A Systematic Review of miRNA and cfDNA as Potential Biomarkers for Liquid Biopsy in Myocarditis and Inflammatory Dilated Cardiomyopathy
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Data Extraction
2.3. Disease Classification
2.4. miRNA Screening
2.5. Final Assessment
3. Results
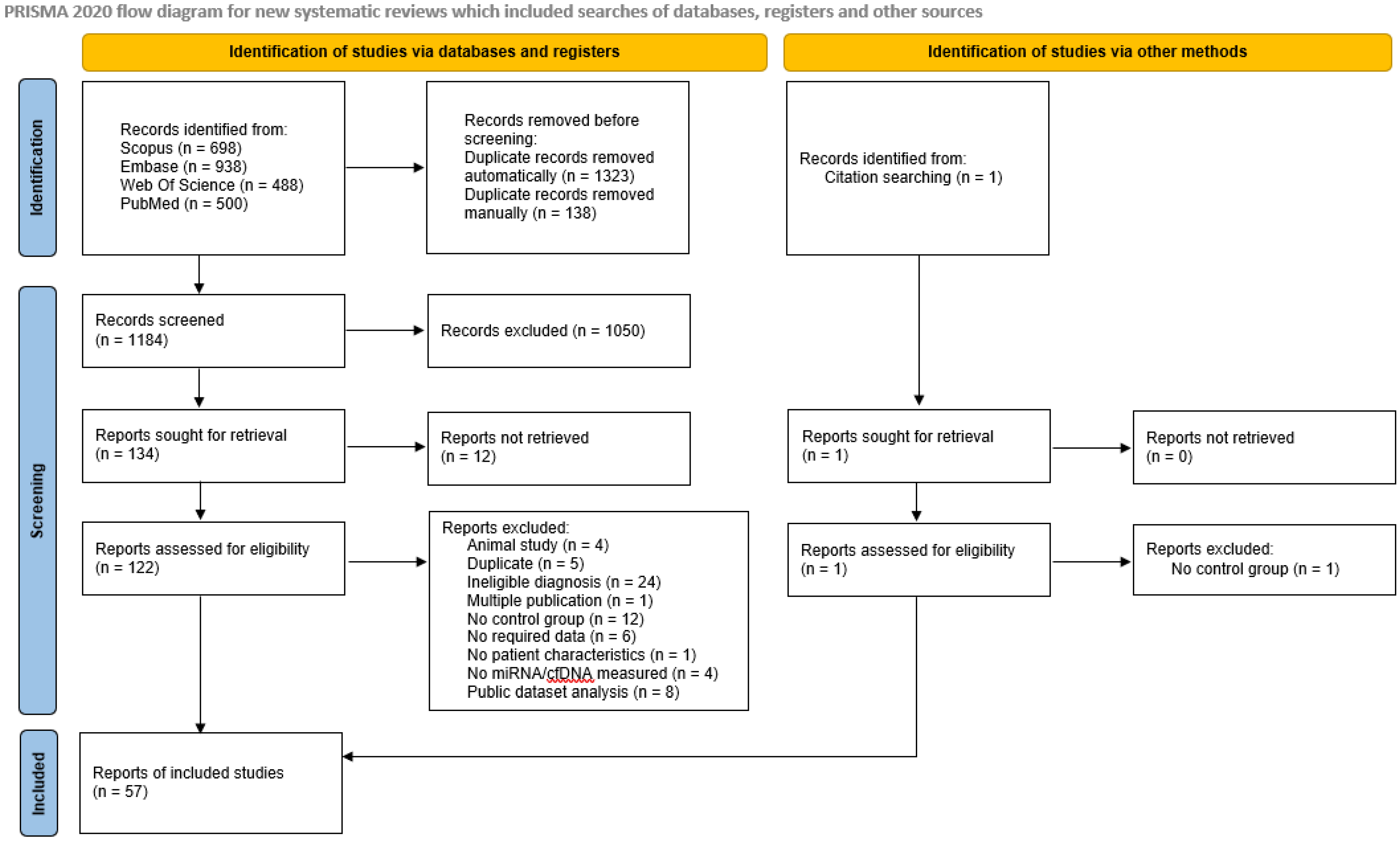
3.1. Identification and Screening Results
3.2. Characteristics of Included Studies
3.3. Results of miRNA Screening and Assessment
4. Discussion
4.1. Best Candidates for Liquid Biopsy Biomarkers
4.1.1. miR-Chr8:96
4.1.2. miR-155
4.1.3. miR-206
4.2. Potential Candidates for Liquid Biopsy Biomarkers
4.3. Possible Roles of cfDNA
4.4. Combining miRNAs
4.5. Recommendations for Future Studies
4.6. Study Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tschöpe, C.; Ammirati, E.; Bozkurt, B.; Caforio, A.L.P.; Cooper, L.T.; Felix, S.B.; Hare, J.M.; Heidecker, B.; Heymans, S.; Hübner, N.; et al. Myocarditis and inflammatory cardiomyopathy: Current evidence and future directions. Nat. Rev. Cardiol. 2021, 18, 169–193. [Google Scholar] [CrossRef] [PubMed]
- McDonagh, T.A.; Metra, M.; Adamo, M.; Gardner, R.S.; Baumbach, A.; Böhm, M.; Burri, H.; Butler, J.; Čelutkienė, J.; Chioncel, O.; et al. 2021 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: Developed by the Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC) with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur. Heart J. 2021, 42, 3599–3726. [Google Scholar] [CrossRef] [PubMed]
- From, A.M.; Maleszewski, J.J.; Rihal, C.S. Current status of endomyocardial biopsy. Mayo Clin. Proc. 2011, 86, 1095–1102. [Google Scholar] [CrossRef] [PubMed]
- Wikipedia Contributors. “Liquid Biopsy,” Wikipedia, The Free Encyclopedia. Available online: https://en.wikipedia.org/w/index.php?title=Liquid_biopsy&oldid=1102304614 (accessed on 14 August 2022).
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- van der Vaart, M.; Pretorius, P.J. The Origin of Circulating Free DNA. Clin. Chem. 2007, 53, 2215. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; Moher, D.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. PRISMA 2020 explanation and elaboration: Updated guidance and exemplars for reporting systematic reviews. BMJ 2021, 372, n160. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Domínguez, R.; Sánchez-Díaz, R.; de la Fuente, H.; Jiménez-Borreguero, L.J.; Matesanz-Marín, A.; Relaño, M.; Jiménez-Alejandre, R.; Linillos-Pradillo, B.; Tsilingiri, K.; Martín-Mariscal, M.L.; et al. A Novel Circulating MicroRNA for the Detection of Acute Myocarditis. N. Engl. J. Med. 2021, 384, 2014–2027. [Google Scholar] [CrossRef]
- Enes Coşkun, M.; Kervancıoğlu, M.; Öztuzcu, S.; Yılmaz Coşkun, F.; Ergün, S.; Başpınar, O.; Kılınç, M.; Temel, L.; Coşkun, M.Y. Plasma microRNA profiling of children with idiopathic dilated cardiomyopathy. Biomarkers 2016, 21, 56–61. [Google Scholar] [CrossRef]
- Fan, K.L.; Zhang, H.F.; Shen, J.; Zhang, Q.; Li, X.L. Circulating microRNAs levels in Chinese heart failure patients caused by dilated cardiomyopathy. Indian Heart J. 2013, 65, 12–16. [Google Scholar] [CrossRef]
- Obradovic, D.; Rommel, K.P.; Blazek, S.; Klingel, K.; Gutberlet, M.; Lücke, C.; Büttner, P.; Thiele, H.; Adams, V.; Lurz, P.; et al. The potential role of plasma miR-155 and miR-206 as circulatory biomarkers in inflammatory cardiomyopathy. ESC Heart Fail. 2021, 8, 1850–1860. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, X.; Wang, D.; Jiang, X.; Zhang, M.; Lv, K. Serum exosome microRNA panel as a noninvasive biomarker for molecular diagnosis of fulminant myocarditis. Mol. Ther.-Methods Clin. Dev. 2021, 20, 142–151. [Google Scholar] [CrossRef]
- Corsten, M.F.; Papageorgiou, A.; Verhesen, W.; Carai, P.; Lindow, M.; Obad, S.; Summer, G.; Coort, S.L.M.; Hazebroek, M.; van Leeuwen, R.; et al. MicroRNA Profiling Identifies MicroRNA-155 as an Adverse Mediator of Cardiac Injury and Dysfunction During Acute Viral Myocarditis. Circ. Res. 2012, 111, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Gumus, G.; Giray, D.; Bobusoglu, O.; Tamer, L.; Karpuz, D.; Hallioglu, O. MicroRNA values in children with rheumatic carditis: A preliminary study. Rheumatol. Int. 2018, 38, 1199–1205. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.L.; Li, M.F.; Cui, F.; Feng, F.; Kong, L.; Zhang, F.H.; Hao, H.; Yin, M.X.; Liu, Y. Altered exosomal miR-181d and miR-30a related to the pathogenesis of CVB3 induced myocarditis by targeting SOCS3. Eur. Rev. Med. Pharm. Sci. 2019, 23, 2208–2215. [Google Scholar] [CrossRef]
- Yang, H.; Shan, L.; Gao, Y.; Li, L.; Xu, G.; Wang, B.; Yin, X.; Gao, C.; Liu, J.; Yang, W. MicroRNA-181b Serves as a Circulating Biomarker and Regulates Inflammation in Heart Failure. Dis. Mrk. 2021, 2021, 4572282. [Google Scholar] [CrossRef] [PubMed]
- Besler, C.; Urban, D.; Watzka, S.; Lang, D.; Rommel, K.-P.; Kandolf, R.; Klingel, K.; Thiele, H.; Linke, A.; Schuler, G.; et al. Endomyocardial miR-133a levels correlate with myocardial inflammation, improved left ventricular function, and clinical outcome in patients with inflammatory cardiomyopathy. Eur. J. Heart Fail. 2016, 18, 1442–1451. [Google Scholar] [CrossRef]
- Besler, C.; Urban, D.; Watzka, S.; Klingel, K.; Kandolf, R.; Schuler, G.; Adams, V.; Lurz, P. Abstract 17607: MicroRNA Expression Profiles in Endomyocardial Biopsies from Patients with Myocarditis–Association with Left Ventricular Function and Clinical Events. Circulation 2015, 132, A17607. [Google Scholar] [CrossRef]
- Glezeva, N.; Moran, B.; Collier, P.; Moravec, C.S.; Phelan, D.; Donnellan, E.; Russell-Hallinan, A.; O’Connor, D.P.; Gallagher, W.M.; Gallagher, J.; et al. Targeted DNA Methylation Profiling of Human Cardiac Tissue Reveals Novel Epigenetic Traits and Gene Deregulation Across Different Heart Failure Patient Subtypes. Circ. Heart Fail. 2019, 12, e005765. [Google Scholar] [CrossRef] [PubMed]
- Kuehl, U.; Lassner, D.; Gast, M.; Stroux, A.; Rohde, M.; Siegismund, C.; Wang, X.; Escher, F.; Gross, M.; Skurk, C.; et al. Differential Cardiac MicroRNA Expression Predicts the Clinical Course in Human Enterovirus Cardiomyopathy. Circ. Heart Fail. 2015, 8, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Corsten, M.F.; Dennert, R.; Jochems, S.; Kuznetsova, T.; Devaux, Y.; Hofstra, L.; Wagner, D.R.; Staessen, J.A.; Heymans, S.; Schroen, B. Circulating MicroRNA-208b and MicroRNA-499 Reflect Myocardial Damage in Cardiovascular Disease. Circ. Cardiovasc. Genet. 2010, 3, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Tili, E.; Croce, C.M.; Michaille, J.J. miR-155: On the crosstalk between inflammation and cancer. Int. Rev. Immunol. 2009, 28, 264–284. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, F.; Xia, H.; Yao, S. MicroRNA-155: Regulation of Immune Cells in Sepsis. Mediat. Inflamm. 2021, 2021, 8874854. [Google Scholar] [CrossRef]
- Zidar, N.; Boštjančič, E.; Glavač, D.; Štajer, D. MicroRNAs, Innate Immunity and Ventricular Rupture in Human Myocardial Infarction. Dis. Mrk. 2011, 31, 247654. [Google Scholar] [CrossRef]
- Zeng, Z.; Wang, K.; Li, Y.; Xia, N.; Nie, S.; Lv, B.; Zhang, M.; Tu, X.; Li, Q.; Tang, T.; et al. Down-regulation of microRNA-451a facilitates the activation and proliferation of CD4(+) T cells by targeting Myc in patients with dilated cardiomyopathy. J. Biol. Chem. 2017, 292, 6004–6013. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-Y.; Lee, H.-C.; Fu, C.-Y.; Ding, Y.-Y.; Chen, J.-S.; Lee, M.-H.; Huang, W.-J.; Tsai, H.-J. miR-1 and miR-206 target different genes to have opposing roles during angiogenesis in zebrafish embryos. Nat. Commun. 2013, 4, 2829. [Google Scholar] [CrossRef] [PubMed]
- Kleeberger, J.A.; Neuser, J.; de Gonzalo-Calvo, D.; Kempf, T.; Bauersachs, J.; Thum, T.; Widder, J.D. microRNA-206 correlates with left ventricular function after transcatheter aortic valve implantation. Am. J. Physiol. Heart Circ. Physiol. 2017, 313, H1261–H1266. [Google Scholar] [CrossRef]
- Yang, Y.; Re, D.P.D.; Nakano, N.; Sciarretta, S.; Zhai, P.; Park, J.; Sayed, D.; Shirakabe, A.; Matsushima, S.; Park, Y.; et al. miR-206 Mediates YAP-Induced Cardiac Hypertrophy and Survival. Circ. Res. 2015, 117, 891–904. [Google Scholar] [CrossRef]
- Yan, Y.; Dang, H.; Zhang, X.; Wang, X.; Liu, X. The protective role of MiR-206 in regulating cardiomyocytes apoptosis induced by ischemic injury by targeting PTP1B. Biosci. Rep. 2020, 40, BSR20191000. [Google Scholar] [CrossRef]
- Zhai, C.; Qian, Q.; Tang, G.; Han, B.; Hu, H.; Yin, D.; Pan, H.; Zhang, S. MicroRNA-206 Protects against Myocardial Ischaemia-Reperfusion Injury in Rats by Targeting Gadd45β. Mol. Cells 2017, 40, 916–924. [Google Scholar] [CrossRef]
- Jin, Y.; Zhou, T.Y.; Cao, J.N.; Feng, Q.T.; Fu, Y.J.; Xu, X.; Yang, C.J. MicroRNA-206 Downregulates Connexin43 in Cardiomyocytes to Induce Cardiac Arrhythmias in a Transgenic Mouse Model. Heart Lung Circ. 2019, 28, 1755–1761. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Lu, Q.; Yang, A.; Rao, J.; Xie, W.; He, C.; Wang, W.; Li, H.; Zhang, Z. MiR-206 regulates the Th17/Treg ratio during osteoarthritis. Mol. Med. 2021, 27, 64. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xie, Y.; Li, L.; Li, X.; Shen, L.; Gong, J.; Zhang, R. MicroRNA-30a Modulates Type I Interferon Responses to Facilitate Coxsackievirus B3 Replication via Targeting Tripartite Motif Protein 25. Front. Immunol. 2020, 11, 603437. [Google Scholar] [CrossRef] [PubMed]
- Aleshcheva, G.; Pietsch, H.; Escher, F.; Schultheiss, H.-P. MicroRNA profiling as a novel diagnostic tool for identification of patients with inflammatory and/or virally induced cardiomyopathies. ESC Heart Fail. 2021, 8, 408–422. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.L.; Lin, L. MiR-155 and miR-148a reduce cardiac injury by inhibiting NF-κB pathway during acute viral myocarditis. Eur. Rev. Med. Pharm. Sci. 2014, 18, 2349–2356. [Google Scholar]
- Liu, Y.L.; Wu, W.; Xue, Y.; Gao, M.; Yan, Y.; Kong, Q.; Pang, Y.; Yang, F. MicroRNA-21 and -146b are involved in the pathogenesis of murine viral myocarditis by regulating TH-17 differentiation. Arch. Virol. 2013, 158, 1953–1963. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.G.; Liu, H.; Zhang, J.B.; Zhang, S.L.; Zhao, L.H.; Liang, W.Q. Upregulated microRNA-214 enhances cardiac injury by targeting ITCH during coxsackievirus infection. Mol. Med. Rep. 2015, 12, 1258–1264. [Google Scholar] [CrossRef]
- Wang, D.; Li, T.; Cui, H.; Zhang, Y. Analysis of the Indicating Value of Cardiac Troponin I, Tumor Necrosis Factor-α, Interleukin-18, Mir-1 and Mir-146b for Viral Myocarditis among Children. Cell. Physiol. Biochem. 2016, 40, 1325–1333. [Google Scholar] [CrossRef]
- Yan, M.; Wang, J.; Wang, S.; Zhang, Y.; Liu, L.; Zhao, H. Expression Levels of MicroRNA-146b and Anti-Cardiac Troponin I in Serum of Children with Viral Myocarditis and Their Clinical Significance. Iran J. Public Health 2021, 50, 510–519. [Google Scholar] [CrossRef]
- Qian, X.; Shah, P.; Agbor-Enoh, S. Noninvasive biomarkers in heart transplant: 2020-2021 year in review. Curr. Opin. Organ. Transpl. 2022, 27, 7–14. [Google Scholar] [CrossRef]
- Zemmour, H.; Planer, D.; Magenheim, J.; Moss, J.; Neiman, D.; Gilon, D.; Korach, A.; Glaser, B.; Shemer, R.; Landesberg, G.; et al. Non-invasive detection of human cardiomyocyte death using methylation patterns of circulating DNA. Nat. Commun. 2018, 9, 1443. [Google Scholar] [CrossRef] [PubMed]
- Calderon-Dominguez, M.; Belmonte, T.; Quezada-Feijoo, M.; Ramos, M.; Calderon-Dominguez, J.; Campuzano, O.; Mangas, A.; Toro, R. Plasma microrna expression profile for reduced ejection fraction in dilated cardiomyopathy. Sci. Rep. 2021, 11, 7517. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.C.; Laperriere, G.; Germain, H. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: From variable nonsense to publication quality data. Sci. Rep. 2017, 7, 2409. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.F.; Korevaar, D.A.; Altman, D.G.; Bruns, D.E.; Gatsonis, C.A.; Hooft, L.; Irwig, L.; Levine, D.; Reitsma, J.B.; de Vet, H.C.W.; et al. STARD 2015 guidelines for reporting diagnostic accuracy studies: Explanation and elaboration. BMJ Open 2016, 6, e012799. [Google Scholar] [CrossRef] [PubMed]
- Roux, J.; Gonzàlez-Porta, M.; Robinson-Rechavi, M. Comparative analysis of human and mouse expression data illuminates tissue-specific evolutionary patterns of miRNAs. Nucleic Acids Res. 2012, 40, 5890–5900. [Google Scholar] [CrossRef]
- Brundin, M.; Wågsäter, D.; Alehagen, U.; Carlhäll, C.-J. Circulating microRNA-29-5p can add to the discrimination between dilated cardiomyopathy and ischaemic heart disease. ESC Heart Fail. 2021, 8, 3865–3874. [Google Scholar] [CrossRef]
- Ishiguro, T.; Hayashi, M.; Fujiwara, W.; Okumura, S.; Yoshinaga, M.; Yamada, R.; Ueda, S.; Ito, T.; Niwa, Y.; Miyazaki, A.; et al. Circulating miR-489 as a potential new biomarker for idiopathic dilated cardiomyopathy. Fujita Med. J. 2021, 7, 18–22. [Google Scholar] [CrossRef]
- Jiao, M.; You, H.-Z.; Yang, X.-Y.; Yuan, H.; Li, Y.-L.; Liu, W.-X.; Jin, M.; Du, J. Circulating microRNA signature for the diagnosis of childhood dilated cardiomyopathy. Sci. Rep. 2018, 8, 724. [Google Scholar] [CrossRef]
- Wang, H.; Chen, F.; Tong, J.; Li, Y.; Cai, J.; Wang, Y.; Li, P.; Hao, Y.; Tian, W.; Lv, Y.; et al. Circulating microRNAs as novel biomarkers for dilated cardiomyopathy. Cardiol. J. 2017, 24, 65–73. [Google Scholar] [CrossRef]
- Chen, J.H.; He, J.; Zhou, R.; Zheng, N. Expression and Significance of Circulating microRNA-29b in Adult Fulminant Myocarditis. Zhongguo Yi Xue Ke Xue Yuan Xue Bao 2022, 44, 102–109. [Google Scholar] [CrossRef]
- Onrat, S.T.; Onrat, E.; Ercan Onay, E.; Yalım, Z.; Avşar, A. The Genetic Determination of the Differentiation Between Ischemic Dilated Cardiomyopathy and Idiopathic Dilated Cardiomyopathy. Genet. Test. Mol. Biomark. 2018, 22, 644–651. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, L.; Sun, H.; Yu, Z.; Liu, X.; Luo, X.; Li, C.; Sun, D.; Li, T. MicroRNA-381 protects myocardial cell function in children and mice with viral myocarditis via targeting cyclooxygenase-2 expression. Exp. Med. 2018, 15, 5510–5516. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xie, Y.; Liu, Y.; Qi, Y.; Tang, C.; Li, X.; Zuo, K.; Sun, D.; Shen, Y.; Pang, D.; et al. Alteration in microRNA-25 expression regulate cardiac function via renin secretion. Exp. Cell Res. 2018, 365, 119–128. [Google Scholar] [CrossRef]
- Rubiś, P.; Totoń-Żurańska, J.; Wiśniowska-Śmiałek, S.; Dziewięcka, E.; Kołton-Wróż, M.; Wołkow, P.; Pitera, E.; Rudnicka-Sosin, L.; Garlitski, A.C.; Gackowski, A.; et al. The relationship between myocardial fibrosis and myocardial microRNAs in dilated cardiomyopathy: A link between mir-133a and cardiovascular events. J. Cell. Mol. Med. 2018, 22, 2514–2517. [Google Scholar] [CrossRef]
- Marketou, M.; Kontaraki, J.; Patrianakos, A.; Kochiadakis, G.; Anastasiou, I.; Fragkiadakis, K.; Plevritaki, A.; Papadaki, S.T.; Chlouverakis, G.; Parthenakis, F. Peripheral Blood MicroRNAs as Potential Biomarkers of Myocardial Damage in Acute Viral Myocarditis. Genes 2021, 12, 420. [Google Scholar] [CrossRef]
- Yu, M.; Liang, W.; Xie, Y.; Long, Q.; Cheng, X.; Liao, Y.-H.; Yuan, J. Circulating miR-185 might be a novel biomarker for clinical outcome in patients with dilated cardiomyopathy. Sci. Rep. 2016, 6, 33580. [Google Scholar] [CrossRef] [PubMed]
- Xia, K.; Zhang, Y.; Sun, D. miR-217 and miR-543 downregulation mitigates inflammatory response and myocardial injury in children with viral myocarditis by regulating the SIRT1/AMPK/NF-κB signaling pathway. Int. J. Mol. Med. 2020, 45, 634–646. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.Y.; Zhao, Z.; Jin, Z. Expression of miR-98 in myocarditis and its influence on transcription of the FAS/FASL gene pair. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef] [PubMed]
- Woulfe, K.C.; Siomos, A.K.; Nguyen, H.; SooHoo, M.; Galambos, C.; Stauffer, B.L.; Sucharov, C.; Miyamoto, S. Fibrosis and Fibrotic Gene Expression in Pediatric and Adult Patients With Idiopathic Dilated Cardiomyopathy. J. Card. Fail. 2017, 23, 314–324. [Google Scholar] [CrossRef]
- Nie, X.; He, M.; Wang, J.; Chen, P.; Wang, F.; Lai, J.; Li, C.; Yu, T.; Zuo, H.; Cui, G.; et al. Circulating miR-4763-3p Is a Novel Potential Biomarker Candidate for Human Adult Fulminant Myocarditis. Mol. Ther. Methods Clin. Dev. 2020, 17, 1079–1087. [Google Scholar] [CrossRef]
- Voellenkle, C.; van Rooij, J.; Cappuzzello, C.; Greco, S.; Arcelli, D.; Di Vito, L.; Melillo, G.; Rigolini, R.; Costa, E.; Crea, F.; et al. MicroRNA signatures in peripheral blood mononuclear cells of chronic heart failure patients. Physiol. Genom. 2010, 42, 420–426. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.B.; Li, R.C.; Xu, M.; Xu, S.M.; Lai, Y.S.; Wu, H.D.; Xie, X.J.; Gao, W.; Ye, H.; Zhang, Y.Y.; et al. Ultrastructural uncoupling between T-tubules and sarcoplasmic reticulum in human heart failure. Cardiovasc. Res. 2013, 98, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Chen, Y.; Du, Y.; Tao, J.; Zhou, Z.; Yang, Z. Serum Exosomal MiR-92b-5p as a Potential Biomarker for Acute Heart Failure Caused by Dilated Cardiomyopathy. Cell. Physiol. Biochem. 2018, 46, 1939–1950. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, S.; Kong, S.W.; Lu, J.; Bisping, E.; Zhang, H.; Allen, P.D.; Golub, T.R.; Pieske, B.; Pu, W.T. Altered microRNA expression in human heart disease. Physiol. Genom. 2007, 31, 367–373. [Google Scholar] [CrossRef]
- Baumgarten, A.; Bang, C.; Tschirner, A.; Engelmann, A.; Adams, V.; von Haehling, S.; Doehner, W.; Pregla, R.; Anker, M.S.; Blecharz, K.; et al. TWIST1 regulates the activity of ubiquitin proteasome system via the miR-199/214 cluster in human end-stage dilated cardiomyopathy. Int. J. Cardiol. 2013, 168, 1447–1452. [Google Scholar] [CrossRef]
- Naga Prasad, S.V.; Gupta, M.K.; Duan, Z.-H.; Surampudi, V.S.K.; Liu, C.-G.; Kotwal, A.; Moravec, C.S.; Starling, R.C.; Perez, D.M.; Sen, S.; et al. A unique microRNA profile in end-stage heart failure indicates alterations in specific cardiovascular signaling networks. PLoS ONE 2017, 12, e0170456. [Google Scholar] [CrossRef][Green Version]
- Hailu, F.T.; Karimpour-Fard, A.; Toni, L.S.; Bristow, M.R.; Miyamoto, S.D.; Stauffer, B.L.; Sucharov, C.C. Integrated analysis of miRNA–mRNA interaction in pediatric dilated cardiomyopathy. Pediatr. Res. 2022, 92, 98–108. [Google Scholar] [CrossRef]
- Jiao, M.; You, H.; Wang, Z.; Gu, Y.; Wang, X.; Liang, Y.; Xiao, Y.; Jin, M. Diagnosis of dilated cardiomyopathy in children based on microRNA sequencing technology. Chin. J. Appl. Clin. Pediatr. 2020, 24, 982–987. (In Chinese) [Google Scholar]
- Baán, J.A.; Varga, Z.V.; Leszek, P.; Kuśmierczyk, M.; Baranyai, T.; Dux, L.; Ferdinandy, P.; Braun, T.; Mendler, L. Myostatin and IGF-I signaling in end-stage human heart failure: A qRT-PCR study. J. Transl. Med. 2015, 13, 1. [Google Scholar] [CrossRef]
- Tao, L.; Yang, L.; Huang, X.; Hua, F.; Yang, X. Reconstruction and Analysis of the lncRNA-miRNA-mRNA Network Based on Competitive Endogenous RNA Reveal Functional lncRNAs in Dilated Cardiomyopathy. Front. Genet. 2019, 10, 1149. [Google Scholar] [CrossRef] [PubMed]
- Satoh, M.; Minami, Y.; Takahashi, Y.; Tabuchi, T.; Nakamura, M. Expression of microRNA-208 is associated with adverse clinical outcomes in human dilated cardiomyopathy. J. Card. Fail. 2010, 16, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Marketou, M.; Kontaraki, J.; Konstantinou, J.; Parthenakis, F.; Nakou, H.; Lempidakis, D.; Vernardos, M.; Fragkiadakis, K.; Loulakakis, M.; Theodosaki, O.; et al. P2579Differential microRNA gene expression levels in patients with acute myocarditis. Eur. Heart J. 2017, 38, ehx502.P2579. [Google Scholar] [CrossRef]
- Marketou, M.; Kontaraki, J.; Konstantinou, J.; Parthenakis, F.; Patrianakos, A.; Nakou, H.; Lempidakis, D.; Vernardos, M.; Fragkiadakis, K.; Loulakakis, M.; et al. P2573MicroRNA-21 gene expression levels in peripheral monocytes reflect myocardial damage in acute myocarditis. Eur. Heart J. 2017, 38, ehx502.P2573. [Google Scholar] [CrossRef]
- Marketou, M.; Kontaraki, J.; Konstantinou, J.; Maragkoudakis, S.; Plevritaki, A.; Lempidakis, D.; Vougia, D.; Fragiadakis, K.; Kassotakis, S.; Theodosaki, O.; et al. P5562Increased microRNA-21 gene expression levels as a biomarker of myocardial damage in acute myocarditis. Eur. Heart J. 2019, 40, ehz746.0506. [Google Scholar] [CrossRef]
- Marketou, M.; Kontaraki, J.; Fragiadakis, K.; Konstantinou, J.; Maragkoudakis, S.; Lempidakis, D.; Plevritaki, A.; Plataki, M.; Papadaki, S.; Vougia, D.; et al. MicroRNA-208b gene expression levels as a biomarkers of left ventricular dysfunction in patients with acute myocarditis. Eur. Heart J. 2020, 41. [Google Scholar] [CrossRef]
- Heggermont, W.A.; Delrue, L.; Dierckx, R.; Dierickx, K.; Verstreken; Goethals, M.; Bartunek, J.; Vanderheyden, M. The MiR 221/ 222 cluster is elevated both in biopsies of patients with myocarditis and idiopathic non ischemic cardiomyopathy but fails to discriminate between both pathologies. Eur. J. Heart Fail. 2018, 28, 23. [Google Scholar]
- The 4th Joint EFLM-UEMS Congress “Laboratory Medicine at the Clinical Interface” Warsaw, Poland, 21th–24th September, 2016. Clin. Chem. Lab. Med. 2016, 54, eA213–eA366. [CrossRef]
- Zaseeva, A.V.; Zhirov, I.V.; Tereschenko, S.N.; Scvortcov, A.A.; Masenko, V.P.; Kochetov, A.G.; Gimadiev, R.R.; Abramov; Lyang, O.V. The role of circulating miR 21, miR 34a, miR 423, miR 208a and miR 499a in ischemic and dilated cardiomyopathy. Eur. J. Heart Fail. 2016, 18, 492. [Google Scholar]
- Akat, K.M.; Mihailovic, A.; Williams, Z.; Brown, M.; Morozov, P.; Takayama, H.; Drosatos, K.; Tuschl, T.; Schulze, P.C. Abstract 10918: High-Throughput Sequencing Analysis of microRNA Profile Dynamics in Patients with Advanced Heart Failure Undergoing Ventricular Assist Device Placement in Comparison to Normal Adult and Fetal Cardiac Expression. Circulation 2011, 124, A10918. [Google Scholar] [CrossRef]
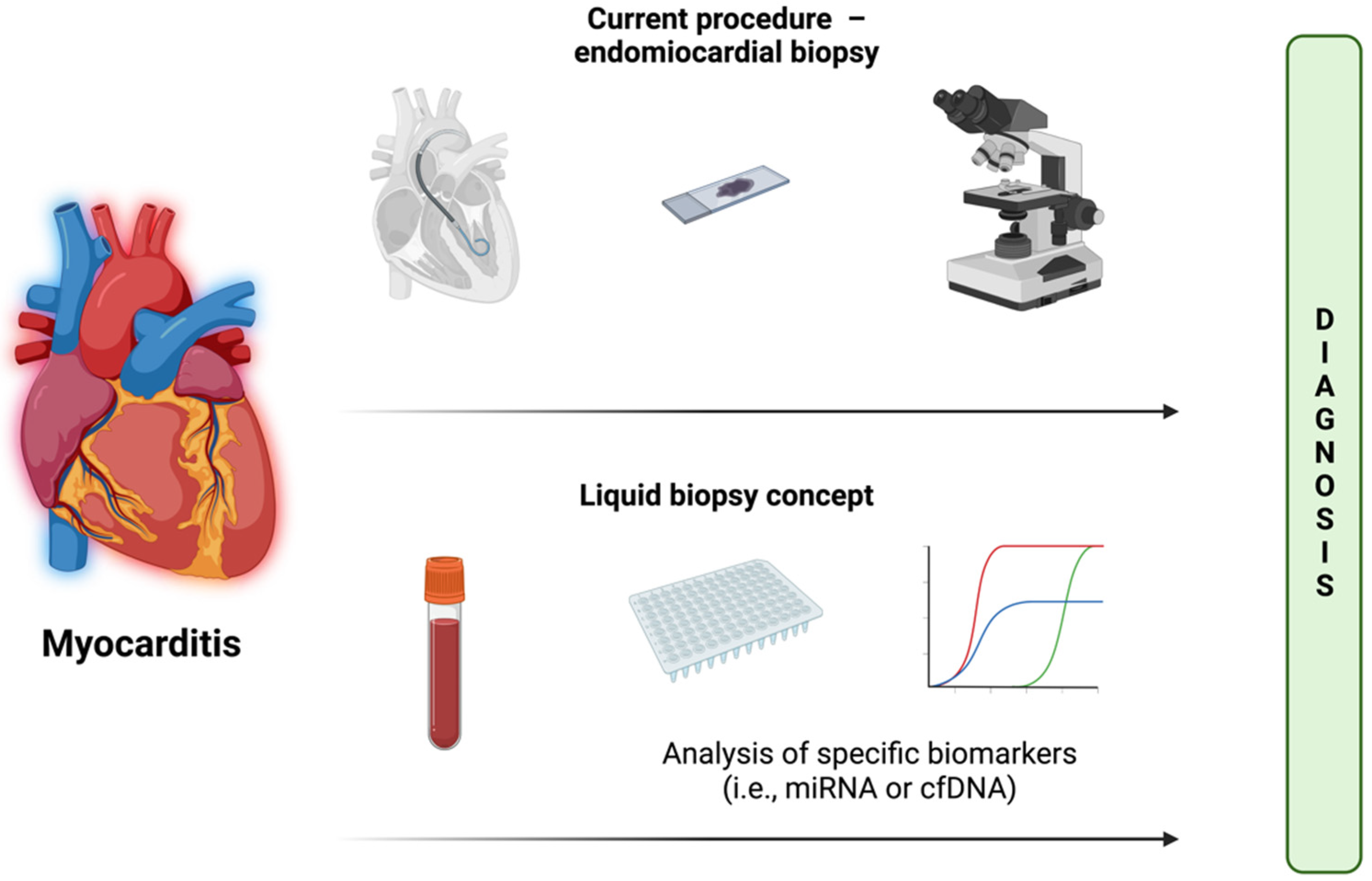

| The article describes a clinical study with measurements of cfDNA level in plasma/serum or miRNA in plasma/serum/myocardial tissue. All types of blood derivatives (peripheral blood mononuclear cells (PBMC), CD4+ cells, or exosomes) were also considered eligible. |
| The article involves a group of patients with myocarditis, inflammatory dilated cardiomyopathy, or non-ischemic idiopathic dilated cardiomyopathy (criteria of disease classification are described below). |
| The article involves a control group (positive i.e., healthy subjects, or negative i.e., patients with myocardial infarction, ischemic dilated cardiomyopathy, or other myocardial disease). |
| The article is a case study or a review. |
| The article describes only an in vitro or animal study. |
| The article does not involve an independent control group. |
| The study group suffers from other serious cardiac diseases (i.e., amyloidosis or infarction). |
| The article is retracted. |
| The article presents only data not useful in the context of this study, for example, there is no possibility of extracting information regarding particular miRNAs. |
| Disease | Criteria |
|---|---|
| Myocarditis (MCI) | The study group was classified into this group when the following clinical diagnosis was met: myocarditis, acute myocarditis, fulminant myocarditis, viral myocarditis, autoimmune myocarditis, or rheumatic carditis. Chagas myocarditis was considered inappropriate. There was no need to provide additional evidence for this diagnosis. |
| Inflammatory Dilated Cardiomyopathy (InfDCM) | The study group was classified into this group when patients met the diagnostic criteria of DCM, ischemic heart disease was excluded, and inflammation was proven with myocardial biopsy. |
| Non-ischemic Idiopathic Dilated Cardiomyopathy (Ni-IDCM) | The study group was classified into this group when patients met the diagnostic criteria of DCM and ischemic heart disease were excluded. The cause of the disease must have been undetermined. |
| Utility as LB Marker | Criteria |
|---|---|
| Very high (5) | Micro RNA was selected in at least one study using a screening method (microarray or miRNAseq) *. Micro RNA was assessed in at least two independent studies or one study involving at least two independent groups. Studies compared miRNA levels in MCI/InfDCM, a healthy control group, and at least two other myocardial diseases. Studies provide valid evidence for patient group homogeneity. At least one study provided a ROC curve for distinguishing MCI/InfDCM from healthy patients or patients with other diseases; AUC > 0.9. If studies compared miRNA expression in other diseases, miRNA must have shown a potential for MCI/InfDCM specificity. |
| High (4) | Micro RNA was selected in at least one study based on a screening method (microarray or miRNAseq) *. Studies compared miRNA levels in MCI/InfDCM, a healthy control group, and at least one myocardial disease. At least one study provided a ROC curve for distinguishing MCI/InfDCM from healthy patients or patients with other diseases; AUC > 0.7/at least one study provides strong evidence for miRNA specificity and sensitivity. If studies compared miRNA expression in other diseases, miRNA must have shown a potential for MCI/InfDCM specificity. |
| Medium (3) | Studies compared miRNA levels in MCI/InfDCM, a healthy control group, and at least one myocardial disease. At least one study provides a statistically significant correlation between miRNA concentration and MCI/InfDCM occurrence. If studies compared expression in MCI/InfDCM with other diseases, miRNA must have shown a potential for MCI/InfDCM specificity. |
| Low (2) | Studies compared miRNA levels in MCI/InfDCM and a healthy control group. At least one study provides a statistically significant correlation between miRNA concentration and MCI/InfDCM occurrence. If studies compared expression in MCI/InfDCM with other diseases, miRNA must have shown a potential for MCI/InfDCM specificity. |
| Very low (1) | Studies did not include evidence for the correlation of miRNA with MCI/InfDCM or miRNA was non-specific. |
| Utility as LB Marker | miRNAs |
|---|---|
| Very high (5) | let-7f, miR-1, miR-27b, miR-142, miR-143, miR-155, miR-223, miR-Chr8:96 |
| High (4) | miR-29b, miR-30a, miR-106a, miR-125b, miR-133a, miR-133b, miR-146a, miR-146b, miR-181b, miR-192, miR-197, miR-206, miR-320a, miR-4763, mmu-miR-93, mmu-miR-379 |
| Medium (3) | miR-15b, miR-16, miR-17, miR-21, miR-27a, miR-125a, miR-151a, miR-185, miR-194, miR-205, miR-208, miR-208a, miR-212, miR-220c, miR-222, miR-342, miR-489, miR-499, miR-660, miR-671 |
| Low (2) | let-7g, miR-7, miR-10b, miR-26a, miR-26b, miR-29, miR-92a, miR-92b, miR-93, miR-98, miR-99b, miR-107, miR-130b, miR-133, miR-135b, miR-147, miR-148a, miR-150, miR-181d, miR-190a, miR-199b, miR-217, miR-221, miR-297, miR-301a, miR-301b, miR-302a, miR-338, miR-339, miR-361, miR-362, miR-363, miR-365a, mir-378, miR-381, miR-422a, miR-423, miR-451-DICER1, miR-451a, miR-454, miR-455, miR-486, miR-495, miR-496, miR-511, miR-518f, miR-520e, miR-543, miR-544, miR-551b, miR-590, miR-595, miR-601, miR-618, miR-770, miR-875, miR-889, miR-1180, miR-1261, miR-1290, miR-3064, miR-3135b, miR-3148, miR-3908, miR-4701, miR-4793, miR-5571, miR-6785, miR-6796, miR-6807, miR-6847, miR-6849, miR-6856, miR-7844 |
| Very low (1) | let-7a, let-7b, let-7c, let-7i, miR-9, miR-10a, miR-16-2, miR-19a, miR-19b, miR-20a, miR-20b, miR-23a, miR-23b, miR-24, miR-24-1, miR-25, miR-28, miR-29a, miR-30b, miR-30c, miR-30e, miR-34a, miR-99a, miR-100, miR-101, miR-103a, miR-122, miR-125, miR-126, miR-132, miR-134, miR-139, miR-140, miR-141, miR-144, miR-145, miR-154, miR-191, miR-193a, miR-195, miR-196a, miR-199a, miR-200c, miR-208b, miR-210, miR-214, miR-215, miR-218, miR-296, miR-323, miR-324, miR-326, miR-365, miR-375, miR-378a, miR-423-5P, miR-449, miR-483, miR-497, miR-499a, miR-502, miR-532, miR-624, miR-629, miR-3940, miR-3960, miR-4821, miR-5010, miR-5088 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lewandowski, P.; Goławski, M.; Baron, M.; Reichman-Warmusz, E.; Wojnicz, R. A Systematic Review of miRNA and cfDNA as Potential Biomarkers for Liquid Biopsy in Myocarditis and Inflammatory Dilated Cardiomyopathy. Biomolecules 2022, 12, 1476. https://doi.org/10.3390/biom12101476
Lewandowski P, Goławski M, Baron M, Reichman-Warmusz E, Wojnicz R. A Systematic Review of miRNA and cfDNA as Potential Biomarkers for Liquid Biopsy in Myocarditis and Inflammatory Dilated Cardiomyopathy. Biomolecules. 2022; 12(10):1476. https://doi.org/10.3390/biom12101476
Chicago/Turabian StyleLewandowski, Piotr, Marcin Goławski, Maciej Baron, Edyta Reichman-Warmusz, and Romuald Wojnicz. 2022. "A Systematic Review of miRNA and cfDNA as Potential Biomarkers for Liquid Biopsy in Myocarditis and Inflammatory Dilated Cardiomyopathy" Biomolecules 12, no. 10: 1476. https://doi.org/10.3390/biom12101476
APA StyleLewandowski, P., Goławski, M., Baron, M., Reichman-Warmusz, E., & Wojnicz, R. (2022). A Systematic Review of miRNA and cfDNA as Potential Biomarkers for Liquid Biopsy in Myocarditis and Inflammatory Dilated Cardiomyopathy. Biomolecules, 12(10), 1476. https://doi.org/10.3390/biom12101476







