Screening Plasma Exosomal RNAs as Diagnostic Markers for Cervical Cancer: An Analysis of Patients Who Underwent Primary Chemoradiotherapy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Blood Samples and Clinical Data
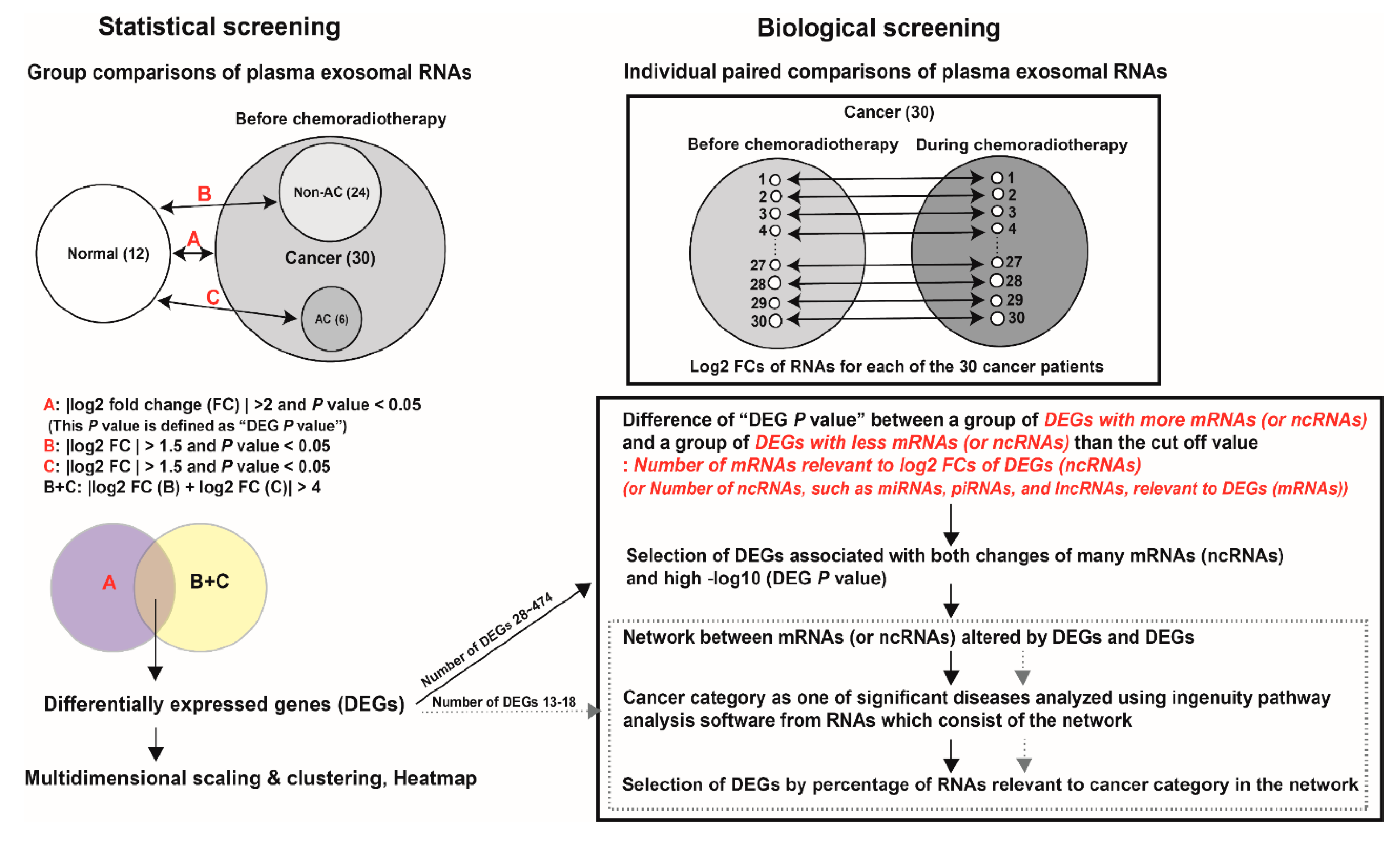
2.2. Screening Process
2.2.1. Statistical Screening
2.2.2. Biological Screening
2.3. Statistical Analysis of Differential RNA Expression
2.4. Multidimensional Scaling and Heatmap Construction
2.5. Network Analysis
2.6. Ingenuity Pathway Analysis
2.7. Receiver Operative Characteristic Analysis
2.8. Table and Boxplots
3. Results
3.1. Clinical Characteristics
3.2. Statistical Screening
3.3. Biological Screening
3.3.1. miRNA
3.3.2. lncRNA
3.3.3. mRNA
3.3.4. snoRNA
3.3.5. piRNA, snRNA, tRNA, and yRNA
3.4. DEG in snoRNA
3.5. Integration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hong, S.; Won, Y.; Park, Y.R.; Jung, K.; Kong, H.; Lee, E.S. Cancer Statistics in Korea: Incidence, Mortality, Survival, and Prevalence in 2017. Cancer Res. Treat. Off. J. Korean Cancer Assoc. 2020, 52, 335. [Google Scholar] [CrossRef]
- Park, Y.; Vongdala, C.; Kim, J.; Ki, M. Changing trends in the incidence (1999–2011) and mortality (1983–2013) of cervical cancer in the Republic of Korea. Epidemiol. Health 2015, 37, e2015024. [Google Scholar] [CrossRef] [Green Version]
- Arbyn, M.; Weiderpass, E.; Bruni, L.; de Sanjosé, S.; Saraiya, M.; Ferlay, J.; Bray, F. Estimates of incidence and mortality of cervical cancer in 2018: A worldwide analysis. Lancet Glob. Health 2020, 8, e191–e203. [Google Scholar] [CrossRef] [Green Version]
- Koshiyama, M. The Effects of the Dietary and Nutrient Intake on Gynecologic Cancers. Healthcare 2019, 7, 88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plummer, M.; Peto, J.; Franceschi, S. International Collaboration of Epidemiological Studies of Cervical C. Time since first sexual intercourse and the risk of cervical cancer. Int. J. Cancer 2012, 130, 2638–2644. [Google Scholar] [CrossRef] [Green Version]
- Min, K.; Lee, Y.J.; Suh, M.; Yoo, C.W.; Lim, M.C.; Choi, J.; Ki, M.; Kim, Y.; Kim, J.; Kim, J.; et al. The Korean guideline for cervical cancer screening. J. Korean Med. Assoc. 2015, 58, 398–407. [Google Scholar] [CrossRef]
- Mayrand, M.; Duarte-Franco, E.; Rodrigues, I.; Walter, S.D.; Hanley, J.; Ferenczy, A.; Ratnam, S.; Coutle, F.; Francois, E.L. Human papillomavirus DNA versus Papanicolaou screening tests for cervical cancer. N. Engl. J. Med. 2007, 357, 1579–1588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, B.; Guo, J.; Sheng, Y.; Wu, G.; Zhao, Y. Human Papillomavirus-Negative Cervical Cancer: A Comprehensive Review. Front. Oncol. 2020, 10, 3007. [Google Scholar] [CrossRef] [PubMed]
- van der Pol, E.; Böing, A.N.; Harrison, P.; Sturk, A.; Nieuwland, R. Classification, Functions, and Clinical Relevance of Extracellular Vesicles. Pharmacol. Rev. 2012, 64, 676–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, B.D.; Wong, W.; Lee, M.M.; Cho, W.C.; Yee, B.K.; Kwan, Y.W.; Tai, W.C. Exosomes in Inflammation and Inflammatory Disease. Proteomics 2019, 19, 1800149. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yu, F.; Ding, H.; Wang, Y.; Li, P.; Wang, K. Emerging Function and Clinical Values of Exosomal MicroRNAs in Cancer. Mol. Ther. Nucleic Acids 2019, 16, 791–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, M.; Hou, L.; Ma, Y.; Zhou, L.; Wang, F.; Cheng, B.; Wang, W.; Lu, B.; Liu, P.; Lu, W.; et al. Exosomal let-7d-3p and miR-30d-5p as diagnostic biomarkers for non-invasive screening of cervical cancer and its precursors. Mol. Cancer 2019, 18, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, T.; Taniuchi, K.; Tsuboi, M.; Sakaguchi, M.; Kohsaki, T.; Okabayashi, T.; Saibara, T. Circulating pancreatic cancer exosomal RNAs for detection of pancreatic cancer. Mol. Oncol. 2019, 13, 212–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, J.; Chen, R.; Zhao, L.; Xu, Y.; Cao, Z.; Xu, H.; Chen, X.; Shi, X.; Zhu, Y.; Lyu, J.; et al. Circulating exosomal mRNA profiling identifies novel signatures for the detection of prostate cancer. Mol. Cancer 2021, 20, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Yu, L.; Mei, Y.; Guarnera, M.; Shen, J.; Li, R.; Liu, Z.; Jiang, F. Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol. Cancer 2010, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Romano, G.; Veneziano, D.; Acunzo, M.; Croce, C.M. Small non-coding RNA and cancer. Carcinogenesis 2017, 38, 485–491. [Google Scholar] [CrossRef] [Green Version]
- Cho, O.; Kim, D.W.; Cheong, J.Y. Plasma Exosomal miRNA Levels after Radiotherapy Are Associated with Early Progression and Metastasis of Cervical Cancer: A Pilot Study. J. Clin. Med. 2021, 10, 2110. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Deng, B.; Zhang, Y.; Zhang, S.; Wen, F.; Miao, Y.; Guo, K. MicroRNA-142-3p inhibits cell proliferation and invasion of cervical cancer cells by targeting FZD7. Tumor Biol. 2015, 36, 8065–8073. [Google Scholar] [CrossRef]
- Li, M.; Li, B.Y.; Xia, H.; Jiang, L.L. Expression of microRNA-142-3p in cervical cancer and its correlation with prognosis. Eur. Rev. Med. Pharm. Sci. 2017, 21, 2346–2350. [Google Scholar]
- Cunha, I.W.; Carvalho, K.C.; Martins, W.K.; Marques, S.M.; Muto, N.H.; Falzoni, R.; Rocha, R.M.; Jr, S.; Simoes, A.C.Q.; Fahham, L.; et al. Identification of genes associated with local aggressiveness and metastatic behavior in soft tissue tumors. Transl. Oncol. 2010, 3, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, H.; Sun, M.; Deng, X.; Wu, X.; Ma, Y.; Li, M.; Shuoa, S.M.; You, Q.; Miao, L. CXCL5/CXCR2 axis in tumor microenvironment as potential diagnostic biomarker and therapeutic target. Cancer Commun. 2020, 40, 69–80. [Google Scholar] [CrossRef] [Green Version]
- Bai, L.; Yao, N.; Qiao, G.; Wu, L.; Ma, X. CXCL5 contributes to the tumorigenicity of cervical cancer and is post-transcriptionally regulated by miR-577. Int. J. Clin. Exp. Pathol. 2020, 13, 2984–2993. [Google Scholar]
- Feng, X.; Zhang, D.; Li, X.; Ma, S.; Zhang, C.; Wang, J.; Li, Y.; Liang, L.; Zhang, P.; Qu, Y.; et al. CXCL5, the upregulated chemokine in patients with uterine cervix cancer, in vivo and in vitro contributes to oncogenic potential of Hela uterine cervix cancer cells. Biomed. Pharmacotherapy 2018, 107, 1496–1504. [Google Scholar] [CrossRef]
- Li, X.; Shu, K.; Wang, Z.; Ding, D. Prognostic significance of KIF2A and KIF20A expression in human cancer: A systematic review and meta-analysis. Medecine 2019, 98, e18040. [Google Scholar] [CrossRef] [PubMed]
- Lei, G.; Xin, X.; Hu, X. Clinical significance of kinesin family member 2A as a facilitating biomarker of disease surveillance and prognostication in cervical cancer patients. Ir. J. Med. Sci. (1971-) 2021. [Google Scholar] [CrossRef] [PubMed]
- Delesque-Touchard, N.; Pendaries, C.; Volle-Challier, C.; Millet, L.; Salel, V.; Herve, C.; Pflieger, A.-M.; Berthou-Soulie, L.; Prades, C.; Sorg, T.; et al. Regulator of G-protein signaling 18 controls both platelet generation and function. PLoS ONE 2014, 9, e113215. [Google Scholar] [CrossRef]
- DeHelian, D.J.; Gupta, S.; Wu, J.; Thorsheim, C.L.; Estevez, B.; Cooper, M.; Litts, K.; Lee-Sundlov, M.M.; Hoffmeister, K.M.; Poncz, M.; et al. RGS10 and RGS18 differentially limit platelet activation, promote platelet production, and prolong platelet survival. Blood 2020, 136, 1773–1782. [Google Scholar] [CrossRef]
- Hu, Y.; Zheng, M.; Wang, S.; Gao, L.; Gou, R.; Liu, O.; Dong, H.; Li, X.; Lin, B. Identification of a five-gene signature of the RGS gene family with prognostic value in ovarian cancer. Genomics 2021, 113, 2134–2144. [Google Scholar] [CrossRef]
- Al-Alwan, M.; Hou, S.; Zhang, T.T.; Makondo, K.; Marshall, A.J. Bam32/DAPP1 promotes B cell adhesion and formation of polarized conjugates with T cells. J. Immunol. 2010, 184, 6961–6969. [Google Scholar] [CrossRef] [Green Version]
- Best, M.G.; Sol, N.; Kooi, I.E.; Tannous, J.; Westerman, B.A.; Rustenburg, F.; Schellen, P.; Verschueren, H.; Post, E.; Koster, J.; et al. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell 2015, 28, 666–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chow, R.D.; Chen, S. Sno-derived RNAs are prevalent molecular markers of cancer immunity. Oncogene 2018, 37, 6442–6462. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Liang, X.Z.; Deng, Y.; Liang, Y.B.; Zhu, X.; Liang, X.Y.; Luo, D.-Z.; Chen, G.; Fang, Y.-Y.; Lan, H.-H.; et al. Prognostic value of small nucleolar RNAs (snoRNAs) for colon adenocarcinoma based on RNA sequencing data. Pathol. Res. Pract. 2020, 216, 152937. [Google Scholar] [CrossRef]
- Gao, L.; Ma, J.; Mannoor, K.; Guarnera, M.A.; Shetty, A.; Zhan, M.; Xing, L.; Stass, S.A.; Jiang, F. Genome-wide small nucleolar RNA expression analysis of lung cancer by next-generation deep sequencing. Int. J. Cancer 2015, 136, E623–E629. [Google Scholar] [CrossRef] [PubMed]
- Crea, F.; Quagliata, L.; Michael, A.; Liu, H.H.; Frumento, P.; Azad, A.A.; Xue, H.; Pikor, L.; Watahiki, A.; Morant, R.; et al. Integrated analysis of the prostate cancer small-nucleolar transcriptome reveals SNORA55 as a driver of prostate cancer progression. Mol. Oncol. 2016, 10, 693–703. [Google Scholar] [CrossRef]
- Roychowdhury, A.; Samadder, S.; Das, P.; Mazumder, D.I.; Chatterjee, A.; Addya, S.; Mondal, R.; Roy, A.; Roychoudhury, S.; Panda, C.K. Deregulation of H19 is associated with cervical carcinoma. Genomics 2020, 112, 961–970. [Google Scholar] [CrossRef]
- Hu, B.; Fan, H.; Lv, X.; Chen, S.; Shao, Z. Prognostic significance of CXCL5 expression in cancer patients: A meta-analysis. Cancer Cell Int. 2018, 18, 68. [Google Scholar] [CrossRef] [Green Version]
- Haemmerle, M.; Stone, R.L.; Menter, D.G.; Afshar-Kharghan, V.; Sood, A.K. The Platelet Lifeline to Cancer: Challenges and Opportunities. Cancer Cell 2018, 33, 965–983. [Google Scholar] [CrossRef] [Green Version]
- Driedonks, T.A.P.; van der Grein, S.G.; Ariyurek, Y.; Buermans, H.P.J.; Jekel, H.; Chow, F.W.N.; Wauben, M.H.M.; Buck, A.H.; AC‘t Hoen, P.; Nolte, E.N.M.; et al. Immune stimuli shape the small non-coding transcriptome of extracellular vesicles released by dendritic cells. Cell. Mol. Life Sci. 2018, 75, 3857–3875. [Google Scholar] [CrossRef] [Green Version]
- Rimer, J.M.; Lee, J.; Holley, C.L.; Crowder, R.J.; Chen, D.L.; Hanson, P.I.; Ory, D.S.; Schaffer, J.E. Long-range function of secreted small nucleolar RNAs that direct 2′-O-methylation. J. Biol. Chem. 2018, 293, 13284–13296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef] [PubMed]
- Cho, O.; Chun, M.; Oh, Y.T.; Noh, O.K.; Chang, S.J.; Ryu, H.S.; Lee, E.J. Prognostic implication of simultaneous anemia and lymphopenia during concurrent chemoradiotherapy in cervical squamous cell carcinoma. Tumour Biol. 2017, 39, 1010428317733144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, O.; Chun, M.; Chang, S.J.; Oh, Y.T.; Noh, O.K. Prognostic Value of Severe Lymphopenia During Pelvic Concurrent Chemoradiotherapy in Cervical Cancer. Anticancer Res. 2016, 36, 3541–3547. [Google Scholar]




| Normal | Cancer | DEG in snoRNA | p | ||
|---|---|---|---|---|---|
| (n = 12) | (n = 30) | No (n = 18) | Yes (n = 12) | ||
| Age (years) | 49.2 ± 11.6 | 49.9 ± 10.1 | 47.9 ± 11.4 | 52.8 ± 7.1 | 0.199 |
| FIGO stage 2018 | 0.627 | ||||
| 5 (16.7%) | 4 (22.2%) | 1 (8.3%) | ||
| 15 (50.0%) | 9 (50.0%) | 6 (50.0%) | ||
| 7 (23.3%) | 4 (22.2%) | 3 (25.0%) | ||
| 3 (10.0%) | 1 (5.6%) | 2 (16.7%) | ||
| Pathology | 0.374 | ||||
| 5 (16.7%) | 4 (22.2%) | 1 (8.3%) | ||
| 1 (3.3%) | 0 (0.0%) | 1 (8.3%) | ||
| 1 (3.3%) | 1 (5.6%) | 0 (0.0%) | ||
| 23 (76.7%) | 13 (72.2%) | 10 (83.3%) | ||
| Radiotherapy field | 0.464 | ||||
| Pelvis | 21 (70.0%) | 14 (77.8%) | 7 (58.3%) | ||
| Pelvis with paraaortic region | 9 (30.0%) | 4 (22.2%) | 5 (41.7%) | ||
| Hemoglobin (g/dl) | |||||
| Pretreatment | 12.1 ± 1.5 | 12.0 ± 1.5 | 12.2 ± 1.6 | 0.78 | |
| Second week during CCRT | 11.1 ± 1.4 | 11.3 ± 1.2 | 10.8 ± 1.7 | 0.336 | |
| Absolute lymphocyte count (cells/μL) | |||||
| Pretreatment | 1754 ± 470 | 1758 ± 451 | 1747 ± 518 | 0.95 | |
| First week after CCRT | 931 ± 393 | 929 ± 281 | 936 ± 546 | 0.966 | |
| Second week after CCRT | 511 [371; 632] | 575 [505; 661] | 384 [323; 466] | 0.008 | |
| Pretreatment tumor marker (ng/mL) | |||||
| Squamous cell carcinoma antigen | 3.7 [0.9; 16.6] | 2.2 [0.8; 4.8] | 13.1 [4.0; 60.6] | 0.016 | |
| Cytokeratin fragment 21-1 | 2.5 [1.8; 10.2] | 2.2 [1.2; 2.8] | 8.4 [2.5; 16.6] | 0.031 | |
| Pretreatment tumor volume (cm3) | 50.5 [18.1; 94.1] | 40.6 [15.2; 94.1] | 61.0 [30.9; 103.3] | 0.346 | |
| RNA | Known Biological Functions | Tissue | Suggested Biological Functions | Exosome |
|---|---|---|---|---|
| miR-142-3p | Tumor suppressor [20] | ↓(CC) [21] | Tumor suppressor | ↓ |
| ARL6IP5 | Tumor suppressor (https://bioinfo.uth.edu/TSGene/ accessed on 1 September 2021) | ↓(STT) [22] | Tumor suppressor | ↓ |
| CXCL5 | Recruits and activates granulocytes and promotes angiogenesis, tumor growth, and metastasis in the tumor microenvironment [23] | ↑(CC) [24,25] | Tumors with exosome-derived CXCL5 use it to facilitate their progression through infiltration of leukocytes in the tumor microenvironment | ↓ |
| KIF2A | Required for cell mitosis [26] | ↑(CC) [27] | Rapid mitosis of cancer cells may promote the absorption of KIF2A from exosomes | ↓ |
| RGS18 | Negative regulator of G protein-coupled receptors and controls platelet activation and production [28,29] | ↑(OC) [30] | Tumors may absorb RGS18 present in exosomes, which can promote thrombogenesis. The reduction of exosomal RGS18 by tumors may promote activated platelets around the primary tumor, which can facilitate tumor growth and invasion. Therefore, dysregulation of RGS18 can result in tumorigenesis through persistent platelet activation | ↓ |
| DAPP1 | Activation of antigen-specific T cells [31] | NA | This may contribute to tumorigenesis through deficiency of tumor-specific immunity | ↓ |
| LINC00989 | Decreases with RGS18 in tumor-educated platelets [32] | ↓(PaC) [32] | The two lncRNAs may facilitate platelet activation in cancer patients via targeting RGS18 | ↓ |
| LOC105374768 | NA | NA | ↓ | |
| SNORD17 | The derived RNA positively correlates with CD8 T cell infiltration in thymoma and stomach cancer [33] | ↑(COC) [34] | Promotion of these snoRNAs present in exosomes may be related to cancer related-lymphopenia | ↑ |
| SCARNA12 | NA | ↑(LC) [35] | ↑ | |
| SNORA6 | The derived RNA negatively correlates with CD8 T cell infiltration in LGG, PC, pancreatic cancer, and HNC [33] | ↑(PC) [36] | ↑ | |
| SNORA12 | NA | ↓(CC) [37] ↑(LC) [35] | ↑ | |
| SCARNA1 | NA | ↑(LC) [35] | ↑ | |
| SNORD97 | NA | ↓(CC) [37] | Promotion of these snoRNAs present in exosomes may be related to decreased lymphocyte activity | ↑ |
| SNORD62 | NA | NA | ↑ | |
| SNORD38A | The derived RNA negatively correlates with CD8 T cell infiltration in HNC, LC, TGCT, and PCPG [33] | ↑(COC) [34] | ↑ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, O.; Kim, D.-W.; Cheong, J.-Y. Screening Plasma Exosomal RNAs as Diagnostic Markers for Cervical Cancer: An Analysis of Patients Who Underwent Primary Chemoradiotherapy. Biomolecules 2021, 11, 1691. https://doi.org/10.3390/biom11111691
Cho O, Kim D-W, Cheong J-Y. Screening Plasma Exosomal RNAs as Diagnostic Markers for Cervical Cancer: An Analysis of Patients Who Underwent Primary Chemoradiotherapy. Biomolecules. 2021; 11(11):1691. https://doi.org/10.3390/biom11111691
Chicago/Turabian StyleCho, Oyeon, Do-Wan Kim, and Jae-Youn Cheong. 2021. "Screening Plasma Exosomal RNAs as Diagnostic Markers for Cervical Cancer: An Analysis of Patients Who Underwent Primary Chemoradiotherapy" Biomolecules 11, no. 11: 1691. https://doi.org/10.3390/biom11111691
APA StyleCho, O., Kim, D.-W., & Cheong, J.-Y. (2021). Screening Plasma Exosomal RNAs as Diagnostic Markers for Cervical Cancer: An Analysis of Patients Who Underwent Primary Chemoradiotherapy. Biomolecules, 11(11), 1691. https://doi.org/10.3390/biom11111691






