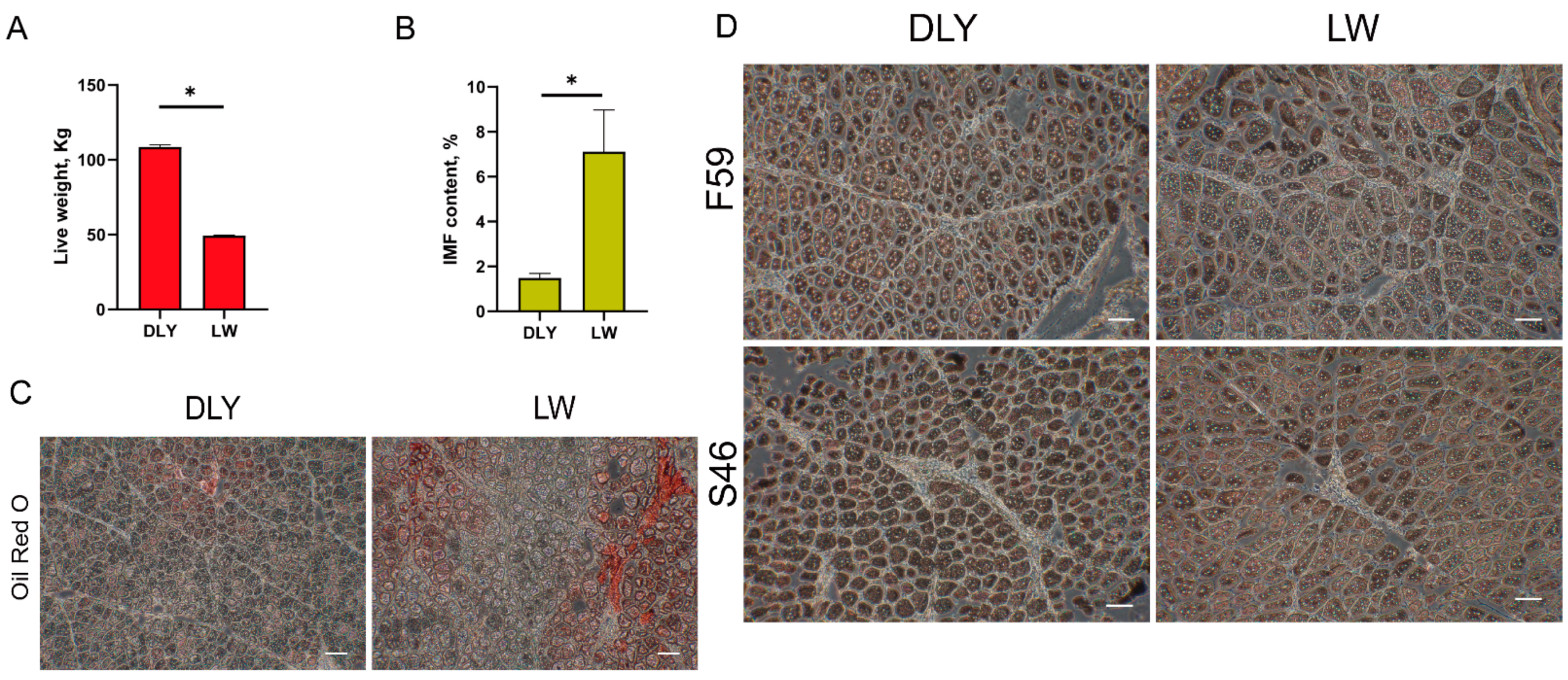
2.1. Animals and Tissue Samples
2.2. IMF Content Measurement
2.3. Total RNA Isolation, Library Preparation, and Sequencing
2.4. Bioinformatic Analysis of DE mRNA, miRNA and lncRNA and Their Interactions
2.5. Immunohistochemistry
2.6. Overexpression and Knockdown Assay
2.7. Cell Culture and Transfection
2.8. RT-qPCR
2.9. Statistical Analysis
3. Results
3.1. Overview of the Transcriptome Sequencing Data of Porcine LD Muscles
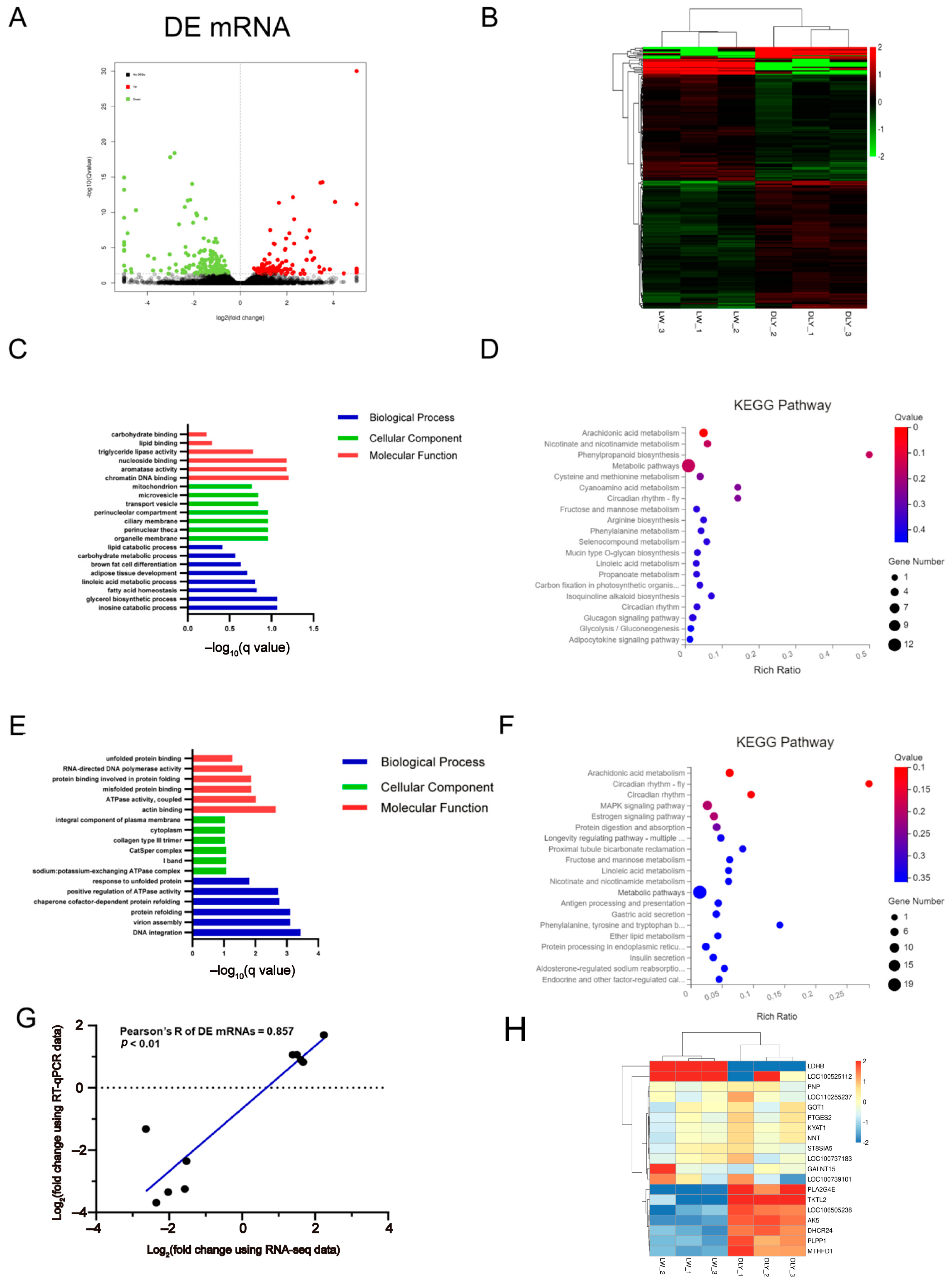
3.2. Differentially Expressed mRNAs in the LD Muscle between LW and DLY Pigs
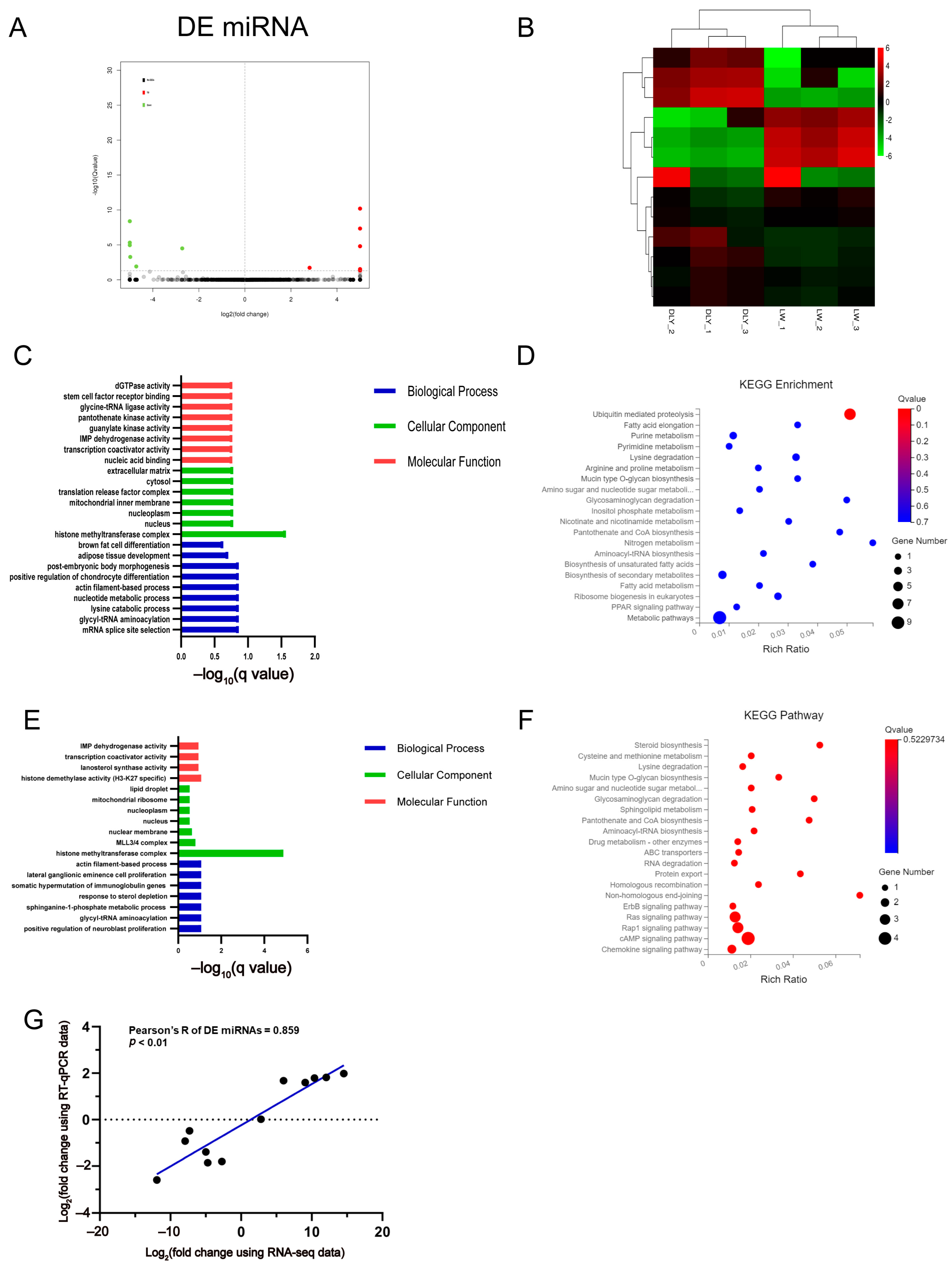
3.3. Differentially Expressed miRNAs in the LD Muscle between LW and DLY Pigs
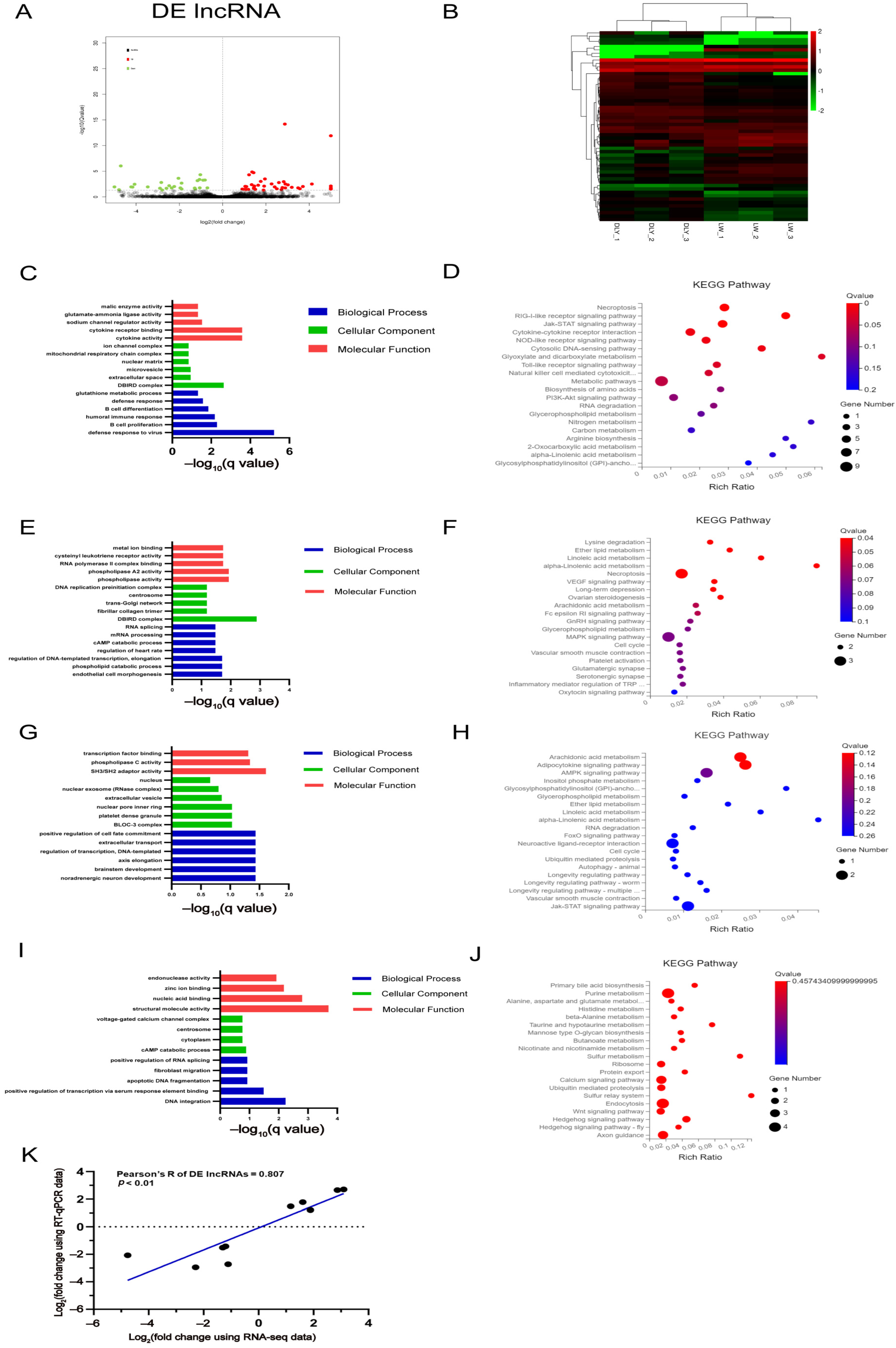
3.4. Differentially Expressed lncRNAs in the LD muscle between LW and DLY Pigs
3.5. Integrated Analysis on DE mRNAs, miRNAs, and lncRNAs in the LD Muscle between LW and DLY Pigs
3.6. The Role of GALNT15 in Lipid Deposition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chang, K.C.; Da Costa, N.; Blackley, R.; Southwood, O.; Evans, G.; Plastow, G.; Wood, J.D.; Richardson, R.I. Relationships of Myosin Heavy Chain Fibre Types to Meat Quality Traits in Traditional and Modern Pigs. Meat Sci. 2003, 64, 93–103. [Google Scholar] [CrossRef]
- Joo, S.T.; Kim, G.D.; Hwang, Y.H.; Ryu, Y.C. Control of Fresh Meat Quality through Manipulation of Muscle Fiber Characteristics. Meat Sci. 2013, 95, 828–836. [Google Scholar] [CrossRef]
- Hausman, G.J.; Basu, U.; Du, M.; Fernyhough-Culver, M.; Dodson, M.V. Intermuscular and Intramuscular Adipose Tissues: Bad vs. Good Adipose Tissues. Adipocyte 2014, 3, 242–255. [Google Scholar] [CrossRef]
- Schiaffino, S.; Reggiani, C. Fiber Types in Mammalian Skeletal Muscles. Physiol. Rev. 2011, 91, 1447–1531. [Google Scholar] [CrossRef]
- Qaisar, R.; Bhaskaran, S.; Van Remmen, H. Muscle Fiber Type Diversification during Exercise and Regeneration. Free Radic. Biol. Med. 2016, 98, 56–67. [Google Scholar] [CrossRef]
- Guo, J.; Shan, T.; Wu, T.; Zhu, L.N.; Ren, Y.; An, S.; Wang, Y. Comparisons of Different Muscle Metabolic Enzymes and Muscle Fiber Types in Jinhua and Landrace Pigs. J. Anim. Sci. 2011, 89, 185–191. [Google Scholar] [CrossRef]
- Criado-Mesas, L.; Ballester, M.; Crespo-Piazuelo, D.; Castelló, A.; Fernández, A.I.; Folch, J.M. Identification of EQTLs Associated with Lipid Metabolism in Longissimus Dorsi Muscle of Pigs with Different Genetic Backgrounds. Sci. Rep. 2020, 10, 9845. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, X.; Jing, X.; He, X.; Wang, L.; Liu, Y.; Liu, D. Transcriptomics Analysis on Excellent Meat Quality Traits of Skeletal Muscles of the Chinese Indigenous Min Pig Compared with the Large White Breed. Int. J. Mol. Sci. 2018, 19, 21. [Google Scholar] [CrossRef]
- Zhao, X.; Mo, D.; Li, A.; Gong, W.; Xiao, S.; Zhang, Y.; Qin, L.; Niu, Y.; Guo, Y.; Liu, X.; et al. Comparative Analyses by Sequencing of Transcriptomes during Skeletal Muscle Development between Pig Breeds Differing in Muscle Growth Rate and Fatness. PLoS ONE 2011, 6, e19774. [Google Scholar] [CrossRef]
- Damon, M.; Wyszynska-Koko, J.; Vincent, A.; Hérault, F.; Lebret, B. Comparison of Muscle Transcriptome between Pigs with Divergent Meat Quality Phenotypes Identifies Genes Related to Muscle Metabolism and Structure. PLoS ONE 2012, 7, e33763. [Google Scholar] [CrossRef]
- Xu, J.; Wang, C.; Jin, E.; Gu, Y.; Li, S.; Li, Q. Identification of Differentially Expressed Genes in Longissimus Dorsi Muscle between Wei and Yorkshire Pigs Using RNA Sequencing. Genes Genom. 2018, 40, 413–421. [Google Scholar] [CrossRef]
- Piórkowska, K.; Żukowski, K.; Ropka-Molik, K.; Tyra, M. Detection of Genetic Variants between Different Polish Landrace and Puławska Pigs by Means of RNA-Seq Analysis. Anim. Genet. 2018, 49, 215–225. [Google Scholar] [CrossRef]
- Li, A.; Mo, D.; Zhao, X.; Jiang, W.; Cong, P.; He, Z.; Xiao, S.; Liu, X.; Chen, Y. Comparison of the Longissimus Muscle Proteome between Obese and Lean Pigs at 180 Days. Mamm. Genome 2013, 24, 72–79. [Google Scholar] [CrossRef]
- Liu, J.; Damon, M.; Guitton, N.; Guisle, I.; Ecolan, P.; Vincent, A.; Cherel, P.; Gondret, F. Differentially-Expressed Genes in Pig Longissimus Muscles with Contrasting Levels of Fat, as Identified by Combined Transcriptomic, Reverse Transcription PCR, and Proteomic Analyses. J. Agric. Food Chem. 2009, 57, 3808–3817. [Google Scholar] [CrossRef]
- Ma, C.; Wang, W.; Wang, Y.; Sun, Y.; Kang, L.; Zhang, Q.; Jiang, Y. TMT-Labeled Quantitative Proteomic Analyses on the Longissimus Dorsi to Identify the Proteins Underlying Intramuscular Fat Content in Pigs. J. Proteom. 2020, 213, 103630. [Google Scholar] [CrossRef]
- Yang, Y.; Yan, J.; Fan, X.; Chen, J.; Wang, Z.; Liu, X.; Yi, G.; Liu, Y.; Niu, Y.; Zhang, L.; et al. The Genome Variation and Developmental Transcriptome Maps Reveal Genetic Differentiation of Skeletal Muscle in Pigs. PLoS Genet. 2021, 17, e1009910. [Google Scholar] [CrossRef]
- Chen, W.; Fang, G.F.; Wang, S.D.; Wang, H.; Zeng, Y. qing Longissimus Lumborum Muscle Transcriptome Analysis of Laiwu and Yorkshire Pigs Differing in Intramuscular Fat Content. Genes Genom. 2017, 39, 759–766. [Google Scholar] [CrossRef]
- Qiu, K.; Xu, D.; Wang, L.; Zhang, X.; Jiao, N.; Gong, L.; Yin, J. Association Analysis of Single-Cell RNA Sequencing and Proteomics Reveals a Vital Role of Ca2+ Signaling in the Determination of Skeletal Muscle Development Potential. Cells 2020, 9, 1045. [Google Scholar] [CrossRef]
- Cech, T.R.; Steitz, J.A. The Noncoding RNA Revolution—Trashing Old Rules to Forge New Ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef]
- Agbu, P.; Carthew, R.W. MicroRNA-Mediated Regulation of Glucose and Lipid Metabolism. Nat. Rev. Mol. Cell Biol. 2021, 22, 425–438. [Google Scholar] [CrossRef]
- Qi, K.; Liu, Y.; Li, C.; Li, X.; Li, X.; Wang, K.; Qiao, R.; Han, X. Construction of CircRNA-Related CeRNA Networks in Longissimus Dorsi Muscle of Queshan Black and Large White Pigs. Mol. Genet. Genom. 2021, 297, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ren, Q.; Hua, L.; Chen, J.; Zhang, J.; Bai, H.; Li, H.; Xu, B.; Shi, Z.; Cao, H.; et al. Comprehensive Analysis of Differentially Expressed MRNA, LncRNA and CircRNA and Their CeRNA Networks in the Longissimus Dorsi Muscle of Two Different Pig Breeds. Int. J. Mol. Sci. 2019, 20, 1107. [Google Scholar] [CrossRef]
- Wang, Y.; Ma, C.; Sun, Y.; Li, Y.; Kang, L.; Jiang, Y. Dynamic Transcriptome and DNA Methylome Analyses on Longissimus Dorsi to Identify Genes Underlying Intramuscular Fat Content in Pigs. BMC Genom. 2017, 18, 780. [Google Scholar] [CrossRef]
- Wang, Y.; Ning, C.; Wang, C.; Guo, J.; Wang, J.; Wu, Y. Genome-Wide Association Study for Intramuscular Fat Content in Chinese Lulai Black Pigs. Asian Australas. J. Anim. Sci. 2019, 32, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. SOAP: Short Oligonucleotide Alignment Program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Sahraeian, S.M.E.; Mohiyuddin, M.; Sebra, R.; Tilgner, H.; Afshar, P.T.; Au, K.F.; Bani Asadi, N.; Gerstein, M.B.; Wong, W.H.; Snyder, M.P.; et al. Gaining Comprehensive Biological Insight into the Transcriptome by Performing a Broad-Spectrum RNA-Seq Analysis. Nat. Commun. 2017, 8, 59. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and Quantifying Mammalian Transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Gene, T.; Consortium, O. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Carbon, S.; Douglass, E.; Good, B.M.; Unni, D.R.; Harris, N.L.; Mungall, C.J.; Basu, S.; Chisholm, R.L.; Dodson, R.J.; Hartline, E.; et al. The Gene Ontology Resource: Enriching a GOld Mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for Linking Genomes to Life and the Environment. Nucleic Acids Res. 2008, 36, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Kachitvichyanukul, V.; Schmeiser, B. Computer Generation of Hypergeometric Random Variates. J. Stat. Comput. Simul. 1985, 22, 127–145. [Google Scholar] [CrossRef]
- Abdi, H. The Bonferonni and Šidák Corrections for Multiple Comparisons. Encycl. Meas. Stat. 2007, 103–107. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis—Bookreview. J. Stat. Softw. 2010, 35, 1–3. [Google Scholar]
- Tafer, H.; Hofacker, I.L. RNAplex: A Fast Tool for RNA-RNA Interaction Search. Bioinformatics 2008, 24, 2657–2663. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, S.; Zhou, Q.; Liao, Y.; Luo, W.; Peng, Z.; Ren, R.; Wang, H. Disturbance of Calcium Homeostasis and Myogenesis Caused by TET2 Deletion in Muscle Stem Cells. Cell Death Discov. 2022, 8, 1–14. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative CT Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Rakus, D.; Gizak, A.; Deshmukh, A.; Wïniewski, J.R. Absolute Quantitative Profiling of the Key Metabolic Pathways in Slow and Fast Skeletal Muscle. J. Proteome Res. 2015, 14, 1400–1411. [Google Scholar] [CrossRef]
- Spiegelman, B.M.; Puigserver, P.; Wu, Z. Regulation of Adipogenesis and Energy Balance by PPARγ and PGC-1. Int. J. Obes. 2000, 24, S8–S10. [Google Scholar] [CrossRef] [PubMed]
- Stechschulte, L.A.; Hinds, T.D.; Khuder, S.S.; Shou, W.; Najjar, S.M.; Sanchez, E.R. FKBP51 Controls Cellular Adipogenesis through P38 Kinase-Mediated Phosphorylation of GRα and PPARγ. Mol. Endocrinol. 2014, 28, 1265–1275. [Google Scholar] [CrossRef] [PubMed]
- Fatica, A.; Bozzoni, I. Long Non-Coding RNAs: New Players in Cell Differentiation and Development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef]
- Gao, S.-Z.; Zhao, S.-M. Physiology, Affecting Factors and Strategies for Control of Pig Meat Intramuscular Fat. Recent Pat. Food. Nutr. Agric. 2009, 1, 59–74. [Google Scholar] [PubMed]
- Cui, J.; Chen, W.; Liu, J.; Xu, T.; Zeng, Y. Study on Quantitative Expression of PPARγ and ADRP in Muscle and Its Association with Intramuscular Fat Deposition of Pig. Springer Plus 2016, 5, 1501. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, J.; Yang, D.D.; Liu, Z.L.; Zeng, Y.Q.; Chen, W. Expression of Lipid Metabolism Genes Provides New Insights into Intramuscular Fat Deposition in Laiwu Pigs. Asian Australas. J. Anim. Sci. 2020, 33, 390–397. [Google Scholar] [CrossRef]
- Elustondo, P.A.; White, A.E.; Hughes, M.E.; Brebner, K.; Pavlov, E.; Kane, D.A. Physical and Functional Association of Lactate Dehydrogenase (LDH) with Skeletal Muscle Mitochondria. J. Biol. Chem. 2013, 288, 25309–25317. [Google Scholar] [CrossRef]
- Liu, X.; Trakooljul, N.; Muráni, E.; Krischek, C.; Schellander, K.; Wicke, M.; Wimmers, K.; Ponsuksili, S. Molecular Changes in Mitochondrial Respiratory Activity and Metabolic Enzyme Activity in Muscle of Four Pig Breeds with Distinct Metabolic Types. J. Bioenerg. Biomembr. 2016, 48, 55–65. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, Z.; Chao, Z.; Li, B.; Li, R.; Jiang, A.; Kim, K.H.; Liu, H. Transcriptome Analysis Reveals the Genetic Basis of Skeletal Muscle Glycolytic Potential Based on a Pig Model. Gene 2021, 766, 145157. [Google Scholar] [CrossRef]
- Liu, L.; Qian, K.; Wang, C. Discovery of Porcine MiRNA-196a/b May Influence Porcine Adipogenesis in Longissimus Dorsi Muscle by MiRNA Sequencing. Anim. Genet. 2017, 48, 175–181. [Google Scholar] [CrossRef]
- Yang, Y.; Cheng, Z.; Zhang, W.; Hei, W.; Lu, C.; Cai, C.; Zhao, Y.; Gao, P.; Guo, X.; Cao, G.; et al. Glutamic-Oxaloacetic Transaminase 1 Regulates Adipocyte Differentiation by Altering Nicotinamide Adenine Dinucleotide Phosphate Content. Anim. Biosci. 2022, 35, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Ho, A.T.V.; Palla, A.R.; Blake, M.R.; Yucel, N.D.; Wang, Y.X.; Magnusson, K.E.G.; Holbrook, C.A.; Kraft, P.E.; Delp, S.L.; Blau, H.M. Prostaglandin E2 is Essential for Efficacious Skeletal Muscle Stem-Cell Function, Augmenting Regeneration & Strength. Proc. Natl. Acad. Sci. USA 2017, 114, 6675–6684. [Google Scholar] [CrossRef] [PubMed]
- Kampjut, D.; Sazanov, L.A. Structure and Mechanism of Mitochondrial Proton-Translocating Transhydrogenase. Nature 2019, 573, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Keren, A.; Tamir, Y.; Bengal, E. The P38 MAPK Signaling Pathway: A Major Regulator of Skeletal Muscle Development. Mol. Cell. Endocrinol. 2006, 252, 224–230. [Google Scholar] [CrossRef]
- Cervera, I.P.; Gabriel, B.M.; Aldiss, P.; Morton, N.M. The Phospholipase A2 Family’s Role in Metabolic Diseases: Focus on Skeletal Muscle. Physiol. Rep. 2021, 9, e14662. [Google Scholar] [CrossRef]
- Dzeja, P.P.; Chung, S.; Faustino, R.S.; Behfar, A.; Terzic, A. Developmental Enhancement of Adenylate Kinase-AMPK Metabolic Signaling Axis Supports Stem Cell Cardiac Differentiation. PLoS ONE 2011, 6, e19300. [Google Scholar] [CrossRef]
- Várkuti, B.H.; Yang, Z.; Kintses, B.; Erdélyi, P.; Bárdos-Nagy, I.; Kovács, A.L.; Hári, P.; Kellermayer, M.; Vellai, T.; Málnási-Csizmadia, A. A Novel Actin Binding Site of Myosin Required for Effective Muscle Contraction. Nat. Struct. Mol. Biol. 2012, 19, 299–306. [Google Scholar] [CrossRef]
- Mirebeau-Prunier, D.; Le Pennec, S.; Jacques, C.; Gueguen, N.; Poirier, J.; Malthiery, Y.; Savagner, F. Estrogen-Related Receptor α and PGC-1-Related Coactivator Constitute a Novel Complex Mediating the Biogenesis of Functional Mitochondria. FEBS J. 2010, 277, 713–725. [Google Scholar] [CrossRef]
- Summermatter, S.; Santos, G.; Pérez-Schindler, J.; Handschin, C. Skeletal Muscle PGC-1α Controls Whole-Body Lactate Homeostasis through Estrogen-Related Receptor α- Dependent Activation of LDH B and Repression of LDH A. Proc. Natl. Acad. Sci. USA 2013, 110, 8738–8743. [Google Scholar] [CrossRef]
- Uldry, M.; Yang, W.; St-Pierre, J.; Lin, J.; Seale, P.; Spiegelman, B.M. Complementary Action of the PGC-1 Coactivators in Mitochondrial Biogenesis and Brown Fat Differentiation. Cell Metab. 2006, 3, 333–341. [Google Scholar] [CrossRef]
- Ceafalan, L.C.; Dobre, M.; Milanesi, E.; Niculae, A.M.; Manole, E.; Gherghiceanu, M.; Hinescu, M.E. Gene Expression Profile of Adhesion and Extracellular Matrix Molecules during Early Stages of Skeletal Muscle Regeneration. J. Cell. Mol. Med. 2020, 24, 10140–10150. [Google Scholar] [CrossRef] [PubMed]
- Stupka, N.; Kintakas, C.; White, J.D.; Fraser, F.W.; Hanciu, M.; Aramaki-Hattori, N.; Martin, S.; Coles, C.; Collier, F.; Ward, A.C.; et al. Versican Processing by a Disintegrin-like and Metalloproteinase Domain with Thrombospondin-1 Repeats Proteinases-5 and-15 Facilitates Myoblast Fusion. J. Biol. Chem. 2013, 288, 1907–1917. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; White, C.; Robert, N.; Rogers, M.B.; Szabo-Rogers, H.L. Localization of Tfap2β, Casq2, Penk, Zic1, and Zic3 Expression in the Developing Retina, Muscle, and Sclera of the Embryonic Mouse Eye. J. Histochem. Cytochem. 2019, 67, 863–871. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, C.; Liu, X.; Kuang, S. Shisa2 Regulates the Fusion of Muscle Progenitors. Stem Cell Res. 2018, 31, 31–41. [Google Scholar] [CrossRef]
- Sautron, V.; Terenina, E.; Gress, L.; Lippi, Y.; Billon, Y.; Larzul, C.; Liaubet, L.; Villa-Vialaneix, N.; Mormède, P. Time Course of the Response to ACTH in Pig: Biological and Transcriptomic Study. BMC Genom. 2015, 16, 961. [Google Scholar] [CrossRef]
- Ruiz-Conca, M.; Gardela, J.; Martínez, C.A.; Wright, D.; López-Bejar, M.; Rodríguez-Martínez, H.; Álvarez-Rodríguez, M. Natural Mating Differentially Triggers Expression of Glucocorticoid Receptor (Nr3c1)-Related Genes in the Preovulatory Porcine Female Reproductive Tract. Int. J. Mol. Sci. 2020, 21, 4437. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Long, X.; Yang, Y.; Pan, H.; Zhang, L.; Guo, Z.; Wang, J.; Lan, J. Identification and Characterization of a Pig FKBP5 Gene with a Novel Expression Pattern in Lymphocytes and Granulocytes. Anim. Biotechnol. 2019, 30, 302–310. [Google Scholar] [CrossRef]
- Messad, F.; Louveau, I.; Koffi, B.; Gilbert, H.; Gondret, F. Investigation of Muscle Transcriptomes Using Gradient Boosting Machine Learning Identifies Molecular Predictors of Feed Efficiency in Growing Pigs. BMC Genom. 2019, 20, 659. [Google Scholar] [CrossRef]
- Lemberger, T.; Desvergne, B.; Wahli, W. Peroxisome Proliferator-Activated Receptors: A Nuclear Receptor Signaling Pathway in Lipid Physiology. Annu. Rev. Cell Dev. Biol. 1996, 12, 335–363. [Google Scholar] [CrossRef]
- Catoire, M.; Alex, S.; Paraskevopulos, N.; Mattijssen, F.; Evers-Van Gogh, I.; Schaart, G.; Jeppesen, J.; Kneppers, A.; Mensink, M.; Voshol, P.J.; et al. Fatty Acid-Inducible ANGPTL4 Governs Lipid Metabolic Response to Exercise. Proc. Natl. Acad. Sci. USA 2014, 111, 1043–1052. [Google Scholar] [CrossRef]
- Tavares, C.D.J.; Aigner, S.; Sharabi, K.; Sathe, S.; Mutlu, B.; Yeo, G.W.; Puigserver, P. Transcriptome-Wide Analysis of PGC-1α-Binding RNAs Identifies Genes Linked to Glucagon Metabolic Action. Proc. Natl. Acad. Sci. USA 2020, 117, 22204–22213. [Google Scholar] [CrossRef]
- Berdeaux, R.; Stewart, R. CAMP Signaling in Skeletal Muscle Adaptation: Hypertrophy, Metabolism, and Regeneration. Am. J. Physiol. Endocrinol. Metab. 2012, 303, E1–E17. [Google Scholar] [CrossRef] [PubMed]
- Bassel-Duby, R.; Olson, E.N. Signaling Pathways in Skeletal Muscle Remodeling. Annu. Rev. Biochem. 2006, 75, 19–37. [Google Scholar] [CrossRef] [PubMed]
- Fischer-Posovszky, P.; Tews, D.; Horenburg, S.; Debatin, K.M.; Wabitsch, M. Differential Function of Akt1 and Akt2 in Human Adipocytes. Mol. Cell. Endocrinol. 2012, 358, 135–143. [Google Scholar] [CrossRef] [PubMed]
- McKinnell, I.W.; Ishibashi, J.; Le Grand, F.; Punch, V.G.J.; Addicks, G.C.; Greenblatt, J.F.; Dilworth, F.J.; Rudnicki, M.A. Pax7 Activates Myogenic Genes by Recruitment of a Histone Methyltransferase Complex. Nat. Cell Biol. 2008, 10, 77–84. [Google Scholar] [CrossRef]
- Rudnicki, M.A.; Le Grand, F.; McKinnell, I.; Kuang, S. The Molecular Regulation of Muscle Stem Cell Function. Cold Spring Harb. Symp. Quant. Biol. 2008, 73, 323–331. [Google Scholar] [CrossRef]
- Wright, A.; Hall, A.; Daly, T.; Fontelonga, T.; Potter, S.; Schafer, C.; Lindsley, A.; Hung, C.; Bodamer, O.; Gussoni, E. Lysine Methyltransferase 2D Regulates Muscle Fiber Size and Muscle Cell Differentiation. FASEB J. 2021, 35, e21955. [Google Scholar] [CrossRef]
- Sun, T.; Fu, M.; Bookout, A.L.; Kliewer, S.A.; Mangelsdorf, D.J. MicroRNA Let-7 Regulates 3T3-L1 Adipogenesis. Mol. Endocrinol. 2009, 23, 925–931. [Google Scholar] [CrossRef]
- Timoneda, O.; Balcells, I.; Córdoba, S.; Lló, A.C.; Sánchez, A. Determination of Reference MicroRNAs for Relative Quantification in Porcine Tissues. PLoS ONE 2012, 7, e44413. [Google Scholar] [CrossRef]
- Ambele, M.A.; Dessels, C.; Durandt, C.; Pepper, M.S. Genome-Wide Analysis of Gene Expression during Adipogenesis in Human Adipose-Derived Stromal Cells Reveals Novel Patterns of Gene Expression during Adipocyte Differentiation. Stem Cell Res. 2016, 16, 725–734. [Google Scholar] [CrossRef]
- Jo, S.H.; Chen, J.; Xu, G.; Grayson, T.B.; Thielen, L.A.; Shalev, A. MiR-204 Controls Glucagon-like Peptide 1 Receptor Expression and Agonist Function. Diabetes 2018, 67, 256–264. [Google Scholar] [CrossRef]
- Tan, Y.; Shen, L.; Gan, M.; Fan, Y.; Cheng, X.; Zheng, T.; Niu, L.; Chen, L.; Jiang, D.; Li, X.; et al. Downregulated MiR-204 Promotes Skeletal Muscle Regeneration. Biomed. Res. Int. 2020, 2020, 3183296. [Google Scholar] [CrossRef]
- Micheli, L.; Leonardi, L.; Conti, F.; Maresca, G.; Colazingari, S.; Mattei, E.; Lira, S.A.; Farioli-Vecchioli, S.; Caruso, M.; Tirone, F. PC4/Tis7/IFRD1 Stimulates Skeletal Muscle Regeneration and is Involved in Myoblast Differentiation as a Regulator of MyoD and NF-ΚB. J. Biol. Chem. 2011, 286, 5691–5707. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, M.; García-Casco, J.M.; Caraballo, C.; Fernández-Barroso, M.Á.; Sánchez-Esquiliche, F.; Gómez, F.; del Carmen Rodríguez, M.; Silió, L. Identification of Candidate Genes and Regulatory Factors Underlying Intramuscular Fat Content Through Longissimus Dorsi Transcriptome Analyses in Heavy Iberian Pigs. Front. Genet. 2018, 9, 608. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, B.; Jiang, A.; Cao, Y.; Hou, L.; Zhang, Z.; Zhang, X.; Liu, H.; Kim, K.H.; Wu, W. Exploring the LncRNAs Related to Skeletal Muscle Fiber Types and Meat Quality Traits in Pigs. Genes 2020, 11, 883. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Zhang, X.; Li, A.; Xie, L.; Miao, X. Genome-Wide Analysis of MRNAs and LncRNAs of Intramuscular Fat Related to Lipid Metabolism in Two Pig Breeds. Cell. Physiol. Biochem. 2018, 50, 2406–2422. [Google Scholar] [CrossRef]
- Sacheck, J.M.; Ohtsuka, A.; McLary, S.C.; Goldberg, A.L. IGF-I Stimulates Muscle Growth by Suppressing Protein Breakdown and Expression of Atrophy-Related Ubiquitin Ligases, Atrogin-1 and MuRF1. Am. J. Physiol. Endocrinol. Metab. 2004, 287, 591–601. [Google Scholar] [CrossRef]
- Fang, H.; Judd, R.L. Adiponectin Regulation and Function. Compr. Physiol. 2018, 8, 1031–1063. [Google Scholar] [CrossRef]
- Shen, B.; Zhao, C.; Wang, Y.; Peng, Y.; Cheng, J.; Li, Z.; Wu, L.; Jin, M.; Feng, H. Aucubin Inhibited Lipid Accumulation and Oxidative Stress via Nrf2/HO-1 and AMPK Signalling Pathways. J. Cell. Mol. Med. 2019, 23, 4063–4075. [Google Scholar] [CrossRef]
- Bouzakri, K.; Zachrisson, A.; Al-Khalili, L.; Zhang, B.B.; Koistinen, H.A.; Krook, A.; Zierath, J.R. SiRNA-Based Gene Silencing Reveals Specialized Roles of IRS-1/Akt2 and IRS-2/Akt1 in Glucose and Lipid Metabolism in Human Skeletal Muscle. Cell Metab. 2006, 4, 89–96. [Google Scholar] [CrossRef]
- Stefan, N.; Vozarova, B.; Del Parigi, A.; Ossowski, V.; Thompson, D.B.; Hanson, R.L.; Ravussin, E.; Tataranni, P.A. The Gln223Arg Polymorphism of the Leptin Receptor in Pima Indians: Influence on Energy Expenditure, Physical Activity and Lipid Metabolism. Int. J. Obes. 2002, 26, 1629–1632. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hou, X.; Wang, L.; Zhao, F.; Wang, L.; Zhang, L. Genome-Wide Expression Profiling of mRNAs, LncRNAs and CircRNAs in Skeletal Muscle of Two Different Pig Breeds. Animals 2021, 11, 3169. [Google Scholar] [CrossRef] [PubMed]

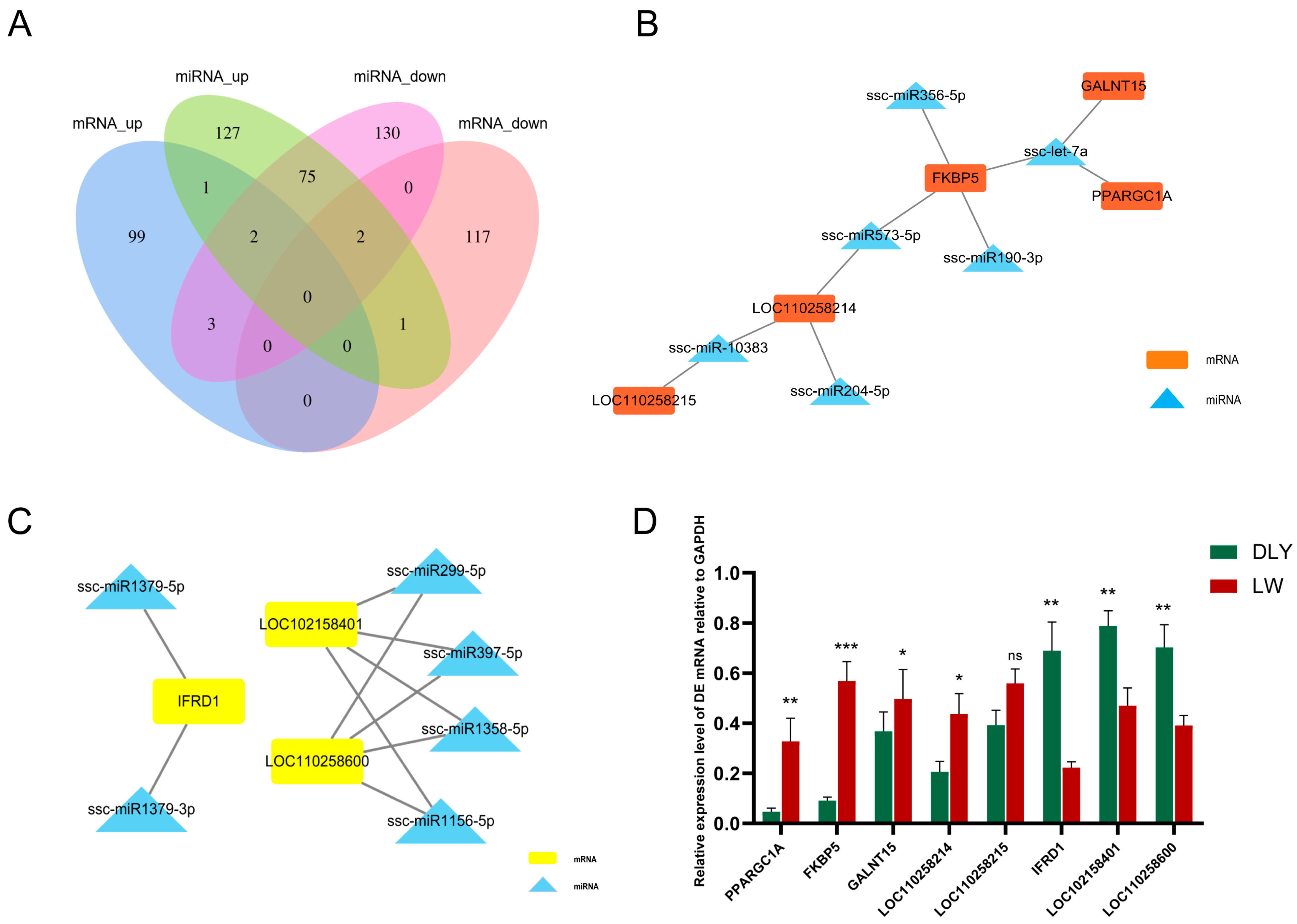
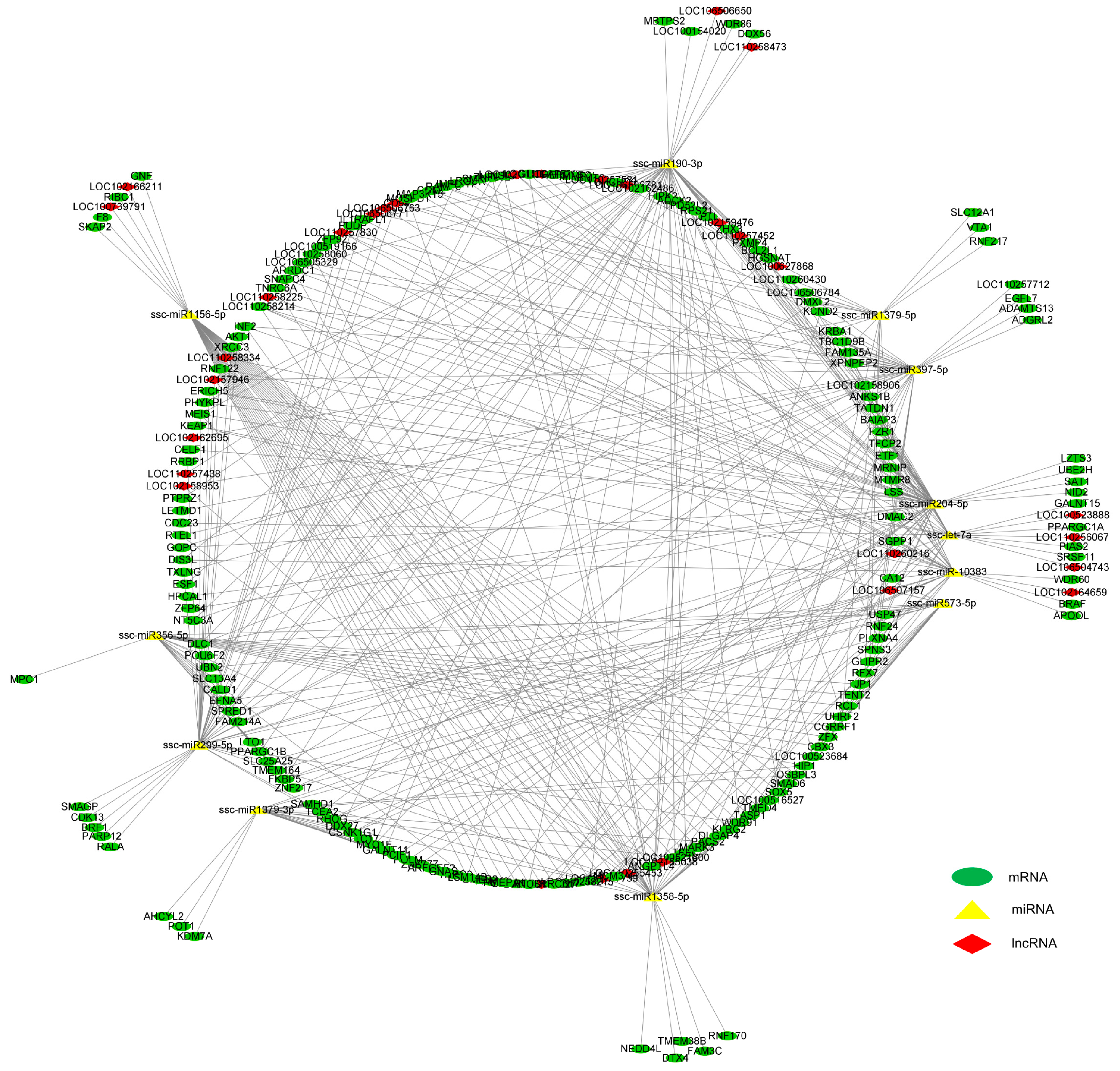
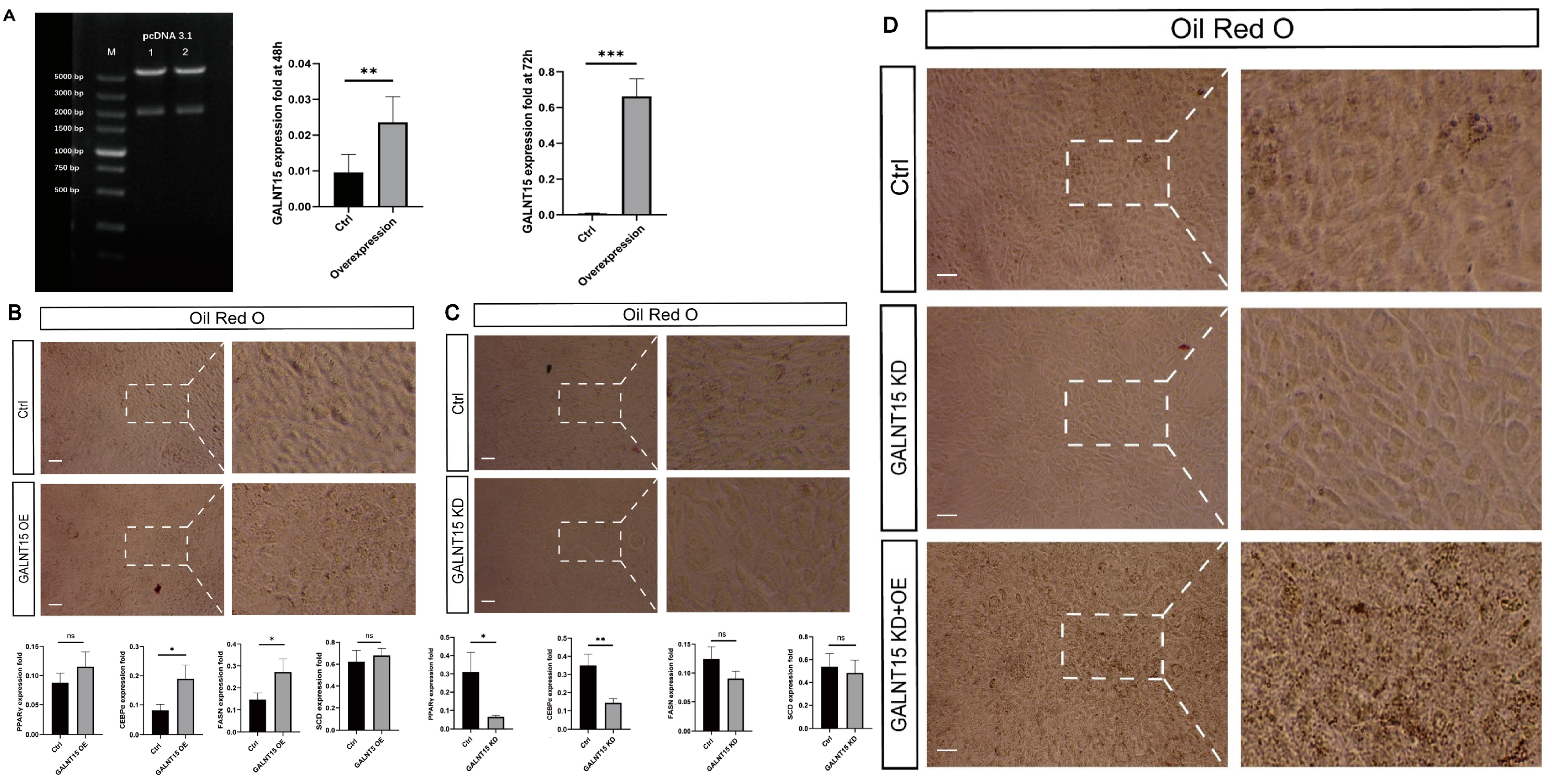
| Sample | DLY_1 | DLY_2 | DLY_3 | LW_1 | LW_2 | LW_3 |
|---|---|---|---|---|---|---|
| Total raw reads (M) | 122.44 | 122.44 | 122.44 | 124.94 | 122.44 | 124.94 |
| Total clean reads (M) | 113.94 | 113.95 | 113.85 | 116.11 | 113.82 | 116.32 |
| Total clean data (Gb) | 11.39 | 11.40 | 11.39 | 11.61 | 11.38 | 11.63 |
| Total mapping (%) | 93.21 | 93.10 | 93.74 | 92.38 | 92.61 | 92.44 |
| Clean reads Q20 (%) | 97.50 | 97.47 | 97.37 | 97.45 | 97.38 | 97.50 |
| Clean reads Q30 (%) | 91.24 | 91.17 | 90.68 | 91.00 | 90.70 | 91.06 |
| Clean reads ratio (%) | 93.06 | 93.07 | 92.98 | 92.93 | 92.96 | 93.11 |
| Sample | DLY_1 | DLY_2 | DLY_3 | LW_1 | LW_2 | LW_3 |
|---|---|---|---|---|---|---|
| Raw Tag Count | 25,165,824 | 25,165,824 | 25,165,824 | 25,165,824 | 25,165,824 | 25,165,824 |
| Clean Tag Count | 23,786,351 | 22,838,322 | 24,113,825 | 24,039,051 | 24,316,156 | 24,243,603 |
| Total mapping (%) | 90.71 | 93.45 | 92.13 | 91.30 | 89.07 | 91.26 |
| Q20 of Clean Tag (%) | 98.70 | 98.60 | 98.60 | 98.50 | 98.70 | 98.70 |
| Percentage of Clean Tag (%) | 94.52 | 90.75 | 95.82 | 95.52 | 96.62 | 96.34 |
| Gene ID | Genes | Log2 (Foldchange) | q Value | DLY Average Read Counts | LW Average Read Counts |
|---|---|---|---|---|---|
| 100621540 | LDHB | 9.667279398 | 6.38 × 10−12 | 0.19 | 158.17 |
| 110258214 | LOC110258214 | 6.986698187 | 5.01 × 10−167 | 391.55 | 49,658.91 |
| 396643 | SSTR3 | 6.043282076 | 0.010101424 | 0.19 | 12.68 |
| 110255206 | LOC110255206 | 5.935360726 | 0.015904385 | 0.19 | 11.72 |
| 100737113 | LOC100737113 | 5.856804446 | 0.015236088 | 0.72 | 41.86 |
| 100737631 | LOC100737631 | 5.78009055 | 0.015904385 | 0.19 | 10.51 |
| 106506226 | LOC106506226 | 5.265941864 | 0.031736179 | 9.43 | 362.74 |
| 102159048 | SYCP2L | 5.181170867 | 0.029172368 | 0.65 | 23.6 |
| 100514305 | STPG2 | 5.095150238 | 0.031736179 | 0.38 | 12.88 |
| 110256818 | LOC110256818 | 4.451349764 | 0.045998639 | 0.71 | 15.63 |
| 100158003 | LOC100158003 | 4.070632429 | 3.88 × 10−12 | 146.32 | 2458.62 |
| 100525112 | LOC100525112 | 3.847920892 | 0.013043917 | 1.94 | 27.88 |
| 110255992 | SPATA32 | 3.596918469 | 0.01550072 | 3.07 | 37.2 |
| 102162178 | LOC102162178 | 3.538200994 | 2.77 × 10−15 | 11 | 127.83 |
| 100514435 | RAB19 | 3.482899896 | 0.036286962 | 2.17 | 24.23 |
| 100513679 | NEK3 | 3.447440519 | 6.95 × 10−15 | 22.96 | 250.51 |
| 100155596 | HRK | 3.420051236 | 0.029848272 | 1.68 | 17.98 |
| 100513483 | DCAF16 | 3.395259004 | 0.005534146 | 2.45 | 25.75 |
| 100156672 | NEK5 | 3.178863345 | 3.65 × 10−4 | 10.29 | 93.2 |
| 100156863 | ZIC3 | 3.11041838 | 6.68 × 10−4 | 12.58 | 108.64 |
| 100126277 | AQP4 | 3.030803942 | 4.32 × 10−5 | 118.82 | 971.11 |
| Gene ID | Genes | Log2 (Foldchange) | q Value | DLY Average Read Counts | LW Average Read Counts |
|---|---|---|---|---|---|
| 110258854 | LOC110258854 | −23.2798 | 1.28 × 10−6 | 108.47 | 0 |
| 106510102 | LOC106510102 | −7.50914 | 5.68 × 10−14 | 172.7 | 0.95 |
| 100737845 | FBXL13 | −7.44403 | 3.85 × 10−6 | 59.49 | 0.34 |
| 106510322 | LOC106510322 | −7.22045 | 2.85 × 10−5 | 25.85 | 0.17 |
| 100517716 | SLC6A2 | −6.05305 | 2.91 × 10−5 | 82.52 | 1.24 |
| 110257759 | LOC110257759 | −5.92607 | 0.003691 | 20.57 | 0.34 |
| 100626756 | ACBD7 | −5.25986 | 9.63 × 10−10 | 158.71 | 4.14 |
| 100157711 | LOC100157711 | −5.16206 | 2.77 × 10−15 | 533.3 | 14.89 |
| 100624179 | COCH | −4.93751 | 0.020584 | 28.48 | 0.93 |
| 397171 | OCA2 | −4.84709 | 1.48 × 10−7 | 183.52 | 6.38 |
| 100621079 | MYH15 | −4.69801 | 0.012088 | 33.34 | 1.28 |
| 100518260 | UNC79 | −4.67715 | 0.038978 | 63.76 | 2.49 |
| 106505804 | LOC106505804 | −4.48474 | 1.50 × 10−10 | 146.11 | 6.53 |
| 100157276 | ADAMTS4 | −3.96958 | 1.96 × 10−4 | 188.52 | 12.03 |
| 100623128 | CCKAR | −3.75171 | 0.017717 | 17.12 | 1.27 |
| 100514811 | PLA2G4E | −3.70605 | 4.49 × 10−4 | 91.7 | 7.03 |
| 100736858 | RPH3A | −3.41325 | 0.017239 | 16.88 | 1.58 |
| 110256933 | LOC110256933 | −3.05582 | 0.009215 | 39.66 | 4.77 |
| 100620172 | IRX5 | −3.03576 | 1.13 × 10−4 | 85.81 | 10.46 |
| 110256218 | LOC110256218 | −3.01193 | 1.96 × 10−18 | 740.73 | 91.83 |
| Genes | Log2 (Foldchange) | q Value | DLY Average Read Counts | LW Average Read Counts |
|---|---|---|---|---|
| ssc-miR1379-5p | 14.53752176 | 6.38 × 10−11 | 0.19 | 286.08 |
| ssc-miR1379-3p | 12.04916787 | 1.55 × 10−5 | 0.19 | 50.99 |
| ssc-miR1156-5p | 10.38801729 | 0.045224024 | 0.19 | 15.71 |
| ssc-miR1358-5p | 9.081557409 | 4.56 × 10−8 | 0.37 | 175.8 |
| ssc-miR397-5p | 6.009154715 | 0.029208088 | 0.37 | 20.77 |
| ssc-miR299-5p | 2.808214579 | 0.018798556 | 136.45 | 869.54 |
| Genes | Log2 (Foldchange) | q Value | DLY Average Read Counts | LW Average Read Counts |
|---|---|---|---|---|
| ssc-miR204-5p | −11.91438513 | 4.70 × 10−6 | 50.8 | 0 |
| ssc-let-7a | −7.912724505 | 4.10 × 10−9 | 68,300.09 | 257.16 |
| ssc-miR190-3p | −7.309917116 | 1.09 × 10−5 | 100.65 | 0.61 |
| ssc-miR573-5p | −4.991746779 | 5.32 × 10−4 | 52.53 | 1.53 |
| ssc-miR356-5p | −4.719263592 | 0.012202205 | 42.26 | 1.5 |
| ssc-miR-10383 | −2.723136638 | 1.40 × 10−7 | 7773.92 | 1083.98 |
| Gene ID | Genes | Log2 (Foldchange) | q Value | DLY Average Read Counts | LW Average Read Counts |
|---|---|---|---|---|---|
| 110257307 | LOC110257307 | 6.435958 | 0.012209 | 0.19 | 16.81 |
| 110258489 | LOC110258489 | 6.250708 | 0.006616 | 0.19 | 14.76 |
| 110255522 | LOC110255522 | 5.833916 | 0.015984 | 0.19 | 10.96 |
| 110259865 | LOC110259865 | 5.703756 | 0.029037 | 0.19 | 10 |
| 102167235 | LOC102167235 | 4.085308 | 0.005753 | 1.42 | 24.06 |
| 102160723 | LOC102160723 | 3.657797 | 0.012209 | 1.65 | 20.77 |
| 110257281 | LOC110257281 | 3.464625 | 0.012209 | 3.22 | 35.57 |
| 102159627 | LOC102159627 | 3.459035 | 0.012206 | 8.95 | 98.4 |
| 102162221 | ZNF648 | 3.094487 | 0.006616 | 16.78 | 143.37 |
| 110259814 | LOC110259814 | 2.94117 | 0.003298 | 8.46 | 64.97 |
| 100157061 | LOC100157061 | 2.868918 | 0.003501 | 7.94 | 58.03 |
| 110260631 | LOC110260631 | 2.860203 | 0.012209 | 7.29 | 52.92 |
| 106509142 | LOC106509142 | 2.824431 | 1.47 × 10−12 | 32.41 | 229.59 |
| 110257827 | LOC110257827 | 2.748534 | 0.00103 | 84.95 | 570.9 |
| 110255857 | LOC110255857 | 2.730341 | 0.021514 | 35.06 | 232.65 |
| 110257909 | LOC110257909 | 2.691043 | 0.00752 | 13.71 | 88.55 |
| 110259137 | LOC110259137 | 2.648392 | 0.029867 | 19.6 | 122.89 |
| 110260388 | LOC110260388 | 2.553643 | 0.005825 | 6 | 35.21 |
| 110260634 | LOC110260634 | 2.47726 | 0.022658 | 9.95 | 55.39 |
| 110257745 | LOC110257745 | 2.349905 | 0.015074 | 6.69 | 34.11 |
| 110256935 | LOC110256935 | 2.211555 | 0.003278 | 46.14 | 213.71 |
| 110259691 | LOC110259691 | 1.911688 | 0.00148 | 48.79 | 183.58 |
| 106505263 | LOC106505263 | 1.887082 | 0.014899 | 19.65 | 72.68 |
| 110255980 | LOC110255980 | 1.606907 | 0.01395 | 32.63 | 99.38 |
| 102163278 | LOC102163278 | 1.424182 | 0.032409 | 26.94 | 72.28 |
| 110261569 | LOC110261569 | 1.380077 | 0.032399 | 60.42 | 157.25 |
| 106507563 | LOC106507563 | 1.30465 | 0.025425 | 70.18 | 173.37 |
| 100622439 | LOC100622439 | 1.170522 | 0.004696 | 241.41 | 543.4 |
| Gene ID | Genes | Log2 (Foldchange) | q Value | DLY Average Read Counts | LW Average Read Counts |
|---|---|---|---|---|---|
| 110259141 | LOC110259141 | −6.21526 | 0.007991 | 12.48 | 0.17 |
| 110260256 | LOC110260256 | −4.7651 | 4.39 × 10−7 | 65.94 | 2.43 |
| 110260878 | LOC110260878 | −4.29011 | 0.012206 | 17.59 | 0.9 |
| 110256028 | LOC110256028 | −4.21142 | 0.002245 | 44.64 | 2.41 |
| 110260798 | LOC110260798 | −4.12382 | 0.00148 | 48.52 | 2.78 |
| 110261211 | LOC110261211 | −3.52091 | 0.011148 | 20.9 | 1.82 |
| 102159118 | LOC102159118 | −3.14539 | 0.032653 | 48.75 | 5.51 |
| 110261331 | LOC110261331 | −2.94541 | 0.012272 | 33.06 | 4.29 |
| 110260773 | LOC110260773 | −2.87334 | 0.001713 | 59.8 | 8.16 |
| 106510214 | LOC106510214 | −2.84826 | 0.027653 | 35.23 | 4.89 |
| 106506422 | LOC106506422 | −2.63706 | 0.011148 | 62.99 | 10.13 |
| 100627270 | LOC100627270 | −2.39084 | 0.014715 | 43.03 | 8.2 |
| 110257276 | LOC110257276 | −2.33742 | 0.030236 | 80.59 | 15.95 |
| 100623270 | LOC100623270 | −2.29408 | 0.009702 | 36.81 | 7.51 |
| 102165609 | LOC102165609 | −2.08699 | 5.73 × 10−4 | 237.9 | 55.99 |
| 110256124 | LOC110256124 | −1.95087 | 0.02857 | 42.02 | 10.87 |
| 106509650 | LOC106509650 | −1.93849 | 0.00275 | 824.96 | 215.22 |
| 106508816 | LOC106508816 | −1.34055 | 0.030236 | 111.44 | 44 |
| 106505720 | LOC106505720 | −1.30212 | 0.041647 | 342.85 | 139.04 |
| 110257277 | LOC110257277 | −1.20335 | 0.006588 | 2752.42 | 1195.28 |
| 110255759 | LOC110255759 | −1.10927 | 0.032409 | 200.91 | 93.13 |
| 110255800 | LOC110255800 | −1.1087 | 0.009503 | 818.25 | 379.43 |
| 102163738 | LOC102163738 | −1.07791 | 0.005457 | 293.6 | 139.08 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Wang, J.; Ma, C.; Wang, W.; Wang, H.; Jiang, Y. Comparative Transcriptomic Analysis of mRNAs, miRNAs and lncRNAs in the Longissimus dorsi Muscles between Fat-Type and Lean-Type Pigs. Biomolecules 2022, 12, 1294. https://doi.org/10.3390/biom12091294
Zhang J, Wang J, Ma C, Wang W, Wang H, Jiang Y. Comparative Transcriptomic Analysis of mRNAs, miRNAs and lncRNAs in the Longissimus dorsi Muscles between Fat-Type and Lean-Type Pigs. Biomolecules. 2022; 12(9):1294. https://doi.org/10.3390/biom12091294
Chicago/Turabian StyleZhang, Jian, Jiying Wang, Cai Ma, Wenlei Wang, Heng Wang, and Yunliang Jiang. 2022. "Comparative Transcriptomic Analysis of mRNAs, miRNAs and lncRNAs in the Longissimus dorsi Muscles between Fat-Type and Lean-Type Pigs" Biomolecules 12, no. 9: 1294. https://doi.org/10.3390/biom12091294
APA StyleZhang, J., Wang, J., Ma, C., Wang, W., Wang, H., & Jiang, Y. (2022). Comparative Transcriptomic Analysis of mRNAs, miRNAs and lncRNAs in the Longissimus dorsi Muscles between Fat-Type and Lean-Type Pigs. Biomolecules, 12(9), 1294. https://doi.org/10.3390/biom12091294





