Coupling Ion Specificity of the Flagellar Stator Proteins MotA1/MotB1 of Paenibacillus sp. TCA20
Abstract
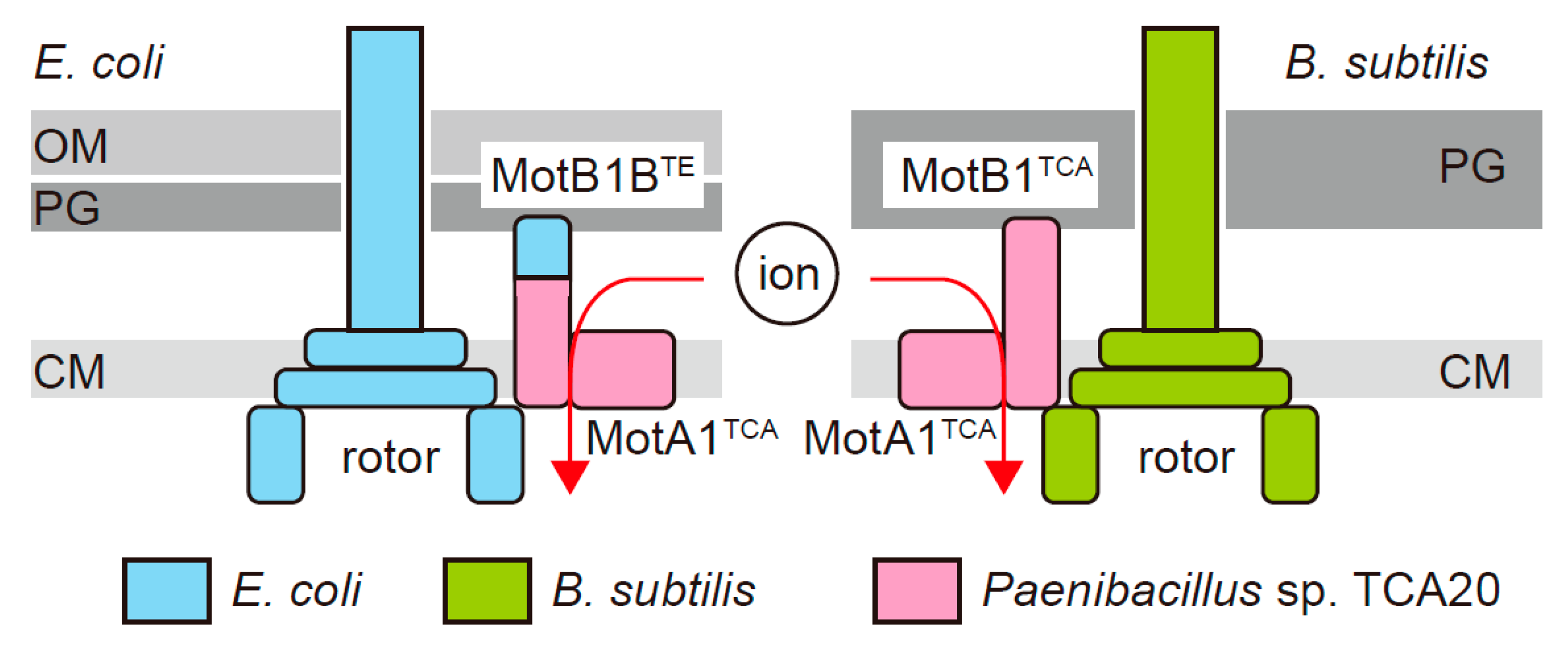
1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids, Growth Conditions, and Media
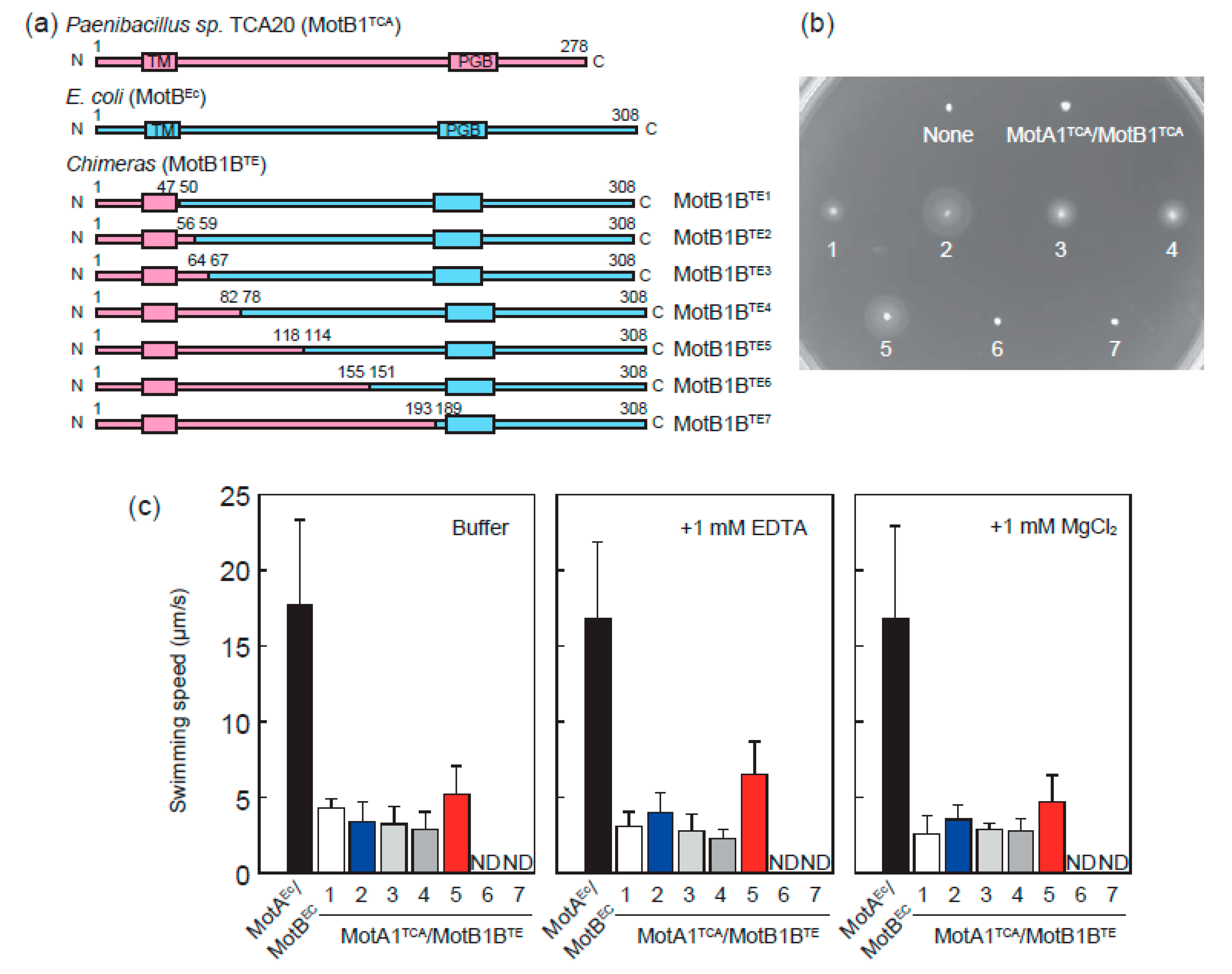
2.2. Construction of Plasmids Encoding Chimera Stator Proteins
2.3. Strain Construction of B. subtilis
2.4. Motility Assays
2.5. Rotation Measurement by Tethered Cell Assays
3. Results
3.1. MotA1TCA and Chimeras between MotB1TCA and MotBEc Function as A Stator in E. coli
3.2. Chimeric Motor in E. coli Is Driven by Protons Rather than Divalent Ions
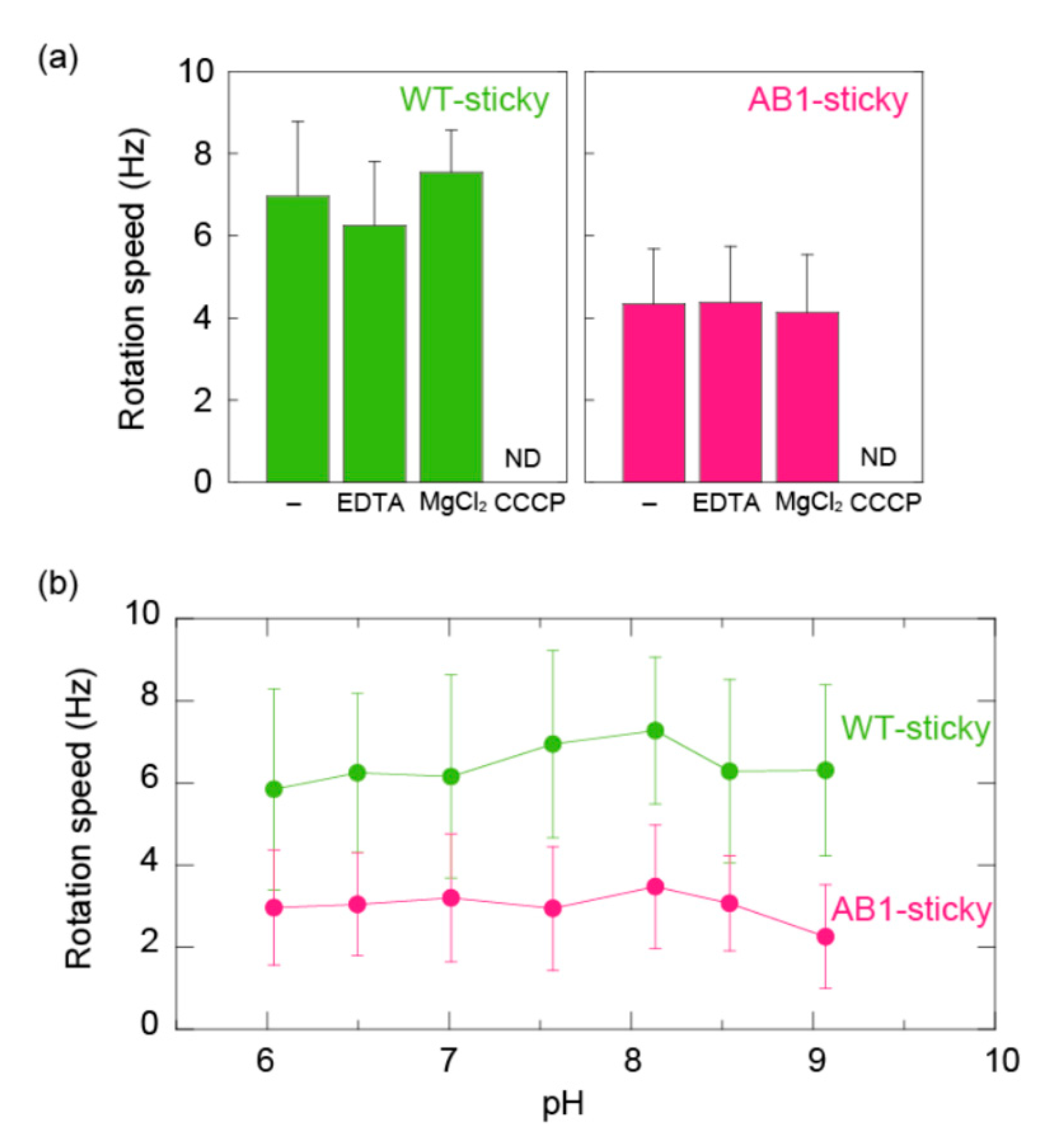
3.3. MotA1TCA/MotB1TCA Couples Monovalent Cations in ΔmotABΔmotPS B. subtilis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Berg, H.C. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 2003, 72, 19–54. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Minamino, T. Flagella-Driven Motility of Bacteria. Biomolecules 2019, 9, 279. [Google Scholar] [CrossRef] [PubMed]
- Sowa, Y.; Berry, R.M. Bacterial flagellar motor. Q. Rev. Biophys. 2008, 41, 103–132. [Google Scholar] [CrossRef]
- Yorimitsu, T.; Homma, M. Na+-driven flagellar motor of Vibrio. Biochim. Biophys. Acta 2001, 1505, 82–93. [Google Scholar] [CrossRef]
- Ito, M.; Hicks, D.B.; Henkin, T.M.; Guffanti, A.A.; Powers, B.D.; Zvi, L.; Uematsu, K.; Krulwich, T.A. MotPS is the stator-force generator for motility of alkaliphilic Bacillus, and its homologue is a second functional Mot in Bacillus subtilis. Mol. Microbiol. 2004, 53, 1035–1049. [Google Scholar] [CrossRef] [PubMed]
- Terahara, N.; Sano, M.; Ito, M. A bacillus flagellar motor that can use both Na+ and K+ as a coupling ion is converted by a single mutation to use only Na+. PLoS ONE 2012, 7, e46248. [Google Scholar] [CrossRef]
- Thormann, K.M.; Paulick, A. Tuning the flagellar motor. Microbiology 2010, 156, 1275–1283. [Google Scholar] [CrossRef]
- Minamino, T.; Terahara, N.; Kojima, S.; Namba, K. Autonomous control mechanism of stator assembly in the bacterial flagellar motor in response to changes in the environment. Mol. Microbiol. 2018, 109, 723–734. [Google Scholar] [CrossRef]
- Asai, Y.; Kawagishi, I.; Sockett, R.E.; Homma, M. Hybrid motor with H+-and Na+-driven components can rotate Vibrio polar flagella by using sodium ions. J. Bacteriol. 1999, 181, 6332–6338. [Google Scholar] [CrossRef]
- Asai, Y.; Kawagishi, I.; Sockett, R.E.; Homma, M. Coupling ion specificity of chimeras between H+-and Na+-driven motor proteins, MotB and PomB, in Vibrio polar flagella. EMBO J. 2000, 19, 3639–3648. [Google Scholar] [CrossRef]
- Asai, Y.; Yakushi, T.; Kawagishi, I.; Homma, M. Ion-coupling determinants of Na+-driven and H+-driven flagellar motors. J. Mol. Biol. 2003, 327, 453–463. [Google Scholar] [CrossRef]
- Takekawa, N.; Nishiyama, M.; Kaneseki, T.; Kanai, T.; Atomi, H.; Kojima, S.; Homma, M. Sodium-driven energy conversion for flagellar rotation of the earliest divergent hyperthermophilic bacterium. Sci. Rep. 2015, 5, 12711. [Google Scholar] [CrossRef]
- Gosink, K.K.; Hase, C.C. Requirements for conversion of the Na+-driven flagellar motor of Vibrio cholerae to the H+-driven motor of Escherichia coli. J. Bacteriol. 2000, 182, 4234–4240. [Google Scholar] [CrossRef] [PubMed]
- Sowa, Y.; Rowe, A.D.; Leake, M.C.; Yakushi, T.; Homma, M.; Ishijima, A.; Berry, R.M. Direct observation of steps in rotation of the bacterial flagellar motor. Nature 2005, 437, 916–919. [Google Scholar] [CrossRef] [PubMed]
- Inoue, Y.; Lo, C.J.; Fukuoka, H.; Takahashi, H.; Sowa, Y.; Pilizota, T.; Wadhams, G.H.; Homma, M.; Berry, R.M.; Ishijima, A. Torque-speed relationships of Na+-driven chimeric flagellar motors in Escherichia coli. J Mol. Biol. 2008, 376, 1251–1259. [Google Scholar] [CrossRef]
- Lo, C.J.; Sowa, Y.; Pilizota, T.; Berry, R.M. Mechanism and kinetics of a sodium-driven bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 2013, 110, E2544–E2551. [Google Scholar] [CrossRef]
- Sowa, Y.; Homma, M.; Ishijima, A.; Berry, R.M. Hybrid-fuel bacterial flagellar motors in Escherichia coli. Proc. Natl. Acad. Sci. USA 2014, 111, 3436–3441. [Google Scholar] [CrossRef]
- Ishida, T.; Ito, R.; Clark, J.; Matzke, N.J.; Sowa, Y.; Baker, M.A.B. Sodium-powered stators of the bacterial flagellar motor can generate torque in the presence of phenamil with mutations near the peptidoglycan-binding region. Mol. Microbiol. 2019, 111, 1689–1699. [Google Scholar] [CrossRef]
- Kojima, S.; Imada, K.; Sakuma, M.; Sudo, Y.; Kojima, C.; Minamino, T.; Homma, M.; Namba, K. Stator assembly and activation mechanism of the flagellar motor by the periplasmic region of MotB. Mol. Microbiol. 2009, 73, 710–718. [Google Scholar] [CrossRef] [PubMed]
- Imazawa, R.; Takahashi, Y.; Aoki, W.; Sano, M.; Ito, M. A novel type bacterial flagellar motor that can use divalent cations as a coupling ion. Sci. Rep. 2016, 6, 19773. [Google Scholar] [CrossRef]
- Lo, C.J.; Leake, M.C.; Pilizota, T.; Berry, R.M. Nonequivalence of membrane voltage and ion-gradient as driving forces for the bacterial flagellar motor at low load. Biophys. J. 2007, 93, 294–302. [Google Scholar] [CrossRef]
- Nozaki, S.; Niki, H. Exonuclease III (XthA) Enforces In Vivo DNA Cloning of Escherichia coli To Create Cohesive Ends. J. Bacteriol. 2019, 201, e00660-18. [Google Scholar] [CrossRef] [PubMed]
- Terahara, N.; Noguchi, Y.; Nakamura, S.; Kami-Ike, N.; Ito, M.; Namba, K.; Minamino, T. Load-and polysaccharide-dependent activation of the Na+-type MotPS stator in the Bacillus subtilis flagellar motor. Sci. Rep. 2017, 7, 46081. [Google Scholar] [CrossRef] [PubMed]
- Kasai, T.; Sowa, Y. Measurements of the Rotation of the Flagellar Motor by Bead Assay. Methods Mol. Biol. 2017, 1593, 185–192. [Google Scholar]
- Chen, X.; Berg, H.C. Solvent-isotope and pH effects on flagellar rotation in Escherichia coli. Biophys. J. 2000, 78, 2280–2284. [Google Scholar] [CrossRef]
- Gabel, C.V.; Berg, H.C. The speed of the flagellar rotary motor of Escherichia coli varies linearly with protonmotive force. Proc. Natl. Acad. Sci. USA 2003, 100, 8748–8751. [Google Scholar] [CrossRef]
- Terahara, N.; Krulwich, T.A.; Ito, M. Mutations alter the sodium versus proton use of a Bacillus clausii flagellar motor and confer dual ion use on Bacillus subtilis motors. Proc. Natl. Acad. Sci. USA 2008, 105, 14359–14364. [Google Scholar] [CrossRef]
- Turner, L.; Ryu, W.S.; Berg, H.C. Real-time imaging of fluorescent flagellar filaments. J. Bacteriol. 2000, 182, 2793–2801. [Google Scholar] [CrossRef]
- Wadhams, G.H.; Armitage, J.P. Making sense of it all: Bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 2004, 5, 1024–1037. [Google Scholar] [CrossRef]
- Terahara, N.; Kodera, N.; Uchihashi, T.; Ando, T.; Namba, K.; Minamino, T. Na+-induced structural transition of MotPS for stator assembly of the Bacillus flagellar motor. Sci. Adv. 2017, 3, eaao4119. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Onoe, S.; Yoshida, M.; Terahara, N.; Sowa, Y. Coupling Ion Specificity of the Flagellar Stator Proteins MotA1/MotB1 of Paenibacillus sp. TCA20. Biomolecules 2020, 10, 1078. https://doi.org/10.3390/biom10071078
Onoe S, Yoshida M, Terahara N, Sowa Y. Coupling Ion Specificity of the Flagellar Stator Proteins MotA1/MotB1 of Paenibacillus sp. TCA20. Biomolecules. 2020; 10(7):1078. https://doi.org/10.3390/biom10071078
Chicago/Turabian StyleOnoe, Sakura, Myu Yoshida, Naoya Terahara, and Yoshiyuki Sowa. 2020. "Coupling Ion Specificity of the Flagellar Stator Proteins MotA1/MotB1 of Paenibacillus sp. TCA20" Biomolecules 10, no. 7: 1078. https://doi.org/10.3390/biom10071078
APA StyleOnoe, S., Yoshida, M., Terahara, N., & Sowa, Y. (2020). Coupling Ion Specificity of the Flagellar Stator Proteins MotA1/MotB1 of Paenibacillus sp. TCA20. Biomolecules, 10(7), 1078. https://doi.org/10.3390/biom10071078




