Comprehensive Genomic Profiling of Cell-Free Circulating Tumor DNA Detects Response to Ribociclib Plus Letrozole in a Patient with Metastatic Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. cfDNA Extraction from Blood Samples
2.2. Genomic DNA Extraction from Blood Samples
2.3. Sequencing and Analysis of cfDNA and gDNA Extracted from Blood Samples
2.4. Sequencing and Analysis of Primary Tumor Genomic DNA
3. Results
3.1. Clinical Case
3.2. Genomic Profiling of Plasma cfDNA
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chaffer, C.L.; Weinberg, R.A. A perspective on cancer cell metastasis. Science 2011, 331, 1559–1564. [Google Scholar] [CrossRef] [PubMed]
- Seyfried, T.N.; Huysentruyt, L.C. On the origin of cancer metastasis. Crit. Rev. Oncog. 2013, 18, 43–73. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Mu, P. Targeting Breast Cancer Metastasis. Breast Cancer 2015, 9, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhu, Y.; Liu, X.; Liao, X.; He, J.; Niu, L. The Clinicopathological features and survival outcomes of patients with different metastatic sites in stage IV breast cancer. BMC Cancer 2019, 19, 1091. [Google Scholar] [CrossRef]
- Karrison, T.G.; Ferguson, D.J.; Meier, P. Dormancy of mammary carcinoma after mastectomy. J. Natl. Cancer Inst. 1999, 91, 80–85. [Google Scholar] [CrossRef]
- Uhr, J.W.; Pantel, K. Controversies in clinical cancer dormancy. Proc. Natl. Acad. Sci. USA 2011, 108, 12396–12400. [Google Scholar] [CrossRef]
- Sosa, M.S.; Bragado, P.; Aguirre-Ghiso, J.A. Mechanisms of disseminated cancer cell dormancy: An awakening field. Nat. Rev. Cancer 2014, 14, 611–622. [Google Scholar] [CrossRef]
- Riggio, A.I.; Varley, K.E.; Welm, A.L. The lingering mysteries of metastatic recurrence in breast cancer. Br. J. Cancer 2021, 124, 13–26. [Google Scholar] [CrossRef]
- Rinnerthaler, G.; Gampenrieder, S.P.; Greil, R. ASCO 2018 highlights: Metastatic breast cancer. Memo 2018, 11, 276–279. [Google Scholar] [CrossRef]
- Caswell-Jin, J.L.; Plevritis, S.K.; Tian, L.; Cadham, C.J.; Xu, C.; Stout, N.K.; Sledge, G.W.; Mandelblatt, J.S.; Kurian, A.W. Change in Survival in Metastatic Breast Cancer with Treatment Advances: Meta-Analysis and Systematic Review. JNCI Cancer Spectr. 2018, 2, pky062. [Google Scholar] [CrossRef]
- Coombes, R.C.; Page, K.; Salari, R.; Hastings, R.K.; Armstrong, A.; Ahmed, S.; Ali, S.; Cleator, S.; Kenny, L.; Stebbing, J.; et al. Personalized detection of circulating tumor DNA antedates breast cancer metastatic recurrence. Clin. Cancer Res. 2019, 25, 4255–4263. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Murillas, I.; Chopra, N.; Comino-Méndez, I.; Beaney, M.; Tovey, H.; Cutts, R.J.; Swift, C.; Kriplani, D.; Afentakis, M.; Hrebien, S.; et al. Assessment of Molecular Relapse Detection in Early-Stage Breast Cancer. JAMA Oncol. 2019, 5, 1473–1478. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.L.; Jansen, L.; Post, W.J.; Bonnema, J.; van de Velde, J.C.; de Bock, G.H. Impact on survival of early detection of isolated breast recurrences after the primary treatment for breast cancer: A meta-analysis. Breast Cancer Res. Treat. 2009, 114, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, S.; Caldas, C. The Implications of Clonal Genome Evolution for Cancer Medicine. N. Engl. J. Med. 2013, 368, 842–851. [Google Scholar] [CrossRef]
- Murtaza, M.; Dawson, S.-J.; Tsui, D.W.Y.; Gale, D.; Forshew, T.; Piskorz, A.M.; Parkinson, C.; Chin, S.-F.; Kingsbury, Z.; Wong, A.S.C.; et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013, 497, 108–112. [Google Scholar] [CrossRef]
- Dang, D.K.; Park, B.H. Circulating tumor DNA: Current challenges for clinical utility. J. Clin. Investig. 2022, 132, e154941. [Google Scholar] [CrossRef]
- Woodhouse, R.; Li, M.; Hughes, J.; Delfosse, D.; Skoletsky, J.; Ma, P.; Meng, W.; Dewal, N.; Milbury, C.; Clark, T.; et al. Clinical and analytical validation of foundation one liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS ONE 2020, 15, e0237802. [Google Scholar] [CrossRef]
- Caputo, V.; De Falco, V.; Ventriglia, A.; Famiglietti, V.; Martinelli, E.; Morgillo, F.; Martini, G.; Corte, C.M.D.; Ciardiello, D.; Poliero, L.; et al. Comprehensive genome profiling by next generation sequencing of circulating tumor DNA in solid tumors: A single academic institution experience. Ther. Adv. Med. Oncol. 2022, 14, 17588359221096878. [Google Scholar] [CrossRef]
- Poh, J.; Ngeow, K.C.; Pek, M.; Tan, K.-H.; Lim, J.S.; Chen, H.; Ong, C.K.; Lim, J.Q.; Lim, S.T.; Lim, C.M.; et al. Analytical and clinical validation of an amplicon-based next generation sequencing assay for ultrasensitive detection of circulating tumor DNA. PLoS ONE 2022, 17, e0267389. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.T.; Mitchell, T.N.; Zehir, A.; Shah, R.H.; Benayed, R.; Syed, A.; Chandramohan, R.; Liu, Z.Y.; Won, H.H.; Scott, S.N.; et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT). J. Mol. Diagn. 2015, 17, 251–264. [Google Scholar] [CrossRef]
- Zehir, A.; Benayed, R.; Shah, R.H.; Syed, A.; Middha, S.; Kim, H.R.; Srinivasan, P.; Gao, J.; Chakravarty, D.; Devlin, S.M.; et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017, 23, 703–713. [Google Scholar] [CrossRef] [PubMed]
- Weigelt, B.; Bi, R.; Kumar, R.; Blecua, P.; Mandelker, D.L.; Geyer, F.C.; Pareja, F.; James, P.A.; kConFab Investigators; Couch, F.J.; et al. The Landscape of Somatic Genetic Alterations in Breast Cancers From ATM Germline Mutation Carriers. J. Natl. Cancer Inst. 2018, 110, 1030–1034. [Google Scholar] [CrossRef] [PubMed]
- Da Cruz Paula, A.; da Silva, E.M.; Segura, S.E.; Pareja, F.; Bi, R.; Selenica, P.; Kim, S.H.; Ferrando, L.; Vahdatinia, M.; Soslow, R.A.; et al. Genomic profiling of primary and recurrent adult granulosa cell tumors of the ovary. Mod. Pathol. 2020, 33, 1606–1617. [Google Scholar] [CrossRef] [PubMed]
- De Mattos-Arruda, L.; Weigelt, B.; Cortes, J.; Won, H.H.; Ng, C.K.Y.; Nuciforo, P.; Bidard, F.-C.; Aura, C.; Saura, C.; Peg, V.; et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: A proof-of-principle. Ann. Oncol. 2014, 25, 1729–1735. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef]
- Cibulskis, K.; Lawrence, M.S.; Carter, S.L.; Sivachenko, A.; Jaffe, D.; Sougnez, C.; Gabriel, S.; Meyerson, M.; Lander, E.S.; Getz, G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013, 31, 213–219. [Google Scholar] [CrossRef]
- Saunders, C.T.; Wong, W.S.W.; Swamy, S.; Becq, J.; Murray, L.J.; Cheetham, R.K. Strelka: Accurate somatic small-variant calling from sequenced tumor–normal sample pairs. Bioinformatics 2012, 28, 1811–1817. [Google Scholar] [CrossRef]
- Koboldt, D.C.; Zhang, Q.; Larson, D.E.; Shen, D.; McLellan, M.D.; Lin, L.; Miller, C.A.; Mardis, E.R.; Ding, L.; Wilson, R.K. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012, 22, 568–576. [Google Scholar] [CrossRef]
- Narzisi, G.; Corvelo, A.; Arora, K.; Bergmann, E.A.; Shah, M.; Musunuri, R.; Emde, A.-K.; Robine, N.; Vacic, V.; Zody, M.C. Genome-wide somatic variant calling using localized colored de Bruijn graphs. Commun. Biol. 2018, 1, 20. [Google Scholar] [CrossRef]
- Narzisi, G.; O’Rawe, J.A.; Iossifov, I.; Fang, H.; Lee, Y.-h.; Wang, Z.; Wu, Y.; Lyon, G.J.; Wigler, M.; Schatz, M.C. Accurate de novo and transmitted indel detection in exome-capture data using microassembly. Nat. Methods 2014, 11, 1033–1036. [Google Scholar] [CrossRef] [PubMed]
- Rimmer, A.; Phan, H.; Mathieson, I.; Iqbal, Z.; Twigg, S.R.F.; WGS500 Consortium; Wilkie, A.O.M.; McVean, G.; Lunter, G. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat. Genet. 2014, 46, 912–918. [Google Scholar] [CrossRef] [PubMed]
- Shen, R.; Seshan, V.E. FACETS: Allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016, 44, e131. [Google Scholar] [CrossRef] [PubMed]
- Pareja, F.; Lee, J.Y.; Brown, D.N.; Piscuoglio, S.; Gularte-Mérida, R.; Selenica, P.; Da Cruz Paula, A.; Arunachalam, S.; Kumar, R.; Geyer, F.C.; et al. The Genomic Landscape of Mucinous Breast Cancer. J. Natl. Cancer Inst. 2019, 111, 737–741. [Google Scholar] [CrossRef]
- Carter, S.L.; Cibulskis, K.; Helman, E.; McKenna, A.; Shen, H.; Zack, T.; Laird, P.W.; Onofrio, R.C.; Winckler, W.; Weir, B.A.; et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 2012, 30, 413–421. [Google Scholar] [CrossRef]
- Golkaram, M.; Salmans, M.L.; Kaplan, S.; Vijayaraghavan, R.; Martins, M.; Khan, N.; Garbutt, C.; Wise, A.; Yao, J.; Casimiro, S.; et al. HERVs establish a distinct molecular subtype in stage II/III colorectal cancer with poor outcome. NPJ Genomic Med. 2021, 6, 13. [Google Scholar] [CrossRef]
- Sherry, S.T.; Ward, M.-H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef]
- Nguyen, B.; Fong, C.; Luthra, A.; Smith, S.A.; DiNatale, R.G.; Nandakumar, S.; Walch, H.; Chatila, W.K.; Madupuri, R.; Kundra, R.; et al. Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell 2022, 185, 563–575.e11. [Google Scholar] [CrossRef]
- Dawson, S.-J.; Tsui, D.W.Y.; Murtaza, M.; Biggs, H.; Rueda, O.M.; Chin, S.-F.; Dunning, M.J.; Gale, D.; Forshew, T.; Mahler-Araujo, B.; et al. Analysis of Circulating Tumor DNA to Monitor Metastatic Breast Cancer. N. Engl. J. Med. 2013, 368, 1199–1209. [Google Scholar] [CrossRef]
- O’Leary, B.; Hrebien, S.; Morden, J.P.; Beaney, M.; Fribbens, C.; Huang, X.; Liu, Y.; Bartlett, C.H.; Koehler, M.; Cristofanilli, M.; et al. Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat. Commun. 2018, 9, 896. [Google Scholar] [CrossRef]
- Hrebien, S.; Citi, V.; Garcia-Murillas, I.; Cutts, R.; Fenwick, K.; Kozarewa, I.; McEwen, R.; Ratnayake, J.; Maudsley, R.; Carr, T.H.; et al. Early ctDNA dynamics as a surrogate for progression-free survival in advanced breast cancer in the BEECH trial. Ann. Oncol. 2019, 30, 945–952. [Google Scholar] [CrossRef] [PubMed]
- Pereira, B.; Chin, S.-F.; Rueda, O.M.; Vollan, H.-K.M.; Provenzano, E.; Bardwell, H.A.; Pugh, M.; Jones, L.; Russell, R.; Sammut, S.-J.; et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 2016, 7, 11479. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Sáez, O.; Chic, N.; Pascual, T.; Adamo, B.; Vidal, M.; González-Farré, B.; Sanfeliu, E.; Schettini, F.; Conte, B.; Brasó-Maristany, F.; et al. Frequency and spectrum of PIK3CA somatic mutations in breast cancer. Breast Cancer Res. 2020, 22, 45. [Google Scholar] [CrossRef]
- Isakoff, S.J.; Engelman, J.A.; Irie, H.Y.; Luo, J.; Brachmann, S.M.; Pearline, R.V.; Cantley, L.C.; Brugge, J.S. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005, 65, 10992–11000. [Google Scholar] [CrossRef] [PubMed]
- Bader, A.G.; Kang, S.; Vogt, P.K. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc. Natl. Acad. Sci. USA 2006, 103, 1475–1479. [Google Scholar] [CrossRef] [PubMed]
- Yates, L.R.; Knappskog, S.; Wedge, D.; Farmery, J.H.R.; Gonzalez, S.; Martincorena, I.; Alexandrov, L.B.; Van Loo, P.; Haugland, H.K.; Lilleng, P.K.; et al. Genomic Evolution of Breast Cancer Metastasis and Relapse. Cancer Cell 2017, 32, 169–184.e7. [Google Scholar] [CrossRef] [PubMed]
- Bardia, A.; Su, F.; Solovieff, N.; Im, S.-A.; Sohn, J.; Lee, K.S.; Campos-Gomez, S.; Jung, K.H.; Colleoni, M.; Vázquez, R.V.; et al. Genomic Profiling of Premenopausal HR+ and HER2- Metastatic Breast Cancer by Circulating Tumor DNA and Association of Genetic Alterations with Therapeutic Response to Endocrine Therapy and Ribociclib. JCO Precis. Oncol. 2021, 5, 1408–1420. [Google Scholar] [CrossRef]
- Topacio, B.R.; Zatulovskiy, E.; Cristea, S.; Xie, S.; Tambo, C.S.; Rubin, S.M.; Sage, J.; Kõivomägi, M.; Skotheim, J.M. Cyclin D-Cdk4,6 Drives Cell-Cycle Progression via the Retinoblastoma Protein’s C-Terminal Helix. Mol. Cell 2019, 74, 758–770.e4. [Google Scholar] [CrossRef] [PubMed]
- Elsheikh, S.; Green, A.R.; Aleskandarany, M.A.; Grainge, M.; Paish, C.E.; Lambros, M.B.K.; Reis-Filho, J.S.; Ellis, I.O. CCND1 amplification and cyclin D1 expression in breast cancer and their relation with proteomic subgroups and patient outcome. Breast Cancer Res. Treat. 2008, 109, 325–335. [Google Scholar] [CrossRef]
- Jeffreys, S.A.; Becker, T.M.; Khan, S.; Soon, P.; Neubauer, H.; de Souza, P.; Powter, B. Prognostic and Predictive Value of CCND1/Cyclin D1 Amplification in Breast Cancer With a Focus on Postmenopausal Patients: A Systematic Review and Meta-Analysis. Front. Endocrinol. 2022, 13, 895729. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Zwijsen, R.M.L.; Wientjens, E.; Klompmaker, R.; der Sman, J.; Bernards, R.; Michalides, R.J.A.M. CDK-independent activation of estrogen receptor by cyclin D1. Cell 1997, 88, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Hortobagyi, G.N.; Stemmer, S.M.; Burris, H.A.; Yap, Y.-S.; Sonke, G.S.; Hart, L.; Campone, M.; Petrakova, K.; Winer, E.P.; Janni, W.; et al. Overall Survival with Ribociclib plus Letrozole in Advanced Breast Cancer. N. Engl. J. Med. 2022, 386, 942–950. [Google Scholar] [CrossRef] [PubMed]
- De Laurentiis, M.; Caputo, R.; Mazza, M.; Mansutti, M.; Masetti, R.; Ballatore, Z.; Torrisi, R.; Michelotti, A.; Zambelli, A.; Ferro, A.; et al. Safety and Efficacy of Ribociclib in Combination with Letrozole in Patients with HR+, HER2− Advanced Breast Cancer: Results from the Italian Subpopulation of Phase 3b CompLEEment-1 Study. Target. Oncol. 2022, 17, 615–625. [Google Scholar] [CrossRef] [PubMed]
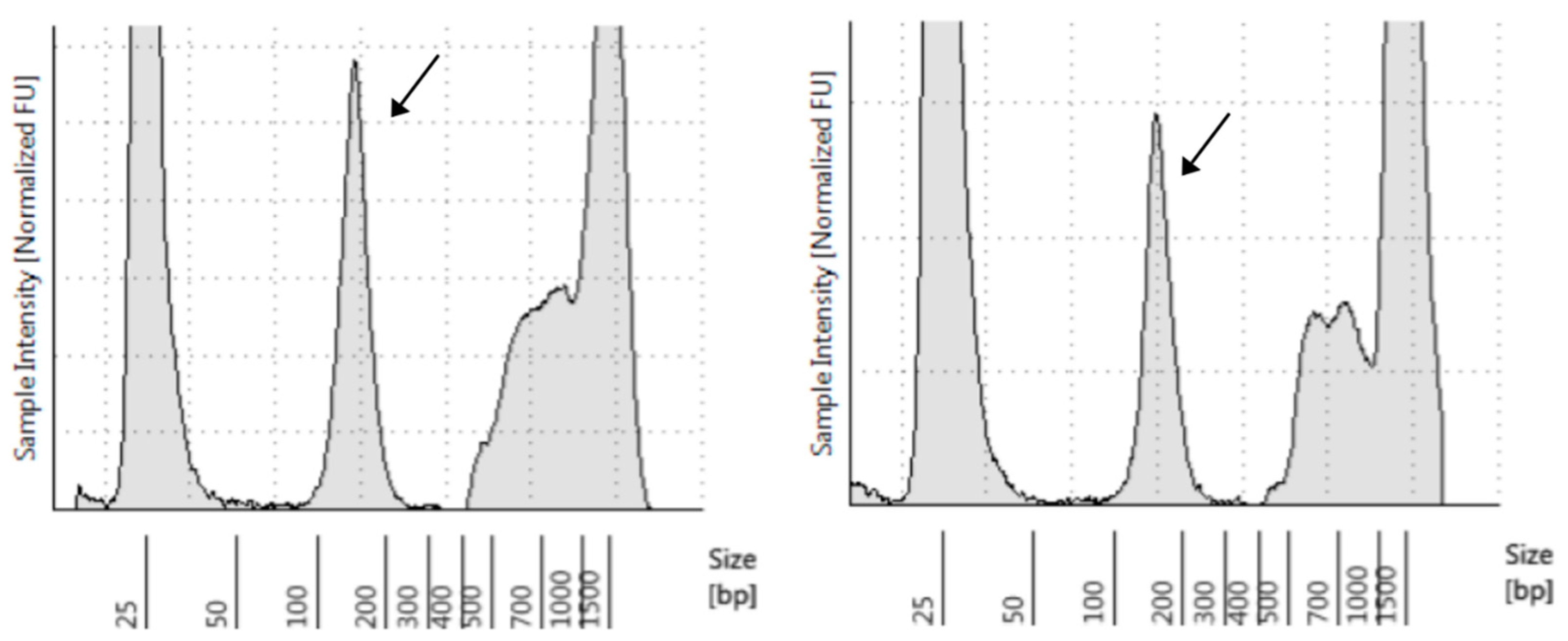
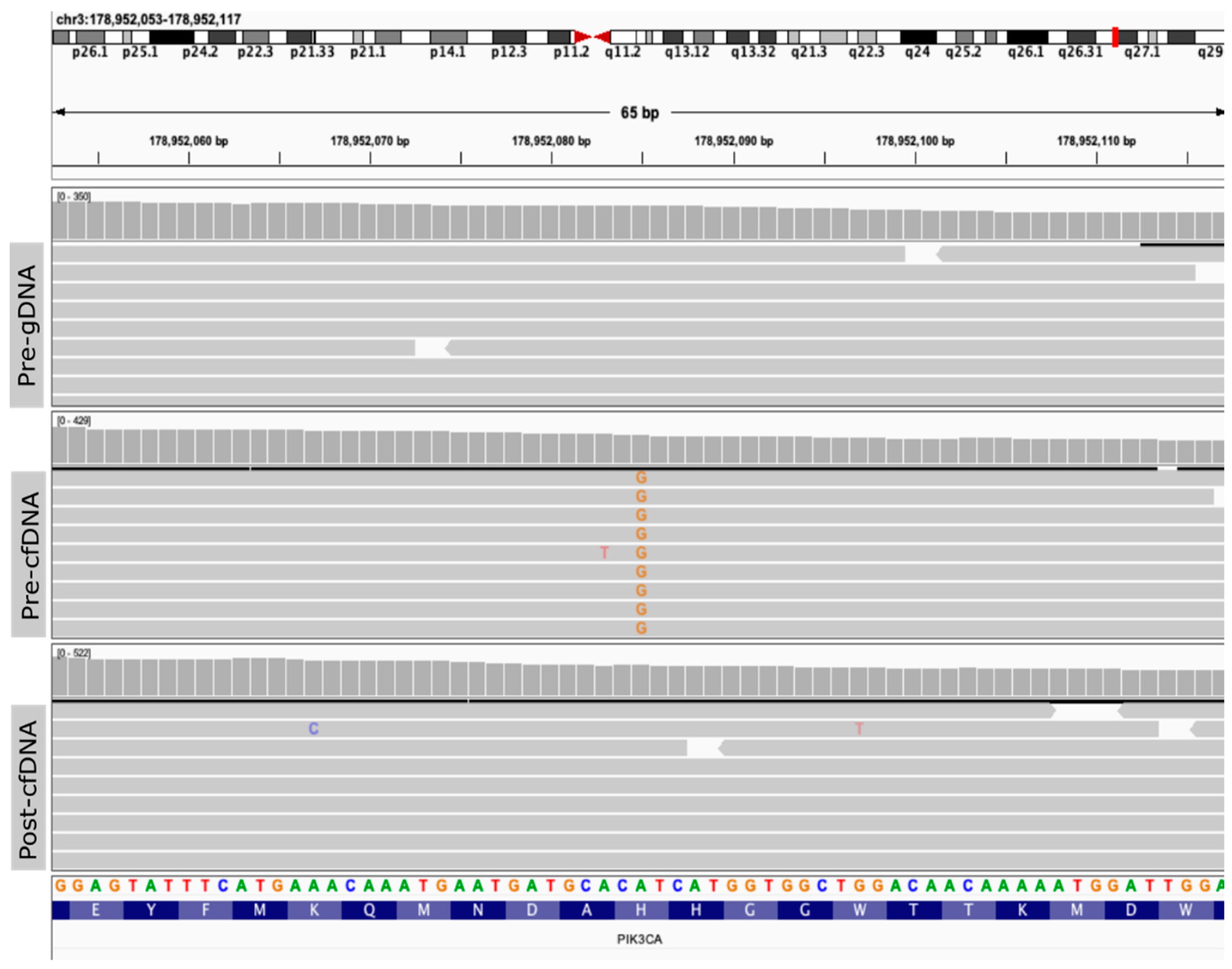
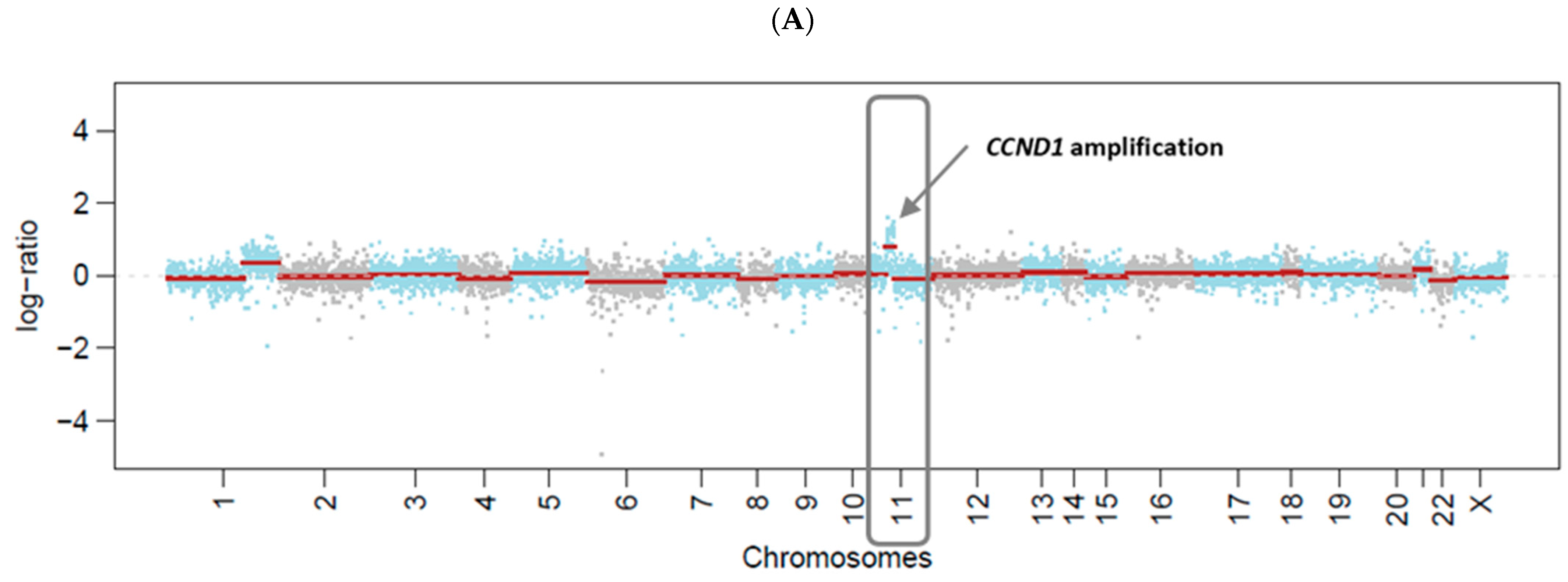

| Target Lesion | Lytic Bone Lesions, Right Iliac with Soft-Tissue Involvement | |||||||
|---|---|---|---|---|---|---|---|---|
| Follow-up date | 19 December 2017 | 13 March 2018 | 5 June 2018 | 27 September 2018 | 4 December 2018 | 11 March 2019 | 4 June 2019 | 29 August 2019 |
| Size | 81 mm | 58 mm | 57 mm | 54 mm | 48 mm | 47 mm | 46 mm | 42 mm |
| Non-target lesion | Lytic bone lesions, lumbar vertebral bodies | |||||||
| Follow-up date | 19 December 2017 | 13 March 2018 | 5 June 2018 | 27 September 2018 | 4 December 2018 | 11 March 2019 | 4 June 2019 | 29 August 2019 |
| Number | Multiple | Multiple | Stable | Stable | Stable | Stable | Stable | Stable |
| Sample ID | Collection Date | Sample Type | Sample Concentration(ng/uL) | Sample Volume(µL) |
|---|---|---|---|---|
| Pre gDNA | 18 September 2017 | Buffy-coat | 70.5 | 40.0 |
| Pre cfDNA | 18 September 2017 | Plasma (4 mL) | 0.7 | 45.0 |
| Post cfDNA | 25 June 2018 | Plasma (4 mL) | 0.3 | 45.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silveira, C.; Sousa, A.C.; Corredeira, P.; Martins, M.; Sousa, A.R.; Da Cruz Paula, A.; Selenica, P.; Brown, D.N.; Golkaram, M.; Kaplan, S.; et al. Comprehensive Genomic Profiling of Cell-Free Circulating Tumor DNA Detects Response to Ribociclib Plus Letrozole in a Patient with Metastatic Breast Cancer. Biomolecules 2022, 12, 1818. https://doi.org/10.3390/biom12121818
Silveira C, Sousa AC, Corredeira P, Martins M, Sousa AR, Da Cruz Paula A, Selenica P, Brown DN, Golkaram M, Kaplan S, et al. Comprehensive Genomic Profiling of Cell-Free Circulating Tumor DNA Detects Response to Ribociclib Plus Letrozole in a Patient with Metastatic Breast Cancer. Biomolecules. 2022; 12(12):1818. https://doi.org/10.3390/biom12121818
Chicago/Turabian StyleSilveira, Catarina, Ana Carla Sousa, Patrícia Corredeira, Marta Martins, Ana Rita Sousa, Arnaud Da Cruz Paula, Pier Selenica, David N. Brown, Mahdi Golkaram, Shannon Kaplan, and et al. 2022. "Comprehensive Genomic Profiling of Cell-Free Circulating Tumor DNA Detects Response to Ribociclib Plus Letrozole in a Patient with Metastatic Breast Cancer" Biomolecules 12, no. 12: 1818. https://doi.org/10.3390/biom12121818
APA StyleSilveira, C., Sousa, A. C., Corredeira, P., Martins, M., Sousa, A. R., Da Cruz Paula, A., Selenica, P., Brown, D. N., Golkaram, M., Kaplan, S., Zhang, S., Liu, L., Weigelt, B., Reis-Filho, J. S., Costa, L., & Carmo-Fonseca, M. (2022). Comprehensive Genomic Profiling of Cell-Free Circulating Tumor DNA Detects Response to Ribociclib Plus Letrozole in a Patient with Metastatic Breast Cancer. Biomolecules, 12(12), 1818. https://doi.org/10.3390/biom12121818








