GFP Fusion to the N-Terminus of MotB Affects the Proton Channel Activity of the Bacterial Flagellar Motor in Salmonella
Abstract
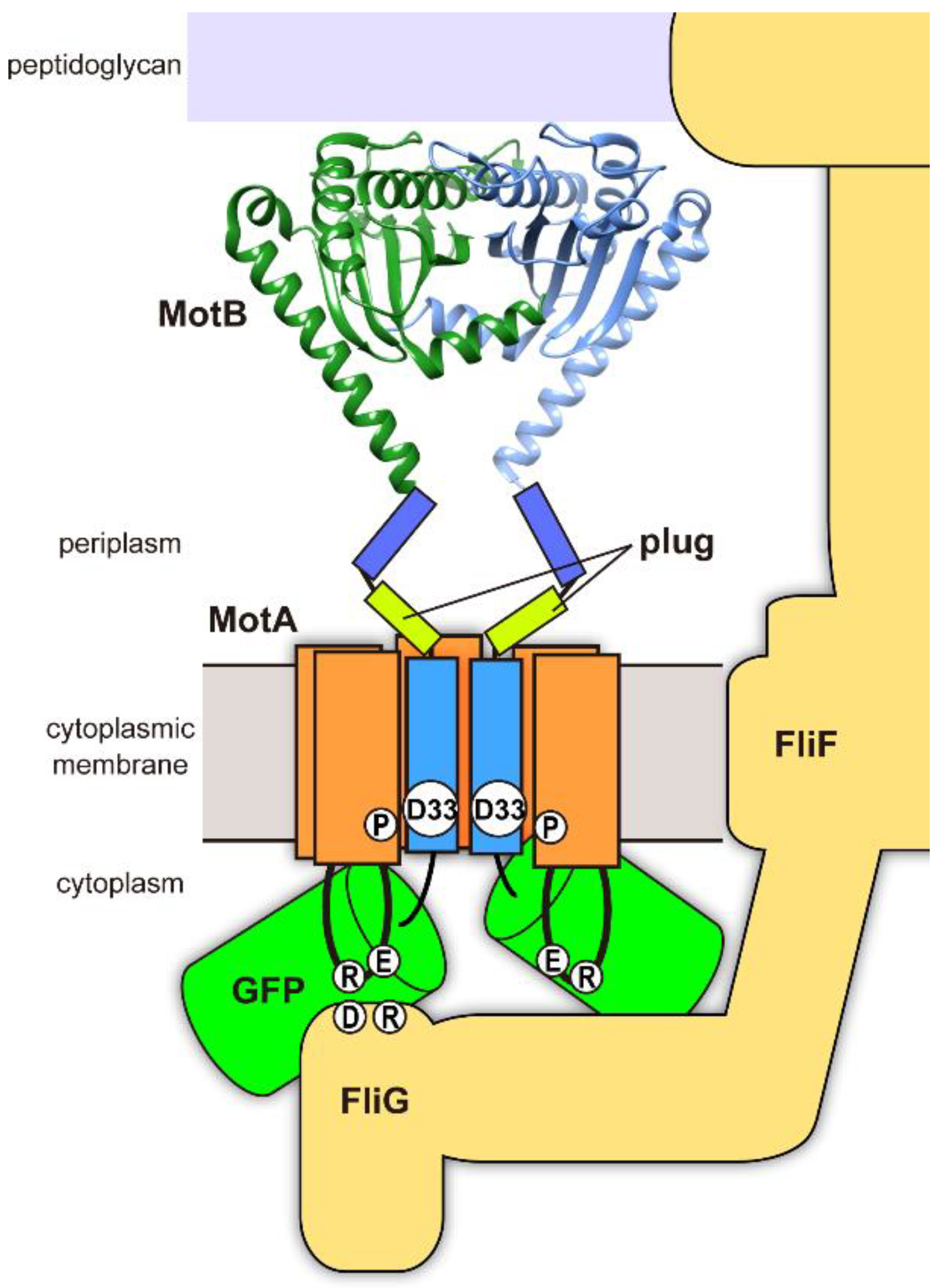
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids and Media
2.2. Cell Growth
2.3. Measurements of Free-Swimming Speeds of Bacterial Cells
2.4. Intracellular pH Measurement
3. Results
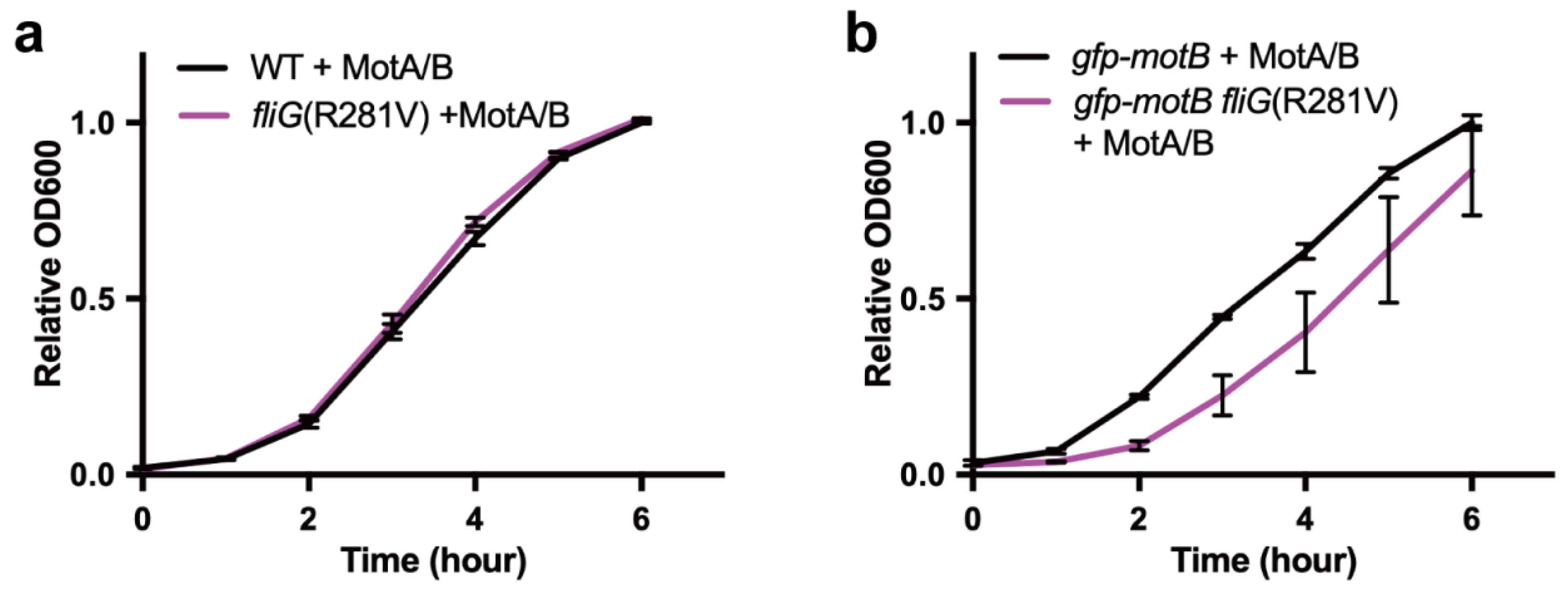
3.1. Effect of MotA/MotB Over-Expression on Cell Growth in gfp-motB fliG(R281V) Strain
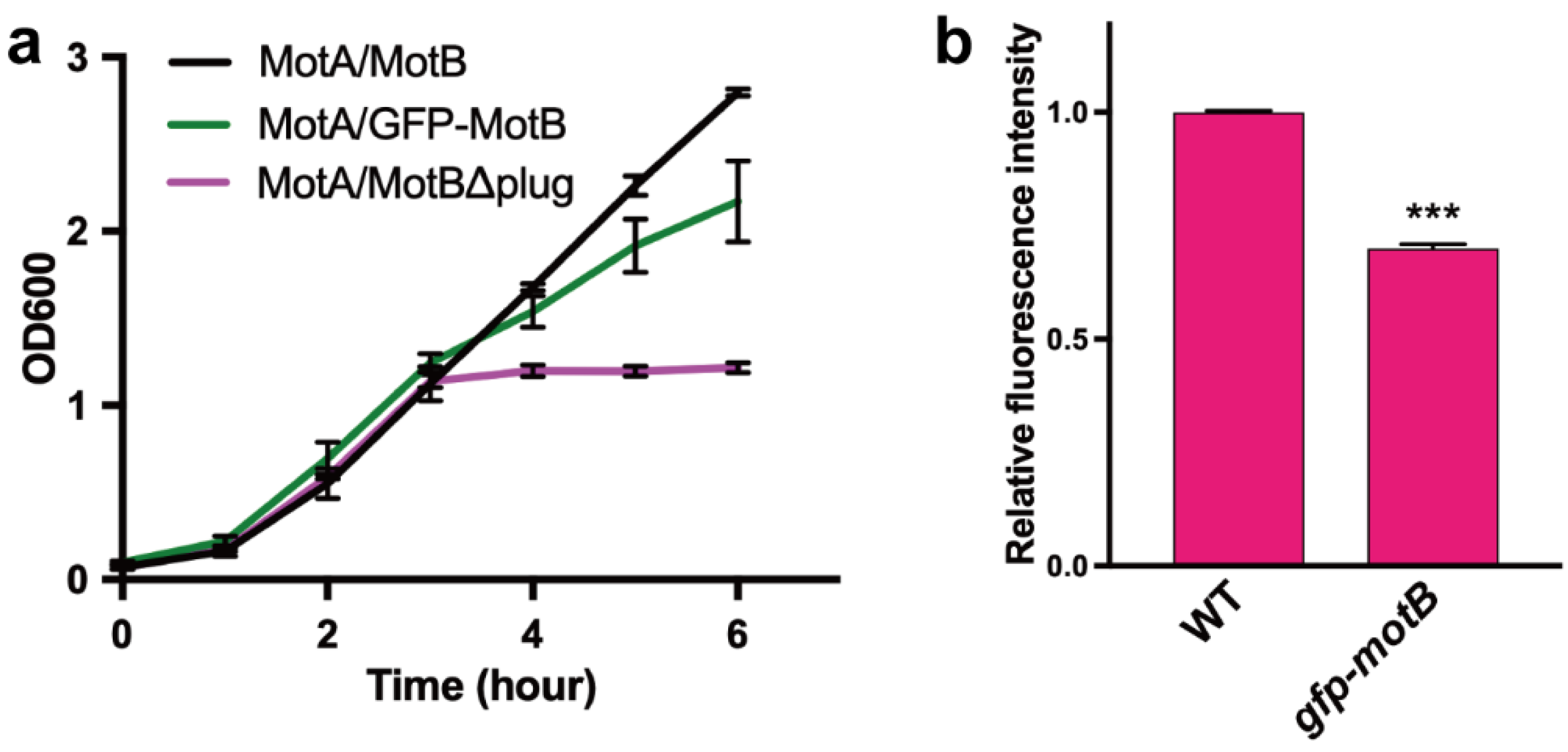
3.2. Effect of the GFP Tagging on the Proton Channel Activity of the MotA/MotB Complex
3.3. Multicopy Effect of Mutant MotA/MotB Complexes on Swimming Motility
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Minamino, T.; Namba, K. Self-assembly and type III protein export of the bacterial flagellum. J. Mol. Microbiol. Biotechnol. 2004, 7, 5–17. [Google Scholar] [CrossRef]
- Minamino, T.; Imada, K. The bacterial flagellar motor and its structural diversity. Trends Microbiol. 2015, 23, 267–274. [Google Scholar] [CrossRef]
- Berg, H.C. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 2003, 72, 19–54. [Google Scholar] [CrossRef]
- Kojima, S.; Blair, D.F. The bacterial flagellar motor: Structure and function of a complex molecular machine. Int. Rev. Cytol. 2004, 233, 93–134. [Google Scholar] [CrossRef] [PubMed]
- Sowa, Y.; Berry, R.M. Bacterial flagellar motor. Q. Rev. Biophys. 2008, 41, 103–132. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, Y.V.; Minamino, T. Structure and function of the bi-directional bacterial flagellar motor. Biomolecules 2014, 4, 217–234. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Minamino, T. Flagella-driven motility of bacteria. Biomolecules 2019, 9, 279. [Google Scholar] [CrossRef]
- Francis, N.R.; Sosinsky, G.E.; Thomas, D.; DeRosier, D.J. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 1994, 235, 1261–1270. [Google Scholar] [CrossRef]
- Minamino, T.; Kinoshita, M.; Namba, K. Directional switching mechanism of the bacterial flagellar motor. Comput. Struct. Biotechnol. J. 2019, 17, 1075–1081. [Google Scholar] [CrossRef]
- Braun, T.F.; Blair, D.F. Targeted disulfide cross-linking of the MotB protein of Escherichia coli: Evidence for two H+ channels in the stator Complex. Biochemistry 2001, 40, 13051–13059. [Google Scholar] [CrossRef]
- Braun, T.F.; Al-Mawsawi, L.Q.; Kojima, S.; Blair, D.F. Arrangement of core membrane segments in the MotA/MotB proton-channel complex of Escherichia coli. Biochemistry 2004, 43, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Kojima, S.; Blair, D.F. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry 2004, 43, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Leake, M.C.; Chandler, J.H.; Wadhams, G.H.; Bai, F.; Berry, R.M.; Armitage, J.P. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 2006, 443, 355–358. [Google Scholar] [CrossRef] [PubMed]
- Reid, S.W.; Leake, M.C.; Chandler, J.H.; Lo, C.J.; Armitage, J.P.; Berry, R.M. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc. Natl. Acad. Sci. USA 2006, 103, 8066–8071. [Google Scholar] [CrossRef] [PubMed]
- Minamino, T.; Terahara, N.; Kojima, S.; Namba, K. Autonomous control mechanism of stator assembly in the bacterial flagellar motor in response to changes in the environment. Mol. Microbiol. 2018, 109, 723–734. [Google Scholar] [CrossRef]
- Zhou, J.; Blair, D.F. Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J. Mol. Biol. 1997, 273, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Lloyd, S.A.; Blair, D.F. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 1998, 95, 6436–6441. [Google Scholar] [CrossRef] [PubMed]
- Sharp, L.L.; Zhou, J.; Blair, D.F. Tryptophan-scanning mutagenesis of MotB, an integral membrane protein essential for flagellar rotation in Escherichia coli. Biochemistry 1995, 34, 9166–9171. [Google Scholar] [CrossRef]
- Zhou, J.; Sharp, L.L.; Tang, H.L.; Lloyd, S.A.; Billings, S.; Braun, T.F.; Blair, D.F. Function of protonatable residues in the flagellar motor of Escherichia coli: A critical role for Asp 32 of MotB. J. Bacteriol. 1998, 180, 2729–2735. [Google Scholar] [CrossRef]
- Kojima, S.; Blair, D.F. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 2001, 40, 13041–13050. [Google Scholar] [CrossRef]
- Che, Y.S.; Nakamura, S.; Kojima, S.; Kami-ike, N.; Namba, K.; Minamino, T. Suppressor analysis of the MotB(D33E) mutation to probe bacterial flagellar motor dynamics coupled with proton translocation. J. Bacteriol. 2008, 190, 6660–6667. [Google Scholar] [CrossRef] [PubMed]
- De Mot, R.; Vanderleyden, J. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol. Microbiol. 1994, 12, 333–334. [Google Scholar] [CrossRef] [PubMed]
- Kojima, S.; Furukawa, Y.; Matsunami, H.; Minamino, T.; Namba, K. Characterization of the periplasmic domain of MotB and implications for its role in the stator assembly of the bacterial flagellar motor. J. Bacteriol. 2008, 190, 3314–3322. [Google Scholar] [CrossRef]
- Kojima, S.; Imada, K.; Sakuma, M.; Sudo, Y.; Kojima, C.; Minamino, T.; Homma, M.; Namba, K. Stator assembly and activation mechanism of the flagellar motor by the periplasmic region of MotB. Mol. Microbiol. 2009, 73, 710–718. [Google Scholar] [CrossRef] [PubMed]
- Hosking, E.R.; Vogt, C.; Bakker, E.P.; Manson, M.D. The Escherichia coli MotAB proton channel unplugged. J. Mol. Biol. 2006, 364, 921–937. [Google Scholar] [CrossRef]
- Morimoto, Y.V.; Che, Y.S.; Minamino, T.; Namba, K. Proton-conductivity assay of plugged and unplugged MotA/B proton channel by cytoplasmic pHluorin expressed in Salmonella. FEBS Lett. 2010, 584, 1268–1272. [Google Scholar] [CrossRef] [PubMed]
- Terahara, N.; Kodera, N.; Uchihashi, T.; Ando, T.; Namba, K.; Minamino, T. Na+-induced structural transition of MotPS for stator assembly of the Bacillus flagellar motor. Sci. Adv. 2017, 3, eaao4119. [Google Scholar] [CrossRef]
- Kojima, S.; Takao, M.; Almira, G.; Kawahara, I.; Sakuma, M.; Homma, M.; Kojima, C.; Imada, K. The helix rearrangement in the periplasmic domain of the flagellar stator B subunit activates peptidoglycan binding and ion influx. Structure 2018, 26, 590–598. [Google Scholar] [CrossRef]
- Togashi, F.; Yamaguchi, S.; Kihara, M.; Aizawa, S.I.; Macnab, R.M. An extreme clockwise switch bias mutation in fliG of Salmonella typhimurium and its suppression by slow-motile mutations in motA and motB. J. Bacteriol. 1997, 179, 2994–3003. [Google Scholar] [CrossRef]
- Muramoto, K.; Macnab, R.M. Deletion analysis of MotA and MotB, components of the force-generating unit in the flagellar motor of Salmonella. Mol. Microbiol. 1998, 29, 1191–1202. [Google Scholar] [CrossRef]
- Fukuoka, H.; Wada, T.; Kojima, S.; Ishijima, A.; Homma, M. Sodium-dependent dynamic assembly of membrane complexes in sodium-driven flagellar motors. Mol. Microbiol. 2009, 71, 825–835. [Google Scholar] [CrossRef] [PubMed]
- Delalez, N.J.; Wadhams, G.H.; Rosser, G.; Xue, Q.; Brown, M.T.; Dobbie, I.M.; Berry, R.M.; Leake, M.C.; Armitage, J.P. Signal-dependent turnover of the bacterial flagellar switch protein FliM. Proc. Natl. Acad. Sci. USA 2010, 107, 11347–11351. [Google Scholar] [CrossRef] [PubMed]
- Fukuoka, H.; Inoue, Y.; Terasawa, S.; Takahashi, H.; Ishijima, A. Exchange of rotor components in functioning bacterial flagellar motor. Biochem. Biophys. Res. Commun. 2010, 394, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, Y.V.; Nakamura, S.; Kami-ike, N.; Namba, K.; Minamino, T. Charged residues in the cytoplasmic loop of MotA are required for stator assembly into the bacterial flagellar motor. Mol. Microbiol. 2010, 78, 1117–1129. [Google Scholar] [CrossRef] [PubMed]
- Lele, P.P.; Hosu, B.G.; Berg, H.C. Dynamics of mechanosensing in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 2013, 110, 11839–11844. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, Y.V.; Nakamura, S.; Hiraoka, K.D.; Namba, K.; Minamino, T. Distinct roles of highly conserved charged residues at the MotA-FliG interface in bacterial flagellar motor rotation. J. Bacteriol. 2013, 195, 474–481. [Google Scholar] [CrossRef]
- Heo, M.; Nord, A.L.; Chamousset, D.; van Rijn, E.; Beaumont, H.J.E.; Pedaci, F. Impact of fluorescent protein fusions on the bacterial flagellar motor. Sci. Rep. 2017, 7, 12583. [Google Scholar] [CrossRef]
- Hara, N.; Namba, K.; Minamino, T. Genetic characterization of conserved charged residues in the bacterial flagellar type III export protein FlhA. PLoS ONE 2011, 6, e22417. [Google Scholar] [CrossRef]
- Minamino, T.; Macnab, R.M. Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 1999, 181, 1388–1394. [Google Scholar] [CrossRef]
- Minamino, T.; Imae, Y.; Oosawa, F.; Kobayashi, Y.; Oosawa, K. Effect of intracellular pH on rotational speed of bacterial flagellar motors. J. Bacteriol. 2003, 185, 1190–1194. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Fujita, H.; Sugata, K.; Taira, T.; Iino, T. Genetic analysis of H2, the structural gene for phase-2 flagellin in Salmonella. J. Gen. Microbiol. 1984, 130, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, K.; Ohto, Y.; Aizawa, S.; Macnab, R.M.; Iino, T. FlgD is a scaffolding protein needed for flagellar hook assembly in Salmonella typhimurium. J. Bacteriol. 1994, 176, 2272–2281. [Google Scholar] [CrossRef]
- Guzman, L.M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995, 177, 4121–4130. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; Ai, H.-W.; Wong, P.; Young, J.D.; Campbell, R.E.; Casey, J.R. Red fuorescent protein pH biosensor to detect concentrative nucleoside transport. J. Biol. Chem. 2009, 284, 20499–20511. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, Y.V.; Namba, K.; Minamino, T. Measurements of fee-swimming speed of motile Salmonella cells in liquid media. Bio-Protoc. 2017, 7, e2093. [Google Scholar] [CrossRef]
- Nakamura, S.; Morimoto, Y.V.; Kudo, S. A lactose fermentation product produced by Lactococcus lactis subsp. lactis acetate inhibits the motility of flagellated pathogenic bacteria. Microbiology 2015, 161, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Kami-ike, N.; Yokota, J.P.; Kudo, S.; Minamino, T.; Namba, K. Effect of intracellular pH on the torque-speed relationship of bacterial proton-driven flagellar motor. J. Mol. Biol. 2009, 386, 332–338. [Google Scholar] [CrossRef]
- Castillo, D.J.; Nakamura, S.; Morimoto, Y.V.; Che, Y.S.; Kami-ike, N.; Kudo, S.; Minamino, T.; Namba, K. The C-terminal periplasmic domain of MotB is responsible for load-dependent control of the number of stators of the bacterial flagellar motor. Biophysics 2013, 9, 173–181. [Google Scholar] [CrossRef]
- Che, Y.S.; Nakamura, S.; Morimoto, Y.V.; Kami-Ike, N.; Namba, K.; Minamino, T. Load-sensitive coupling of proton translocation and torque generation in the bacterial flagellar motor. Mol. Microbiol. 2014, 91, 175–184. [Google Scholar] [CrossRef]
- Pourjaberi, S.N.S.; Terahara, N.; Namba, K.; Minamino, T. The role of a cytoplasmic loop of MotA in load-dependent assembly and disassembly dynamics of the MotA/B stator complex in the bacterial flagellar motor. Mol. Microbiol. 2017, 106, 646–658. [Google Scholar] [CrossRef]

| Strain or Plasmid | Relevant Characteristics | Reference |
|---|---|---|
| Salmonella | ||
| SJW1103 | Wild-type for motility and chemotaxis | [41] |
| SJW1368 | ∆(cheW-flhD); master operon mutant | [42] |
| YVM003 | gfp-motB | [34] |
| YVM034 | gfp-motB fliG(R281V) | [36] |
| YVM036 | motA(E98K) gfp-motB fliG(R281V) | [36] |
| YVM046 | fliG(R281V) | [36] |
| YVM047 | motA(E98K) | [36] |
| YVM048 | motA(E98K) fliG(R281V) | [36] |
| Plasmid | ||
| pBAD24 | Expression vector | [43] |
| pYC20 | pBAD24/MotA+MotB | [26] |
| pYC20(E98K) | pBAD24/MotA(E98K)+MotB | [34] |
| pYC109 | pBAD24/MotA+MotB(∆52–71) | [26] |
| pYVM042 | pBAD24/MotA+GFP-MotB | This study |
| pYVM042(E98K) | pBAD24/MotA(E98K)+GFP-MotB | This study |
| pBAD-mNectarine | pBAD/His-mNectarine (addgene #21717) | [44] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morimoto, Y.V.; Namba, K.; Minamino, T. GFP Fusion to the N-Terminus of MotB Affects the Proton Channel Activity of the Bacterial Flagellar Motor in Salmonella. Biomolecules 2020, 10, 1255. https://doi.org/10.3390/biom10091255
Morimoto YV, Namba K, Minamino T. GFP Fusion to the N-Terminus of MotB Affects the Proton Channel Activity of the Bacterial Flagellar Motor in Salmonella. Biomolecules. 2020; 10(9):1255. https://doi.org/10.3390/biom10091255
Chicago/Turabian StyleMorimoto, Yusuke V., Keiichi Namba, and Tohru Minamino. 2020. "GFP Fusion to the N-Terminus of MotB Affects the Proton Channel Activity of the Bacterial Flagellar Motor in Salmonella" Biomolecules 10, no. 9: 1255. https://doi.org/10.3390/biom10091255
APA StyleMorimoto, Y. V., Namba, K., & Minamino, T. (2020). GFP Fusion to the N-Terminus of MotB Affects the Proton Channel Activity of the Bacterial Flagellar Motor in Salmonella. Biomolecules, 10(9), 1255. https://doi.org/10.3390/biom10091255







