PMEPA1 Gene Isoforms: A Potential Biomarker and Therapeutic Target in Prostate Cancer
Abstract
1. Introduction
2. Discovery of PMEPA1 Gene Isoforms
3. Structure and Biochemical Features of PMEPA1 Isoforms
4. Evolution and Architecture of PMEPA1 Gene and Its Isoforms
5. Expression of PMEPA1 Isoforms
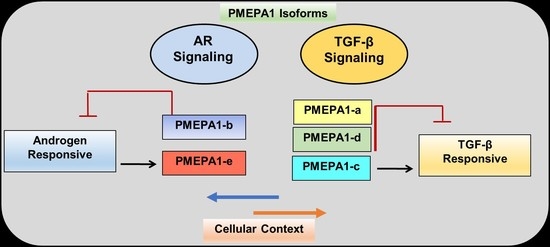
6. PMEPA1-b Isoform Inhibits AR Signaling
7. PMEPA1 Isoforms (a and d) Inhibit TGF-β Signaling
8. PMEPA1 Isoforms: Potential New Biomarkers and Hormone Therapies
9. Future Research Directions
10. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
DoD Disclaimer
References
- Sharad, S.; Dillman, A.A.; Sztupinszki, Z.M.; Szallasi, Z.; Rosner, I.; Cullen, J.; Srivastava, S.; Srinivasan, A.; Li, H. Characterization of unique PMEPA1 gene splice variants (isoforms d and e) from RNA Seq profiling provides novel insights into prognostic evaluation of prostate cancer. Oncotarget 2020, 11, 362–377. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.L.; Shi, Y.; Petrovics, G.; Sun, C.; Makarem, M.; Zhang, W.; Sesterhenn, I.A.; McLeod, D.G.; Sun, L.; Moul, J.W.; et al. PMEPA1, an androgen-regulated NEDD4-binding protein, exhibits cell growth inhibitory function and decreased expression during prostate cancer progression. Cancer Res. 2003, 63, 4299–4304. [Google Scholar] [PubMed]
- Xu, L.L.; Su, Y.P.; Labiche, R.; Segawa, T.; Shanmugam, N.; McLeod, D.G.; Moul, J.W.; Srivastava, S. Quantitative expression profile of androgen-regulated genes in prostate cancer cells and identification of prostate-specific genes. Int. J. Cancer 2001, 92, 322–328. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Mohamed, A.A.; Sharad, S.; Umeda, E.; Song, Y.; Young, D.; Petrovics, G.; McLeod, D.G.; Sesterhenn, I.A.; Sreenath, T.; et al. Silencing of PMEPA1 accelerates the growth of prostate cancer cells through AR, NEDD4 and PTEN. Oncotarget 2015, 6, 15137–15149. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.L.; Shanmugam, N.; Segawa, T.; Sesterhenn, I.A.; McLeod, D.G.; Moul, J.W.; Srivastava, S. A novel androgen-regulated gene, PMEPA1, located on chromosome 20q13 exhibits high level expression in prostate. Genomics 2000, 66, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Rae, F.K.; Hooper, J.D.; Nicol, D.L.; Clements, J.A. Characterization of a novel gene, STAG1/PMEPA1, upregulated in renal cell carcinoma and other solid tumors. Mol. Carcinog. 2001, 32, 44–53. [Google Scholar] [CrossRef]
- Giannini, G.; Ambrosini, M.I.; Di Marcotullio, L.; Cerignoli, F.; Zani, M.; MacKay, A.R.; Screpanti, I.; Frati, L.; Gulino, A. EGF- and cell-cycle-regulated STAG1/PMEPA1/ERG1.2 belongs to a conserved gene family and is overexpressed and amplified in breast and ovarian cancer. Mol. Carcinog. 2003, 38, 188–200. [Google Scholar] [CrossRef]
- Anazawa, Y.; Arakawa, H.; Nakagawa, H.; Nakamura, Y. Identification of STAG1 as a key mediator of a p53-dependent apoptotic pathway. Oncogene 2004, 23, 7621–7627. [Google Scholar] [CrossRef]
- Nakano, N.; Itoh, S.; Watanabe, Y.; Maeyama, K.; Itoh, F.; Kato, M. Requirement of TCF7L2 for TGF-beta-dependent transcriptional activation of the TMEPAI gene. J. Biol. Chem. 2010, 85, 38023–38033. [Google Scholar] [CrossRef]
- Azami, S.; Vo Nguyen, T.T.; Watanabe, Y.; Kato, M. Cooperative induction of transmembrane prostate androgen induced protein TMEPAI/PMEPA1 by transforming growth factor-β and epidermal growth factor signaling. Biochem. Biophys. Res. Commun. 2015, 456, 580–585. [Google Scholar] [CrossRef]
- Sharad, S.; Sztupinszki, Z.M.; Chen, Y.; Kuo, C.; Ravindranath, L.; Szallasi, Z.; Petrovics, G.; Sreenath, T.L.; Dobi, A.; Rosner, I.L.; et al. Analysis of PMEPA1 Isoforms (a and b) as Selective Inhibitors of Androgen and TGF-β Signaling Reveals Distinct Biological and Prognostic Features in Prostate Cancer. Cancers (Basel) 2019, 11, 1995. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Ding, K.; Luo, T.; Xu, R.; Zhang, X.; Huang, B.; Chen, A.; Zhang, D.; Miletic, H.; Bjerkvig, R.; et al. PMEPA1 isoform a drives progression of glioblastoma by promoting protein degradation of the Hippo pathway kinase LATS1. Oncogene 2020, 39, 1125–1139. [Google Scholar] [CrossRef] [PubMed]
- Brunschwig, E.B.; Wilson, K.; Mack, D.; Dawson, D.; Lawrence, E.; Willson, J.K.; Lu, S.; Nosrati, A.; Rerko, R.M.; Swinler, S.; et al. PMEPA1, a transforming growth factor-beta-induced marker of terminal colonocyte differentiation whose expression is maintained in primary and metastatic colon cancer. Cancer Res. 2003, 63, 1568–1575. [Google Scholar] [PubMed]
- Vo Nguyen, T.T.; Watanabe, Y.; Shiba, A.; Noguchi, M.; Itoh, S.; Kato, M. TMEPAI/PMEPA1 enhances tumorigenic activities in lung cancer cells. Cancer Sci. 2014, 105, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Nelson, P.S.; Clegg, N.; Arnold, H.; Ferguson, C.; Bonham, M.; White, J.; Hood, L.; Lin, B. The program of androgen-responsive genes in neoplastic prostate epithelium. Proc. Natl. Acad. Sci. USA 2002, 99, 11890–11895. [Google Scholar] [CrossRef]
- Reichling, T.; Goss, K.H.; Carson, D.J.; Holdcraft, R.W.; Ley-Ebert, C.; Witte, D.; Aronow, B.J.; Groden, J. Transcriptional profiles of intestinal tumors in Apc(Min) mice are unique from those of embryonic intestine and identify novel gene targets dysregulated in human colorectal tumors. Cancer Res. 2005, 65, 166–176. [Google Scholar]
- Singha, P.K.; Yeh, I.T.; Venkatachalam, M.A.; Saikumar, P. Transforming growth factor-beta (TGF-beta)-inducible gene TMEPAI converts TGF-beta from a tumor suppressor to a tumor promoter in breast cancer. Cancer Res. 2010, 70, 6377–6383. [Google Scholar] [CrossRef]
- Liu, R.; Zhou, Z.; Huang, J.; Chen, C. PMEPA1 promotes androgen receptor-negative prostate cell proliferation through suppression the Smad3/4-c-Myc-p21 Cip 1 signaling pathway. J. Pathol. 2011, 223, 683–694. [Google Scholar] [CrossRef]
- Fournier, P.G.; Juárez, P.; Jiang, G.; Clines, G.A.; Niewolna, M.; Kim, H.S.; Walton, H.W.; Peng, X.H.; Liu, Y.; Mohammad, K.S.; et al. The TGF-β Signaling Regulator PMEPA1 Suppresses Prostate Cancer Metastases to Bone. Cancer Cell 2015, 27, 809–821. [Google Scholar] [CrossRef]
- Hu, Y.; He, K.; Wang, D.; Yuan, X.; Liu, Y.; Ji, H.; Song, J. TMEPAI regulates EMT in lung cancer cells by modulating the ROS and IRS-1 signaling pathways. Carcinogenesis 2013, 34, 1764–1772. [Google Scholar] [CrossRef]
- Itoh, S.; Itoh, F. TMEPAI family: Involvement in regulation of multiple signaling pathways. J. Biochem. 2018, 164, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xu, L.L.; Masuda, K.; Raymundo, E.; McLeod, D.G.; Dobi, A.; Srivastava, S. A feedback loop between the androgen receptor and a NEDD4-binding protein, PMEPA1, in prostate cancer cells. J. Biol. Chem. 2008, 283, 28988–28995. [Google Scholar] [CrossRef] [PubMed]
- Peterson, T.A.; Adadey, A.; Santana-Cruz, I.; Sun, Y.; Winder, A.; Kann, M.G. DMDM: Domain Mapping of Disease Mutations. Bioinformatics 2000, 26, 2458–2459. [Google Scholar] [CrossRef] [PubMed]
- Sharad, S.; Ravindranath, L.; Haffner, M.C.; Li, H.; Yan, W.; Sesterhenn, I.A.; Chen, Y.; Ali, A.; Srinivasan, A.; McLeod, D.G.; et al. Methylation of the PMEPA1 gene, a negative regulator of the androgen receptor in prostate cancer. Epigenetics 2014, 9, 918–927. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, X.; Lai, C.; Zhang, H.; Lai, M. PMEPA1 induces EMT via a non-canonical TGF-β signalling in colorectal cancer. J. Cell. Mol. Med. 2019, 23, 3603–3615. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Dobi, A.; Shaheduzzaman, S.; Gao, C.L.; Masuda, K.; Li, H.; Drukier, A.; Gu, Y.; Srikantan, V.; Rhim, J.S.; et al. Characterization of the androgen receptor in a benign prostate tissue-derived human prostate epithelial cell line: RC-165N/human telomerase reverse transcriptase. Prostate Cancer Prostatic Dis. 2007, 10, 30–38. [Google Scholar] [CrossRef][Green Version]
- Itoh, S.; Thorikay, M.; Kowanetz, M.; Moustakas, A.; Itoh, F.; Heldin, C.H.; ten Dijke, P. Elucidation of Smad requirement in transforming growth factor-beta type I receptor-induced responses. J. Biol. Chem. 2003, 278, 3751–3761. [Google Scholar] [CrossRef]
- Singha, P.K.; Pandeswara, S.; Geng, H.; Lan, R.; Venkatachalam, M.A.; Saikumar, P. TGF-β induced TMEPAI/PMEPA1 inhibits canonical Smad signaling through R-Smad sequestration and promotes non-canonical PI3K/Akt signaling by reducing PTEN in triple negative breast cancer. Genes Cancer 2014, 5, 320–336. [Google Scholar]
- Sun, J.; Keim, C.D.; Wang, J.; Kazadi, D.; Oliver, P.M.; Rabadan, R.; Basu, U. E3-ubiquitin ligase Nedd4 determines the fate of AID-associated RNA polymerase II in B cells. Genes Dev. 2013, 27, 1821–1833. [Google Scholar] [CrossRef]
- Fouladkou, F.; Landry, T.; Kawabe, H.; Neeb, A.; Lu, C.; Brose, N.; Stambolic, V.; Rotin, D. The ubiquitin ligase Nedd4-1 is dispensable for the regulation of PTEN stability and localization. Proc. Natl. Acad. Sci. USA 2008, 105, 8585–8590. [Google Scholar] [CrossRef]
- Watanabe, Y.; Itoh, S.; Goto, T.; Ohnishi, E.; Inamitsu, M.; Itoh, F.; Satoh, K.; Wiercinska, E.; Yang, W.; Shi, L.; et al. TMEPAI, a transmembrane TGF-beta-inducible protein, sequesters Smad proteins from active participation in TGF-beta signaling. Mol. Cell 2010, 37, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Abdelaziz, M.; Watanabe, Y.; Kato, M. PMEPA1/TMEPAI knockout impairs tumour growth and lung metastasis in MDA-MB-231 cells without changing monolayer culture cell growth. J. Biochem. 2019, 165, 411–414. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Jing, L.; Li, Y.; Li, Y.; Luo, S.; Wang, S.; Zhou, J.; Liu, Z.; Diao, A. TMEPAI inhibits TGF-β by promoting lysosome degradation of TGF-β receptor and contributes to lung cancer development. Cell Signal. 2014, 26, 2030–2039. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guo, A.; Feng, Y.; Zhang, Y.; Wang, J.; Jing, L.; Yan, Y.; Jing, L.; Liu, Z.; Ma, L.; et al. Sp1 transcription factor promotes TMEPA1 gene expression and contributes to cell proliferation. Cell Proliferation 2016, 49, 710–719. [Google Scholar] [CrossRef]
- Paller, C.; Pu, H.; Begemann, D.E.; Wade, C.A.; Hensley, P.J.; Kyprianou, N. TGF-β receptor I inhibitor enhances response to enzalutamide in a pre-clinical model of advanced prostate cancer. Prostate 2019, 79, 31–43. [Google Scholar] [CrossRef]
- Hornberg, E.; Ylitalo, E.B.; Crnalic, S.; Antti, H.; Stattin, P.; Wildmark, A.; Bergh, A.; Wikstrom, P. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PLoS ONE 2011, 6, e19059. [Google Scholar] [CrossRef]





| Distinct PMEPA1 Motifs | Residues | Binding Domains | |
|---|---|---|---|
| PPPY | 158–161 (C-Terminal) | WW consensus binding sequences | Xu et al., 2003 [2] |
| PPTY | 229–232 (C-Terminal) | WW consensus binding sequences | |
| PPNR | 186–189 (C-Terminal) | Smad | Liu et al., 2011 [18] |
| PPRP | 112–115 (C-Terminal) | PXXP consensus binding SH3 domains | Watanabe et al., 2010 [24] |
| PTYP | 135–138 (C-Terminal) | PXXP consensus binding SH3 domains | Giannini et al. 2003 [7] |
| PCPP | 205–208 (C-Terminal) | PXXP consensus binding SH3 domains | |
| Other Predicted Motifs | |||
| YPYL | 138–141 (C-Terminal) | ||
| YSEV | 232–235 (C-Terminal) | ||
| di-lucine | 255–256 (C-Terminal) | ||
| Other Predicted Potential Casein kinase II and Protein Kinase C Phosphorylation Site | Rae et al., 2001 [6]; Brunschwig et al., 2003 [13] | ||
| S74, S77, Y137, T217, Y219, S221, Y232, Y239 and S250 | |||
| Residue Number | Polymorphism | Reference |
|---|---|---|
| 3 | SER → ARG | Peterson et al., 2010 [23] Brunschwig et al., 2003 [13] |
| 75 | TRP → ARG | |
| 128 | GLU → ASP | |
| 179 | THR → ASN | |
| 220 | SER → GLY | |
| 228 | ALA → PRO |
| Cell Line | Signaling | PMEPA1-a | PMEPA1-b | PMEPA1-c | PMEPA1-d | PMEPA1-e |
|---|---|---|---|---|---|---|
| LNCaP | AR (+) Androgen Sensitive | (++++) | (++) | (++) | (+) | (-) |
| VCaP | AR (+) Androgen Sensitive | (+++++) | (+++) | (++) | (+) | (+) |
| LAPC4 | AR (+) Androgen Sensitive | (++) | (+++) | (++) | (+) | (+) |
| DU145 | AR (-) TGF-β Signaling (+) | (++) | (-) | (++) | (+) | (-) |
| PC3 | AR (-) TGF-β Signaling (+) | (++) | (-) | (++) | (+) | (-) |
| C4-2B | AR (+) Androgen Independent | (+++++) | (+) | (++) | (+) | (-) |
| CWR22v1 | AR (+) Androgen Independent | (++) | (+) | (++) | (+) | (-) |
| BPH-1 | AR (-) | (++) | (-) | (++) | (++) | (-) |
| PrEC | AR (-) | (+++) | (-) | (++) | (+) | (-) |
| Interacting Protein Partner | Domains/Motifs Involved in Binding | PMEPA1 Isoform | Reference |
|---|---|---|---|
| NEED4 | PY motifs PPPY and PPTY are required to bind WW domains | PMEPA1-b | Xu et al., 2003 [2] |
| AR | Tet-Off-induced PMEPA1 protein interacts with endogenous AR protein through NEED4 | PMEPA1-b | Li et al., 2008 [22] |
| Smad 2 and 3 | SIM domain | PMEPA1-a | Watanabe et al., 2010 [24]; Liu et al., 2011 [18] |
| Yes-associated protein YAP65 | SH3-motifs and WW-binding domains | PMEPA1-a | Giannini et al., 2003 [7] |
| GRB-2 | SH3-motifs and WW-binding domains | PMEPA1-a |
| Residue Number | Nature of Mutation | Functional Consequences | PMEPA1 Isoform | References |
|---|---|---|---|---|
| 161 | Y → A | Impairs interaction with NEDD4 protein | PMEPA1-b | Xu et al., 2003 [2]; Li et al., 2008 [22] |
| 232 | Y → A | Impairs interaction with NEDD4 protein | PMEPA1-b | |
| 161 and 232 | Y → A/Y → A | Impairs polyubiquitination of AR | PMEPA1-b | |
| 178–181 | PPNR → AAAA | Blocks nuclear translocation of Smad2 upon TGF-b stimulation | PMEPA1-a | Watanabe et al., 2010 [32] |
| >1–171 * | Deletion | Due to lack of Smad2-binding domain unable to block TGF-b receptor | PMEPA1-a | |
| >1–204 * | Deletion | Asn171- Ser204 domain is required for Smad2 interaction | PMEPA1-a |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharad, S.; Dobi, A.; Srivastava, S.; Srinivasan, A.; Li, H. PMEPA1 Gene Isoforms: A Potential Biomarker and Therapeutic Target in Prostate Cancer. Biomolecules 2020, 10, 1221. https://doi.org/10.3390/biom10091221
Sharad S, Dobi A, Srivastava S, Srinivasan A, Li H. PMEPA1 Gene Isoforms: A Potential Biomarker and Therapeutic Target in Prostate Cancer. Biomolecules. 2020; 10(9):1221. https://doi.org/10.3390/biom10091221
Chicago/Turabian StyleSharad, Shashwat, Albert Dobi, Shiv Srivastava, Alagarsamy Srinivasan, and Hua Li. 2020. "PMEPA1 Gene Isoforms: A Potential Biomarker and Therapeutic Target in Prostate Cancer" Biomolecules 10, no. 9: 1221. https://doi.org/10.3390/biom10091221
APA StyleSharad, S., Dobi, A., Srivastava, S., Srinivasan, A., & Li, H. (2020). PMEPA1 Gene Isoforms: A Potential Biomarker and Therapeutic Target in Prostate Cancer. Biomolecules, 10(9), 1221. https://doi.org/10.3390/biom10091221