Insight into Calcium-Binding Motifs of Intrinsically Disordered Proteins
Abstract
:1. Introduction
2. Materials and Methods
2.1. General Protein and Peptide Production
2.2. aSN Purification
2.3. ANAC046172-338 Purification
2.4. NHE1680-815 Production and Purification
2.5. DSS1 Purification
2.6. ProTα Purification
2.7. NMR Assignments
2.8. NMR Data Acquisition and Analysis
2.9. O-Cresolphthalein Complexone Measurements
2.10. Diffusion Ordered NMR Spectroscopy
2.11. Sequence Properties
3. Results
3.1. Negatively Charged IDPs Interact with Calcium in the Mid-Micromolar Range
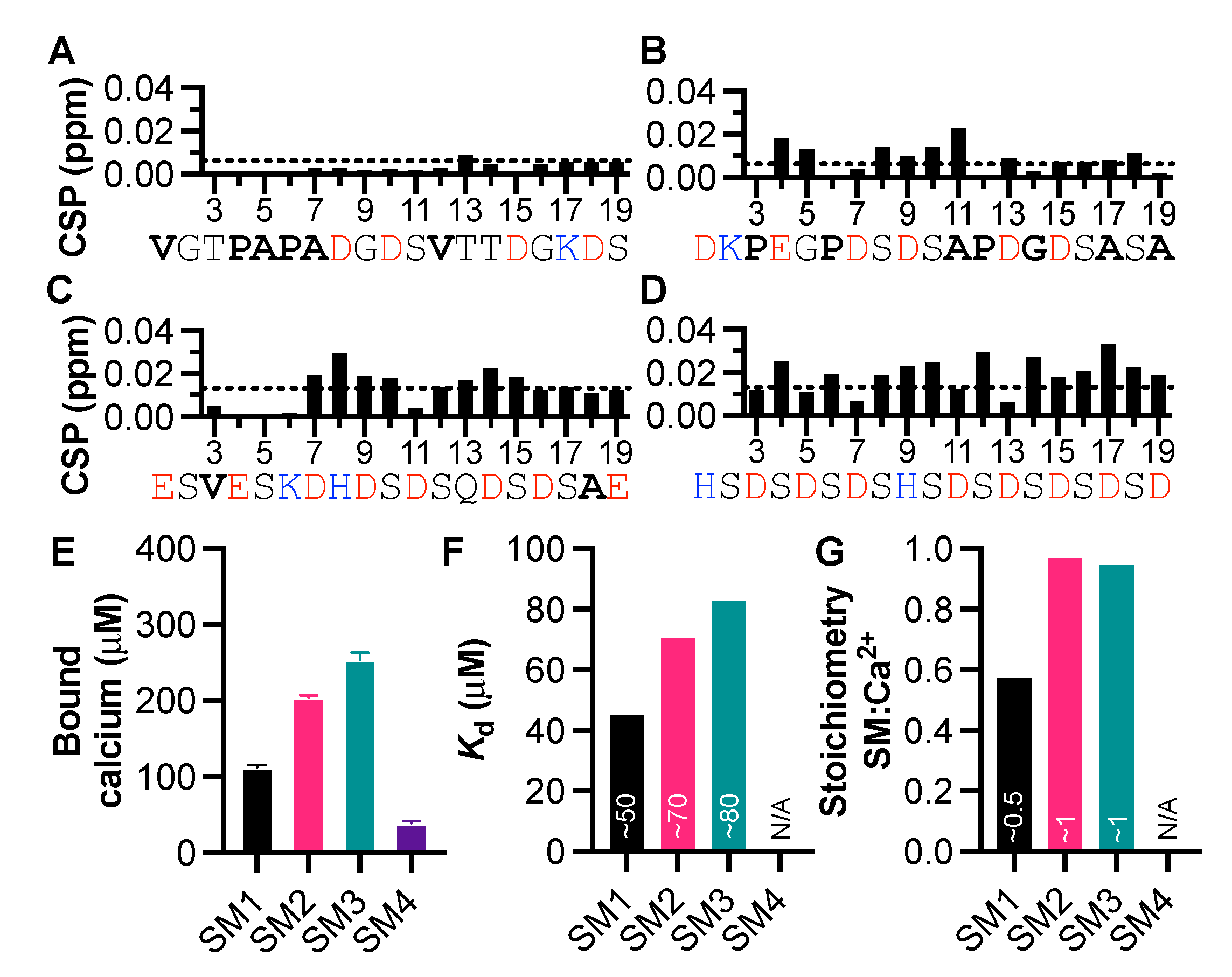
3.2. Peptides from the Starmaker Sequence Reveal the Importance of Negative Charge Distribution in Calcium Binding
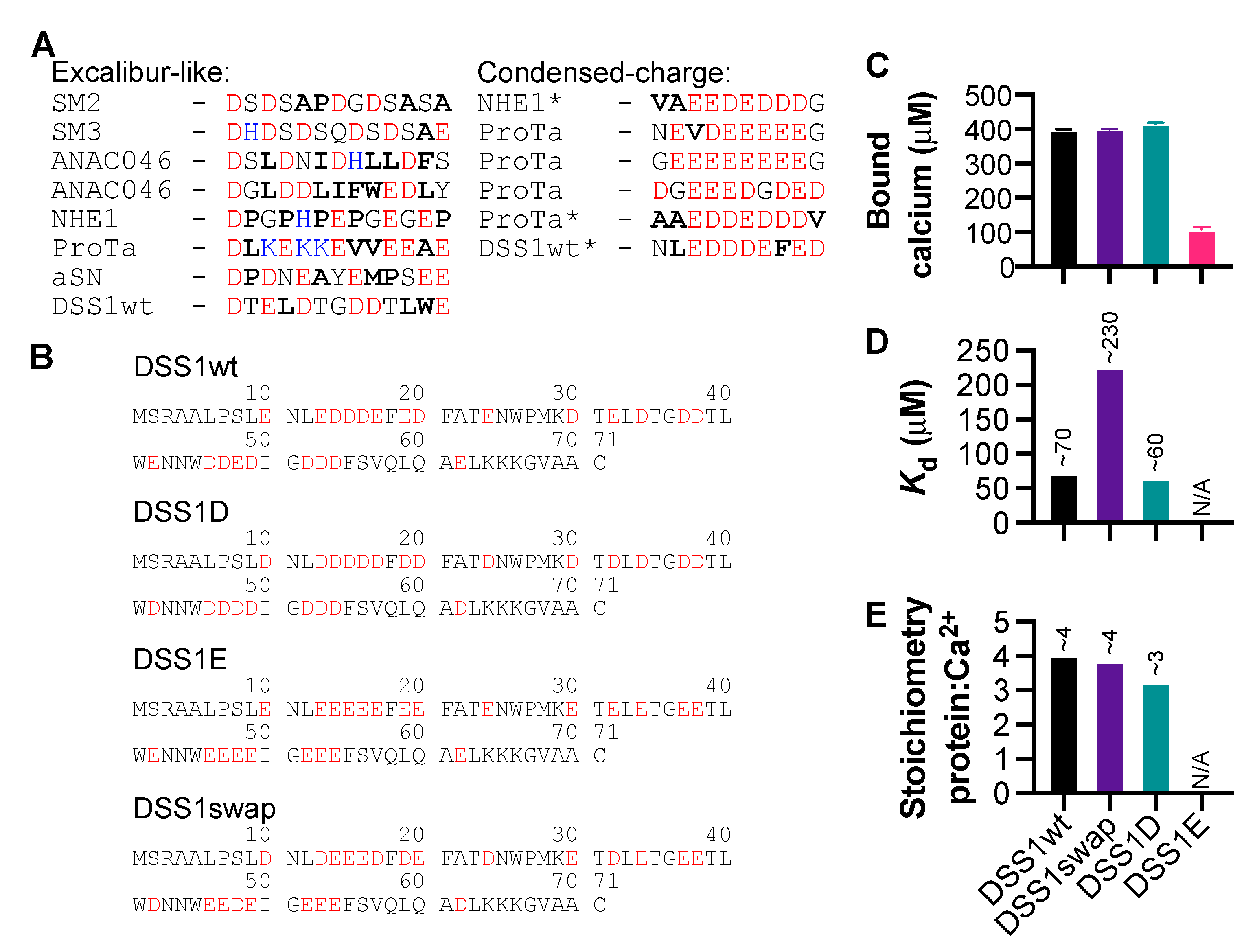
3.3. The Significance of Aspartic Acid in Calcium-Binding IDPs
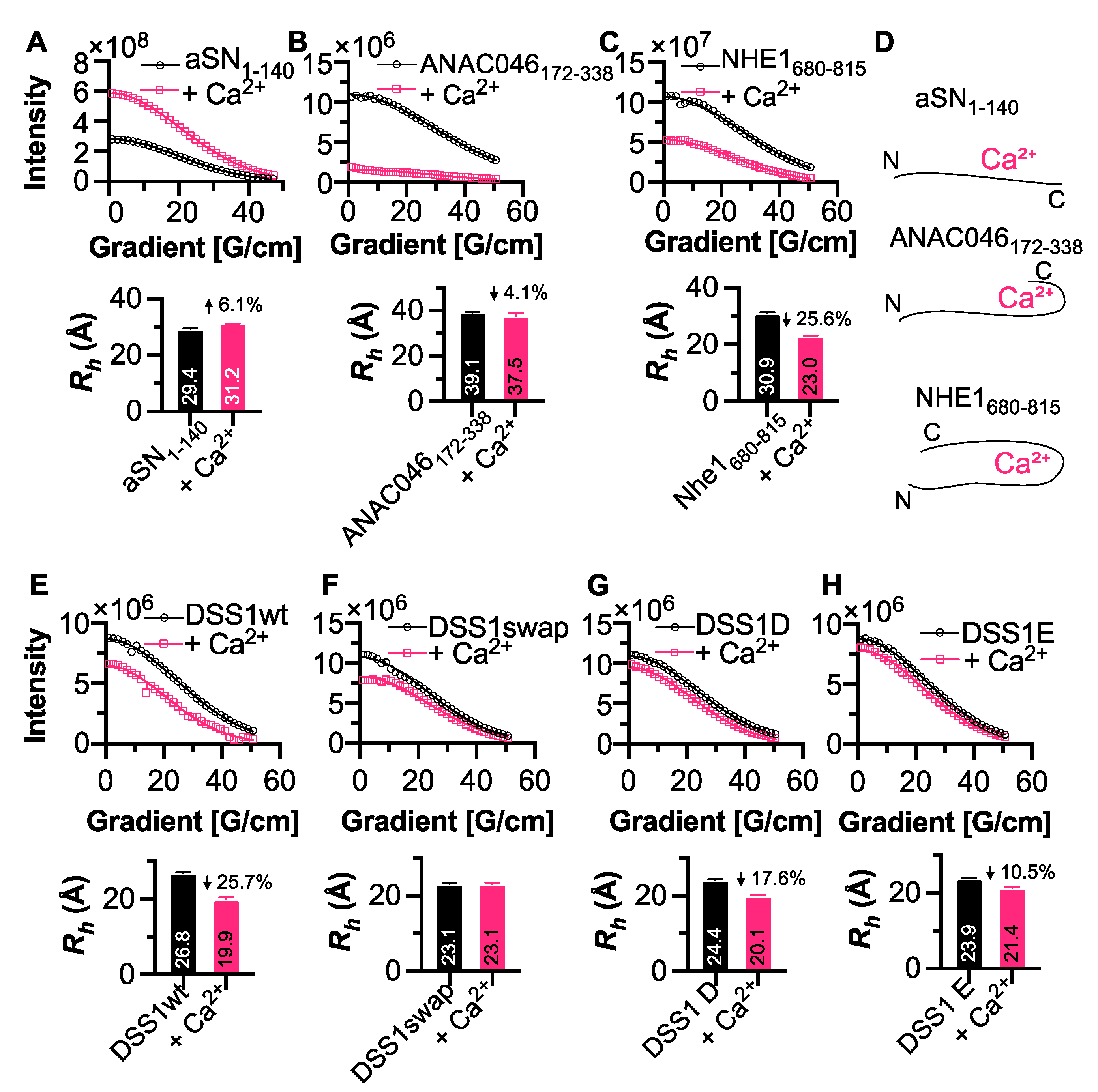
3.4. Conformational Changes Induced by Calcium Interaction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mao, A.H.; Crick, S.L.; Vitalis, A.; Chicoine, C.L.; Pappu, R.V. Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc. Natl. Acad. Sci. USA 2010, 107, 8183–8188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, R.M.; Obradovi, Z.; Mathura, V.; Braun, W.; Garner, E.C.; Young, J.; Takayama, S.; Brown, C.J.; Dunker, A.K. The protein non-folding problem: Amino acid determinants of intrinsic order and disorder. Pac. Symp. Biocomput. 2001, 6, 89–100. [Google Scholar] [CrossRef] [Green Version]
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins Struct. Funct. Genet. 2000, 41, 415–427. [Google Scholar] [CrossRef]
- Uversky, V.N. Intrinsically disordered proteins and their “Mysterious” (meta) physics. Front. Phys. 2019, 7, 10. [Google Scholar] [CrossRef] [Green Version]
- Martin, E.W.; Mittag, T. Relationship of Sequence and Phase Separation in Protein Low-Complexity Regions. Biochemistry 2018, 57, 2478–2487. [Google Scholar] [CrossRef]
- Seiffert, P.; Bugge, K.; Nygaard, M.; Haxholm, G.W.; Martinsen, J.H.; Pedersen, M.N.; Arleth, L.; Boomsma, W.; Kragelund, B.B. Orchestration of signaling by structural disorder in class 1 cytokine receptors. Cell Commun. Signal. 2020, 18, 132. [Google Scholar] [CrossRef]
- Camacho-Zarco, A.R.; Kalayil, S.; Maurin, D.; Salvi, N.; Delaforge, E.; Milles, S.; Jensen, M.R.; Hart, D.J.; Cusack, S.; Blackledge, M. Molecular basis of host-adaptation interactions between influenza virus polymerase PB2 subunit and ANP32A. Nat. Commun. 2020, 11, 3656. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Dalal, Y.; Papoian, G.A. Binding Dynamics of Disordered Linker Histone H1 with a Nucleosomal Particle. J. Mol. Biol. 2021, 433, 166881. [Google Scholar] [CrossRef]
- Borgia, A.; Borgia, M.B.; Bugge, K.; Kissling, V.M.; Heidarsson, P.O.; Fernandes, C.B.; Sottini, A.; Soranno, A.; Buholzer, K.J.; Nettels, D.; et al. Extreme disorder in an ultrahigh-affinity protein complex. Nature 2018, 555, 61–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, H.; Zhou, B.R.; Bai, Y. Binding Affinity and Function of the Extremely Disordered Protein Complex Containing Human Linker Histone H1.0 and Its Chaperone ProTα. Biochemistry 2018, 57, 6645–6648. [Google Scholar] [CrossRef]
- Bulyk, M.L.; Huang, X.; Choo, Y.; Church, G.M. Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc. Natl. Acad. Sci. USA 2001, 98, 7158–7163. [Google Scholar] [CrossRef] [Green Version]
- Hartwig, A. Role of magnesium in genomic stability. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2001, 475, 113–121. [Google Scholar] [CrossRef]
- Berridge, M.J.; Lipp, P.; Bootman, M.D. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 2000, 1, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Kirberger, M.; Qiu, F.; Chen, G.; Yang, J.J. Towards predicting Ca2+-binding sites with different coordination numbers in proteins with atomic resolution. Proteins 2009, 75, 787–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagur, R.; Hajnóczky, G. Intracellular Ca2+ Sensing: Its Role in Calcium Homeostasis and Signaling. Mol. Cell 2017, 66, 780–788. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Zhou, Y.; Yang, W.; Butters, R.; Lee, H.W.; Li, S.; Castiblanco, A.; Brown, E.M.; Yang, J.J. Identification and dissection of Ca2+-binding sites in the extracellular domain of Ca2+-sensing receptor. J. Biol. Chem. 2007, 282, 19000–19010. [Google Scholar] [CrossRef] [Green Version]
- Lewit-Bentley, A.; Réty, S. EF-hand calcium-binding proteins. Curr. Opin. Struct. Biol. 2000, 10, 637–643. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, W.; Kirberger, M.; Lee, H.W.; Ayalasomayajula, G.; Yang, J.J. Prediction of EF-hand calcium-binding proteins and analysis of bacterial EF-hand proteins. Proteins Struct. Funct. Genet. 2006, 65, 643–655. [Google Scholar] [CrossRef]
- Rigden, D.J.; Galperin, M.Y. The DxDxDG motif for calcium binding: Multiple structural contexts and implications for evolution. J. Mol. Biol. 2004, 343, 971–984. [Google Scholar] [CrossRef]
- Rigden, D.J.; Jedrzejas, M.J.; Galperin, M.Y. An extracellular calcium-binding domain in bacteria with a distant relationship to EF-hands. FEMS Microbiol. Lett. 2003, 221, 103–110. [Google Scholar] [CrossRef] [Green Version]
- Hoyer, E.; Knöppel, J.; Liebmann, M.; Steppert, M.; Raiwa, M.; Herczynski, O.; Hanspach, E.; Zehner, S.; Göttfert, M.; Tsushima, S.; et al. Calcium binding to a disordered domain of a type III-secreted protein from a coral pathogen promotes secondary structure formation and catalytic activity. Sci. Rep. 2019, 9, 7115. [Google Scholar] [CrossRef] [Green Version]
- Kapłon, T.M.; Rymarczyk, G.; Nocula-Ługowska, M.; Jakób, M.; Kochman, M.; Lisowski, M.; Szewczuk, Z.; Ozyhar, A. Starmaker exhibits properties of an intrinsically disordered protein. Biomacromolecules 2008, 9, 2118–2125. [Google Scholar] [CrossRef]
- Holehouse, A.S.; Das, R.K.; Ahad, J.N.; Richardson, M.O.G.; Pappu, R.V. CIDER: Resources to Analyze Sequence-Ensemble Relationships of Intrinsically Disordered Proteins. Biophys. J. 2017, 112, 16–21. [Google Scholar] [CrossRef] [Green Version]
- Wojtas, M.; Hołubowicz, R.; Poznar, M.; Maciejewska, M.; Ozyhar, A.; Dobryszycki, P. Calcium Ion Binding Properties and the Effect of Phosphorylation on the Intrinsically Disordered Starmaker Protein. Biochemistry 2015, 54, 6525–6534. [Google Scholar] [CrossRef] [PubMed]
- Han, J.Y.; Choi, T.S.; Kim, H.I. Molecular Role of Ca2+ and Hard Divalent Metal Cations on Accelerated Fibrillation and Interfibrillar Aggregation of α-Synuclein. Sci. Rep. 2018, 8, 1895. [Google Scholar] [CrossRef] [Green Version]
- Pedersen, C.P.; Seiffert, P.; Brakti, I.; Bugge, K. Production of intrinsically disordered proteins for biophysical studies: Tips and tricks. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2020; Volume 2141, pp. 195–209. [Google Scholar]
- Singh, K.K.; Graether, S.P. Expression and purification of an intrinsically disordered protein. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2020; Volume 2141, pp. 181–194. [Google Scholar]
- Cholak, E.; Bugge, K.; Khondker, A.; Gauger, K.; Pedraz-Cuesta, E.; Pedersen, M.E.; Bucciarelli, S.; Vestergaard, B.; Pedersen, S.F.; Rheinstädter, M.C.; et al. Avidity within the N-terminal anchor drives α-synuclein membrane interaction and insertion. FASEB J. 2020, 34, 7462–7482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kjaergaard, M.; Nørholm, A.B.; Hendus-Altenburger, R.; Pedersen, S.F.; Poulsen, F.M.; Kragelund, B.B. Temperature-dependent structural changes in intrinsically disordered proteins: Formation of α-helices or loss of polyproline II? Protein Sci. 2010, 19, 1555–1564. [Google Scholar] [CrossRef] [PubMed]
- Nørholm, A.B.; Hendus-Altenburger, R.; Bjerre, G.; Kjaergaard, M.; Pedersen, S.F.; Kragelund, B.B. The intracellular distal tail of the Na+/H+ exchanger NHE1 is intrinsically disordered: Implications for NHE1 trafficking. Biochemistry 2011, 50, 3469–3480. [Google Scholar] [CrossRef]
- Müller-Späth, S.; Soranno, A.; Hirschfeld, V.; Hofmann, H.; Rüegger, S.; Reymond, L.; Nettels, D.; Schuler, B. Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc. Natl. Acad. Sci. USA 2010, 107, 14609–14614. [Google Scholar] [CrossRef] [Green Version]
- Schenstrøm, S.M.; Rebula, C.A.; Tatham, M.H.; Hendus-Altenburger, R.; Jourdain, I.; Hay, R.T.; Kragelund, B.B.; Hartmann-Petersen, R. Expanded Interactome of the Intrinsically Disordered Protein Dss1. Cell Rep. 2018, 25, 862–870. [Google Scholar] [CrossRef]
- Kazimierczuk, K.; Orekhov, V.Y. Accelerated NMR spectroscopy by using compressed sensing. Angew. Chem. Int. Ed. 2011, 50, 5556–5559. [Google Scholar] [CrossRef] [PubMed]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef]
- Vranken, W.F.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, M.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins Struct. Funct. Genet. 2005, 59, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Connerty, H.V.; Briggs, A.R. Determination of serum calcium by means of orthocresolphthalein complexone. Am. J. Clin. Pathol. 1966, 45, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.H.; Chen, A.; Johnson, C.S. An Improved Diffusion-Ordered Spectroscopy Experiment Incorporating Bipolar-Gradient Pulses. J. Magn. Reson. Ser. A 1995, 115, 260–264. [Google Scholar] [CrossRef]
- Prestel, A.; Bugge, K.; Staby, L.; Hendus-Altenburger, R.; Kragelund, B.B. Characterization of Dynamic IDP Complexes by NMR Spectroscopy, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2018; Volume 611, ISBN 9780128156490. [Google Scholar]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Fauvet, B.; Mbefo, M.K.; Fares, M.B.; Desobry, C.; Michael, S.; Ardah, M.T.; Tsika, E.; Coune, P.; Prudent, M.; Lion, N.; et al. α-Synuclein in central nervous system and from erythrocytes, mammalian cells, and Escherichia coli exists predominantly as disordered monomer. J. Biol. Chem. 2012, 287, 15345–15364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cast, K.; Damaschun, H.; Eckert, K.; Schulze-Forster, K.; Rainer Maurer, H.; Muller-Frohne, M.; Zirwer, D.; Czamecki, J.; Damaschun, G. Prothymosin α: A Biologically Active Protein with Random Coil Conformation. Biochemistry 1995, 34, 13211–13218. [Google Scholar] [CrossRef]
- Lautenschläger, J.; Stephens, A.D.; Fusco, G.; Ströhl, F.; Curry, N.; Zacharopoulou, M.; Michel, C.H.; Laine, R.; Nespovitaya, N.; Fantham, M.; et al. C-terminal calcium binding of α-synuclein modulates synaptic vesicle interaction. Nat. Commun. 2018, 9, 712. [Google Scholar] [CrossRef] [Green Version]
- O’Shea, C.; Kryger, M.; Stender, E.G.P.; Kragelund, B.B.; Willemoës, M.; Skriver, K. Protein intrinsic disorder in Arabidopsis NAC transcription factors: Transcriptional activation by ANAC013 and ANAC046 and their interactions with RCD1. Biochem. J. 2015, 465, 281–294. [Google Scholar] [CrossRef]
- Chichkova, N.V.; Evstafieva, A.G.; Lyakhov, I.G.; Tsvetkov, A.S.; Smirnova, T.A.; Karapetian, R.N.; Karger, E.M.; Vartapetian, A.B. Divalent metal cation binding properties of human prothymosin. Eur. J. Biochem. 2000, 267, 4745–4752. [Google Scholar] [CrossRef]
- Grzybowska, E.A. Calcium-binding proteins with disordered structure and their role in secretion, storage, and cellular signaling. Biomolecules 2018, 8, 42. [Google Scholar] [CrossRef] [Green Version]
- Kalmar, L.; Homola, D.; Varga, G.; Tompa, P. Structural disorder in proteins brings order to crystal growth in biomineralization. Bone 2012, 51, 528–534. [Google Scholar] [CrossRef]
- Poznar, M.; Hołubowicz, R.; Wojtas, M.; Gapiński, J.; Banachowicz, E.; Patkowski, A.; Ożyhar, A.; Dobryszycki, P. Structural properties of the intrinsically disordered, multiple calcium ion-binding otolith matrix macromolecule-64 (OMM-64). Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 1358–1371. [Google Scholar] [CrossRef] [PubMed]
- Redwan, E.; Xue, B.; Almehdar, H.; Uversky, V. Disorder in Milk Proteins: Caseins, Intrinsically Disordered Colloids. Curr. Protein Pept. Sci. 2015, 16, 228–242. [Google Scholar] [CrossRef]
- Sotomayor-Pérez, A.C.; Ladant, D.; Chenal, A. Calcium-induced folding of intrinsically disordered Repeat-in-Toxin (RTX) motifs via changes of protein charges and oligomerization states. J. Biol. Chem. 2011, 286, 16997–17004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nara, M.; Morii, H.; Tanokura, M. Coordination to divalent cations by calcium-binding proteins studied by FTIR spectroscopy. Biochim. Biophys. Acta Biomembr. 2013, 1828, 2319–2327. [Google Scholar] [CrossRef] [Green Version]
- Mizuguchi, M.; Nara, M.; Kawano, K.; Nitta, K. FT-XR study of the Ca2+-binding to bovine α-lactalbumin. Relationships between the type of coordination and characteristics of the bands due to the Asp COO- groups in the Ca2+-binding site. FEBS Lett. 1997, 417, 153–156. [Google Scholar] [CrossRef] [Green Version]
- Olsen, J.G.; Teilum, K.; Kragelund, B.B. Behaviour of intrinsically disordered proteins in protein–protein complexes with an emphasis on fuzziness. Cell. Mol. Life Sci. 2017, 74, 3175–3183. [Google Scholar] [CrossRef]
- Garapati, S.; Monteith, W.; Wilson, C.; Kostenko, A.; Kenney, J.M.; Danell, A.S.; Burns, C.S. Zn2+-binding in the glutamate-rich region of the intrinsically disordered protein prothymosin-alpha. J. Biol. Inorg. Chem. 2018, 23, 1255–1263. [Google Scholar] [CrossRef]
- Pedersen, J.T.; Teilum, K.; Heegaard, N.H.H.; Østergaard, J.; Adolph, H.W.; Hemmingsen, L. Rapid formation of a preoligomeric peptide-metal-peptide complex following copper(II) binding to amyloid β peptides. Angew. Chem. Int. Ed. 2011, 50, 2532–2535. [Google Scholar] [CrossRef]
- Cragnell, C.; Staby, L.; Lenton, S.; Kragelund, B.B.; Skepö, M. Dynamical oligomerisation of histidine rich intrinsically disordered proteins is regulated through zinc-histidine interactions. Biomolecules 2019, 9, 168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faller, P.; Hureau, C.; La Penna, G. Metal ions and intrinsically disordered proteins and peptides: From Cu/Zn amyloid-β to general principles. Acc. Chem. Res. 2014, 47, 2252–2259. [Google Scholar] [CrossRef] [PubMed]
- Dutka, T.L.; Cole, L.; Lamb, G.D. Calcium phosphate precipitation in the sarcoplasmic reticulum reduces action potential-mediated Ca2+ release in mammalian skeletal muscle. Am. J. Physiol. Cell Physiol. 2005, 289, 1502–1512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kristian, T.; Pivovarova, N.B.; Fiskum, G.; Andrews, S.B. Calcium-induced precipitate formation in brain mitochondria: Composition, calcium capacity, and retention. J. Neurochem. 2007, 102, 1346–1356. [Google Scholar] [CrossRef] [Green Version]
- Stathopulos, P.B.; Li, G.Y.; Plevin, M.J.; Ames, J.B.; Ikura, M. Stored Ca2+ depletion-induced oligomerization of stromal interaction molecule 1 (STIM1) via the EF-SAM region: An initiation mechanism for capacitive Ca2+ entry. J. Biol. Chem. 2006, 281, 35855–35862. [Google Scholar] [CrossRef] [Green Version]
- Teilum, K.; Olsen, J.G.; Kragelund, B.B. On the specificity of protein-protein interactions in the context of disorder. Biochem. J. 2021, 478, 2035–2050. [Google Scholar] [CrossRef]
- Peran, I.; Mittag, T. Molecular structure in biomolecular condensates. Curr. Opin. Struct. Biol. 2020, 60, 17–26. [Google Scholar] [CrossRef]
- Lyon, A.S.; Peeples, W.B.; Rosen, M.K. A framework for understanding the functions of biomolecular condensates across scales. Nat. Rev. Mol. Cell Biol. 2021, 22, 215–235. [Google Scholar] [CrossRef]
- Dignon, G.L.; Best, R.B.; Mittal, J. Biomolecular phase separation: From molecular driving forces to macroscopic properties. Annu. Rev. Phys. Chem. 2020, 71, 53–75. [Google Scholar] [CrossRef] [Green Version]
- Mayfield, J.E.; Pollak, A.J.; Worby, C.A.; Xu, J.C.; Tandon, V.; Newton, A.C.; Dixon, J.E. Ca2+-dependent liquid-liquid phase separation underlies intracellular Ca2+ stores. bioRxiv 2021. [Google Scholar] [CrossRef]

| Protein | % D, E 1 | % K, R 1 | NCPR 1 | κ 1 | Hydropathy 1 | pI | MW |
|---|---|---|---|---|---|---|---|
| (kDa) | |||||||
| aSN96-140 | 33.3 | 6.7 | −0.27 | 0.17 | 2.93 | 3.76 | 5.09 |
| ANAC046172-338 | 10.1 | 4.2 | −0.07 | 0.2 | 4.11 | 4.66 | 17.97 |
| NHE1680-815 | 19.3 | 9.6 | −0.1 | 0.27 | 3.65 | 4.38 | 14.51 |
| DSS1wt | 32.4 | 7 | −0.25 | 0.39 | 3.53 | 3.67 | 8.1 |
| ProTa | 48.7 | 9 | −0.4 | 0.42 | 2.53 | 3.66 | 12.2 |
| Peptide | Sequence | % D, E 1 | % K, R 1 | NCPR 1 | κ 1 | Hydropathy 1 | pI | MW (kDa) |
|---|---|---|---|---|---|---|---|---|
| SM1 | VGTPAPADGDSVTTDGKDS | 21.1 | 5.3 | −0.16 | 0.22 | 3.76 | 3.77 | 1.79 |
| SM2 | DKPEGPDSDSAPDGDSASA | 31.6 | 5.3 | −0.26 | 0.16 | 3.01 | 3.61 | 1.82 |
| SM3 | ESVESKDHDSDSQDSDSAE | 42.1 | 5.3 | −0.37 | 0.16 | 2.53 | 3.80 | 2.07 |
| SM4 | HSDSDSDSHSDSDSDSDSD | 42.1 | 0 | −0.42 | 0.06 | 2.31 | 3.69 | 2.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Newcombe, E.A.; Fernandes, C.B.; Lundsgaard, J.E.; Brakti, I.; Lindorff-Larsen, K.; Langkilde, A.E.; Skriver, K.; Kragelund, B.B. Insight into Calcium-Binding Motifs of Intrinsically Disordered Proteins. Biomolecules 2021, 11, 1173. https://doi.org/10.3390/biom11081173
Newcombe EA, Fernandes CB, Lundsgaard JE, Brakti I, Lindorff-Larsen K, Langkilde AE, Skriver K, Kragelund BB. Insight into Calcium-Binding Motifs of Intrinsically Disordered Proteins. Biomolecules. 2021; 11(8):1173. https://doi.org/10.3390/biom11081173
Chicago/Turabian StyleNewcombe, Estella A., Catarina B. Fernandes, Jeppe E. Lundsgaard, Inna Brakti, Kresten Lindorff-Larsen, Annette E. Langkilde, Karen Skriver, and Birthe B. Kragelund. 2021. "Insight into Calcium-Binding Motifs of Intrinsically Disordered Proteins" Biomolecules 11, no. 8: 1173. https://doi.org/10.3390/biom11081173
APA StyleNewcombe, E. A., Fernandes, C. B., Lundsgaard, J. E., Brakti, I., Lindorff-Larsen, K., Langkilde, A. E., Skriver, K., & Kragelund, B. B. (2021). Insight into Calcium-Binding Motifs of Intrinsically Disordered Proteins. Biomolecules, 11(8), 1173. https://doi.org/10.3390/biom11081173







