MDA-MB-231 Breast Cancer Cells Resistant to Pleurocidin-Family Lytic Peptides Are Chemosensitive and Exhibit Reduced Tumor-Forming Capacity
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Cell Culture
2.2. Reagents
2.3. Animals
2.4. MTT Assay
2.5. Peptide Binding Assay
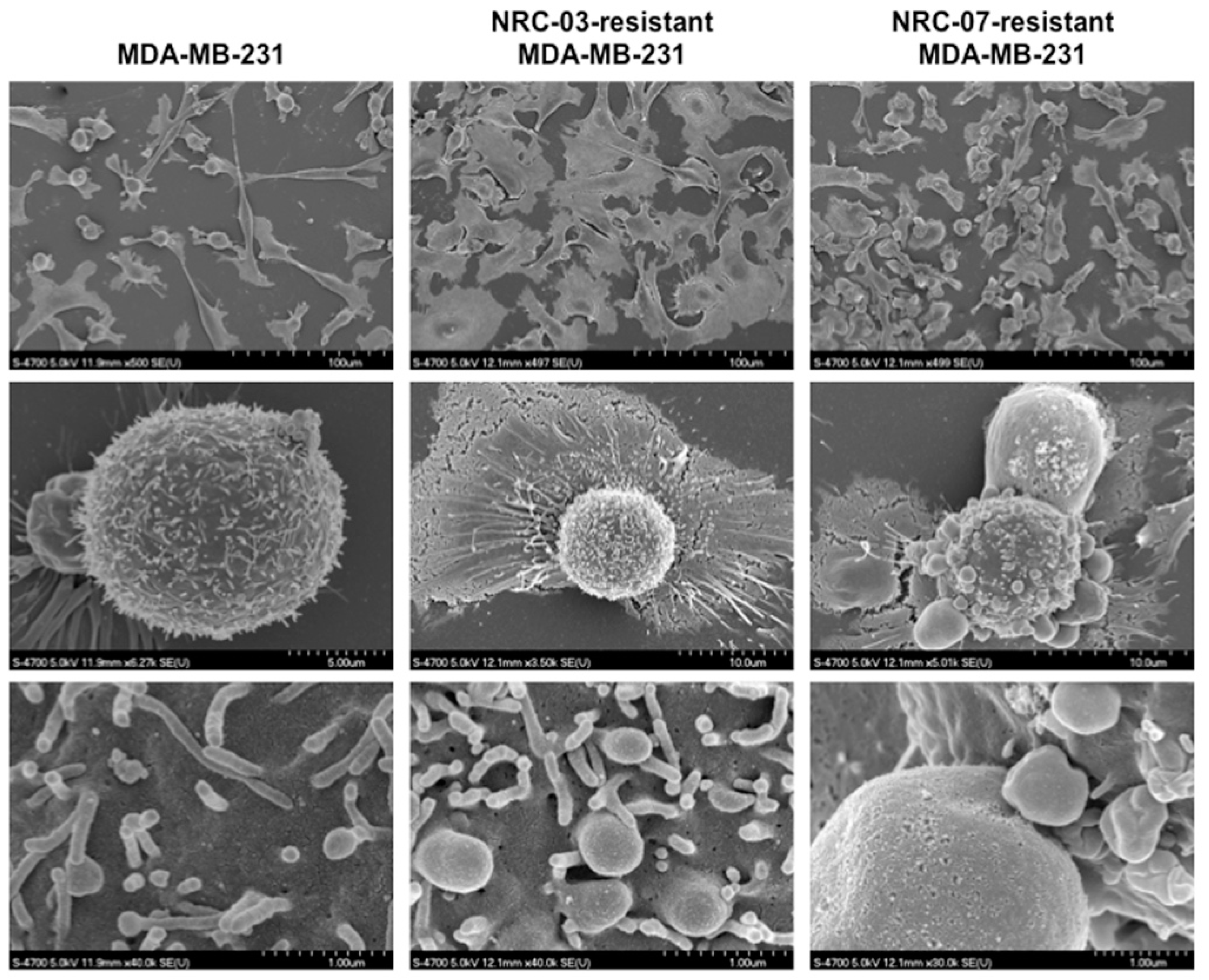
2.6. Scanning Electron Microscopy
2.7. RNA Sequencing Sample Preparation and Analysis
2.8. Breast Cancer Xenografts
2.9. Statistical Analysis
3. Results
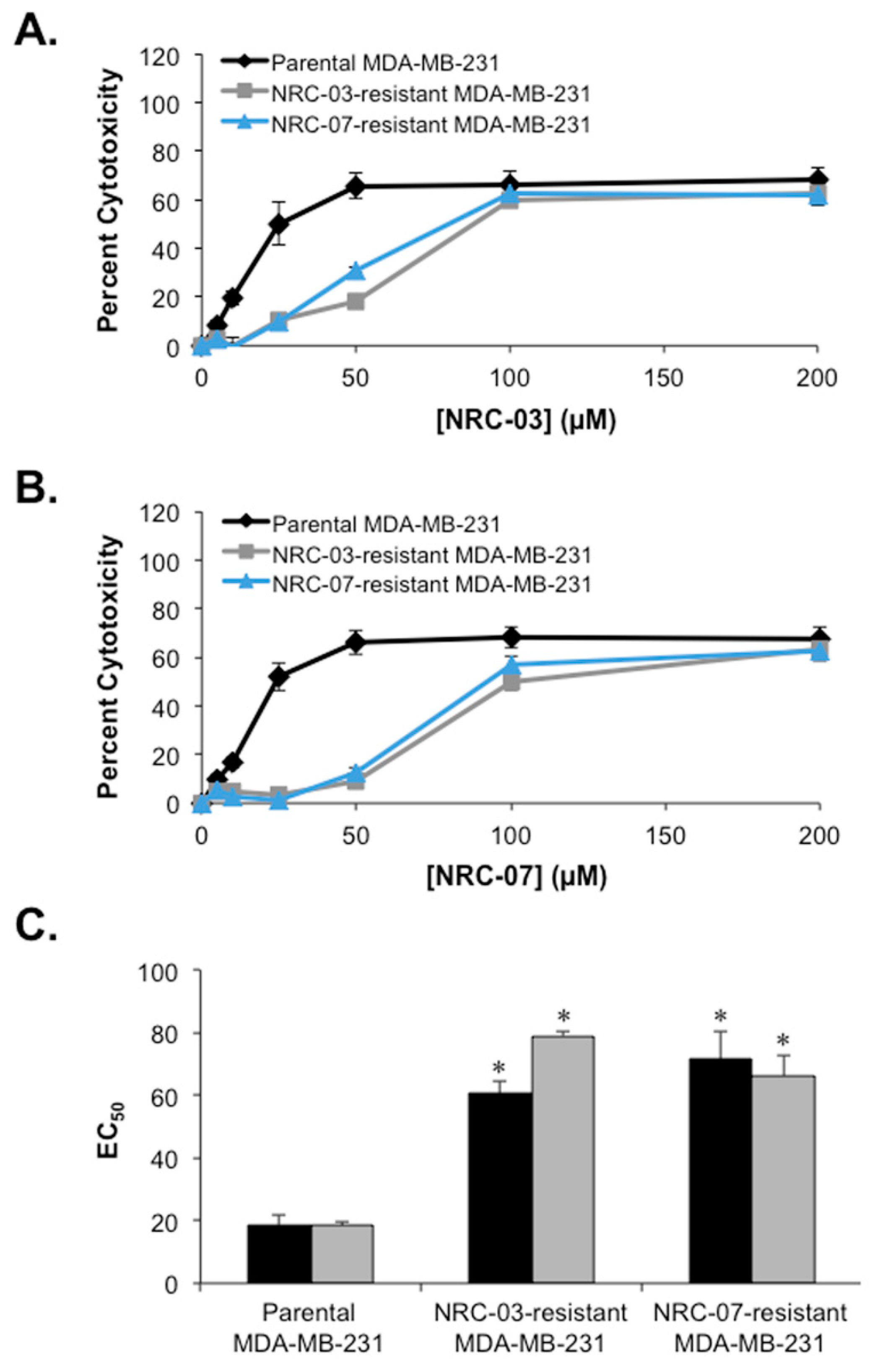
3.1. Continuous Exposure to Either NRC-03 or NRC-07 Results in Low-Level Resistance of Breast Cancer Cells to These Pleurocidins
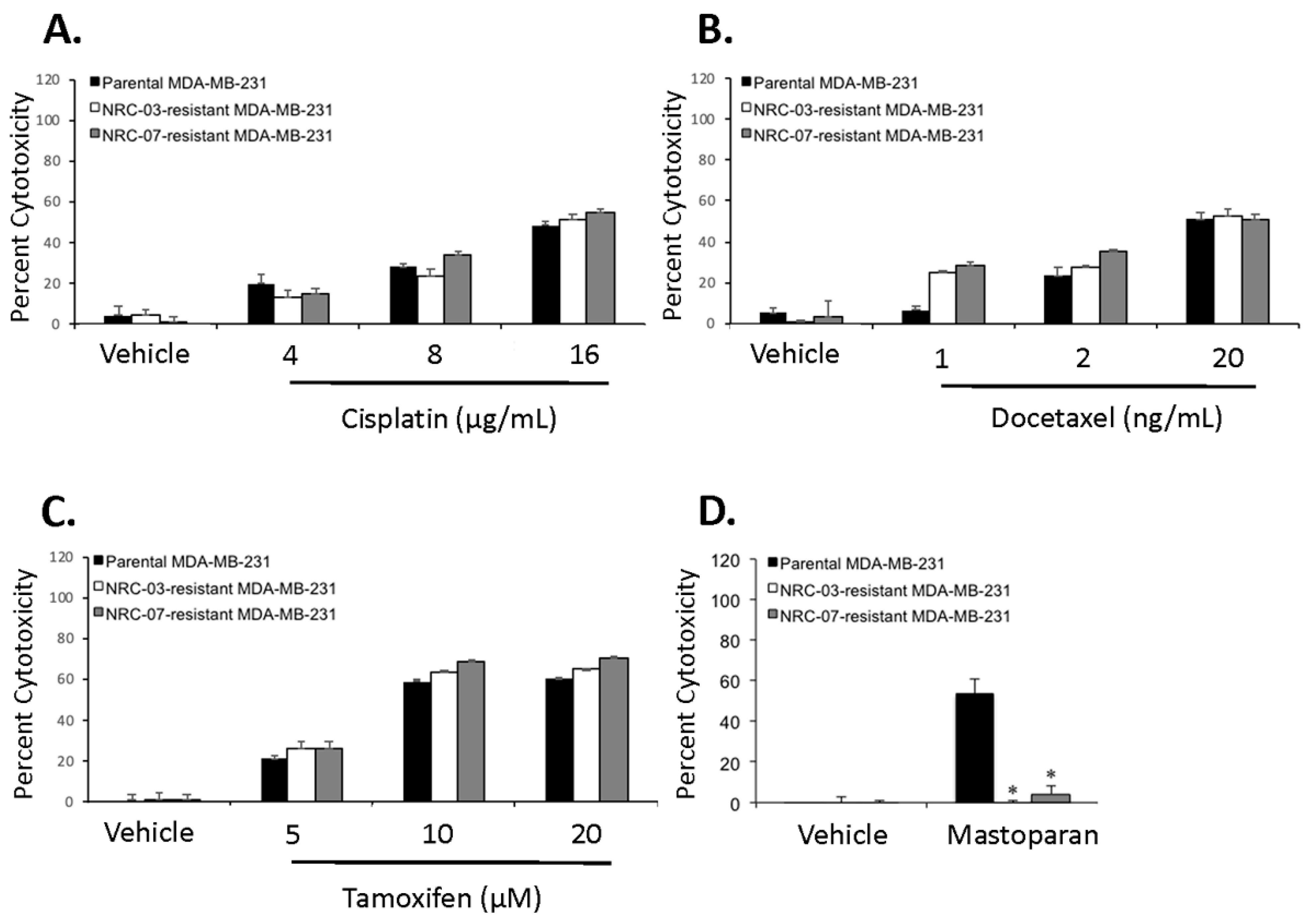
3.2. NRC-03-Resistant and NRC-07-Resistant Breast Cancer Cells Are Susceptible to Chemotherapeutic Drugs but Refractory to an Unrelated DAA Peptide
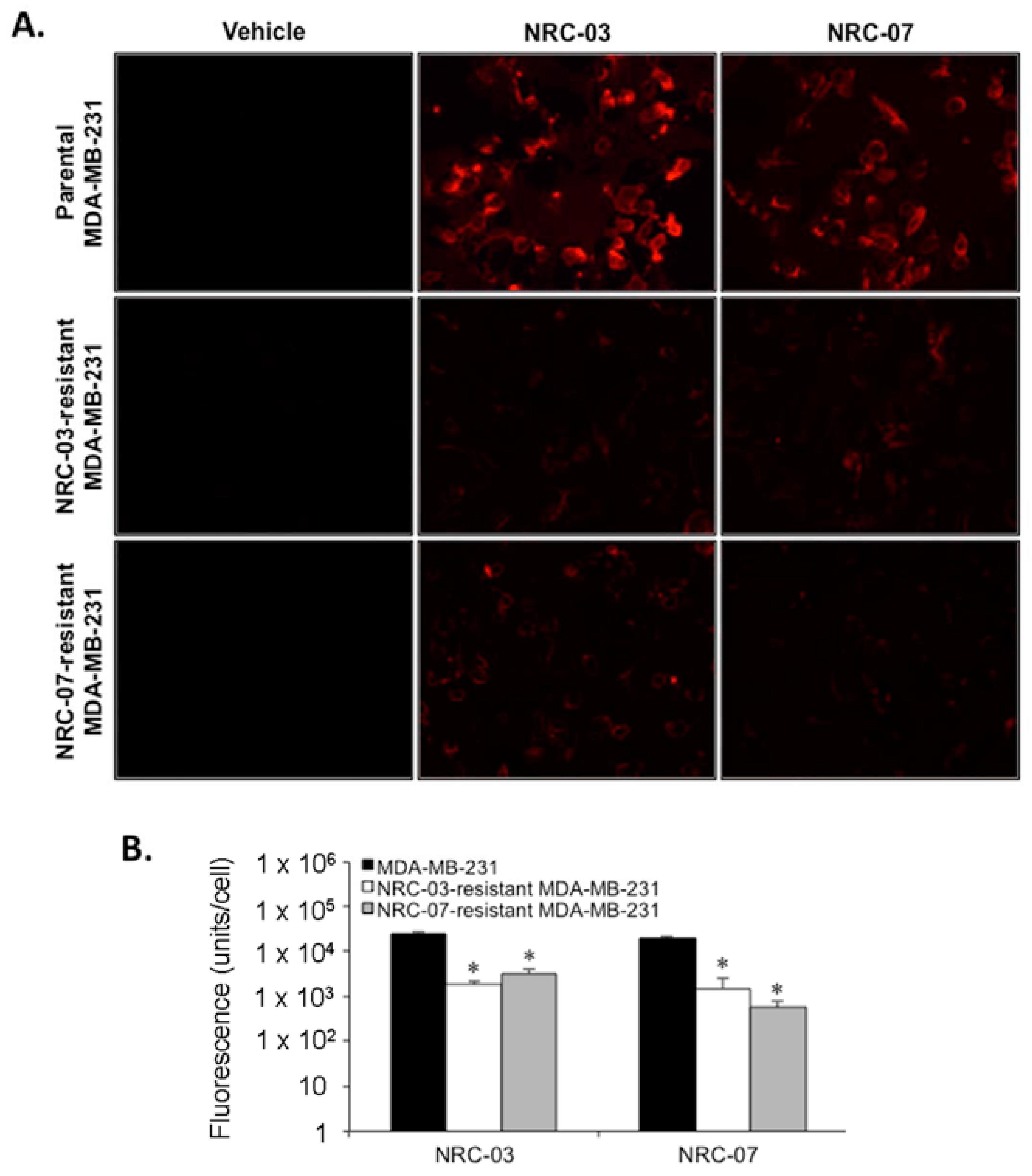
3.3. Breast Cancer Cell Resistance to NRC-03 and NRC-07 Is Associated With Reduced Peptide Binding and Altered Cell Morphology
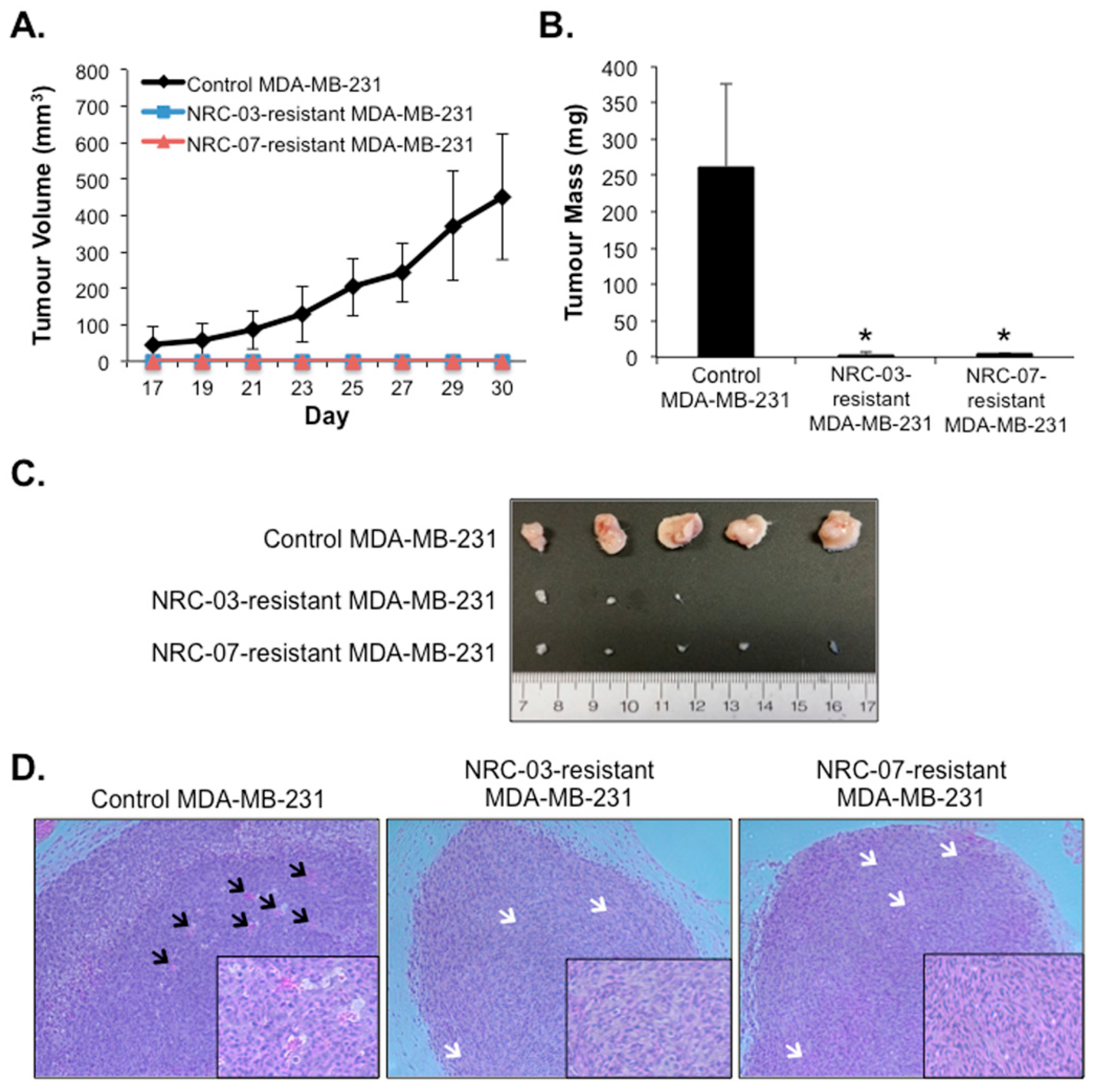
3.4. NRC-03-Resistant and NRC-07-Resistant Breast Cancer Cell-Derived Xenografts Exhibit Impaired Growth and Angiogenesis in Immune-Deficient Mice
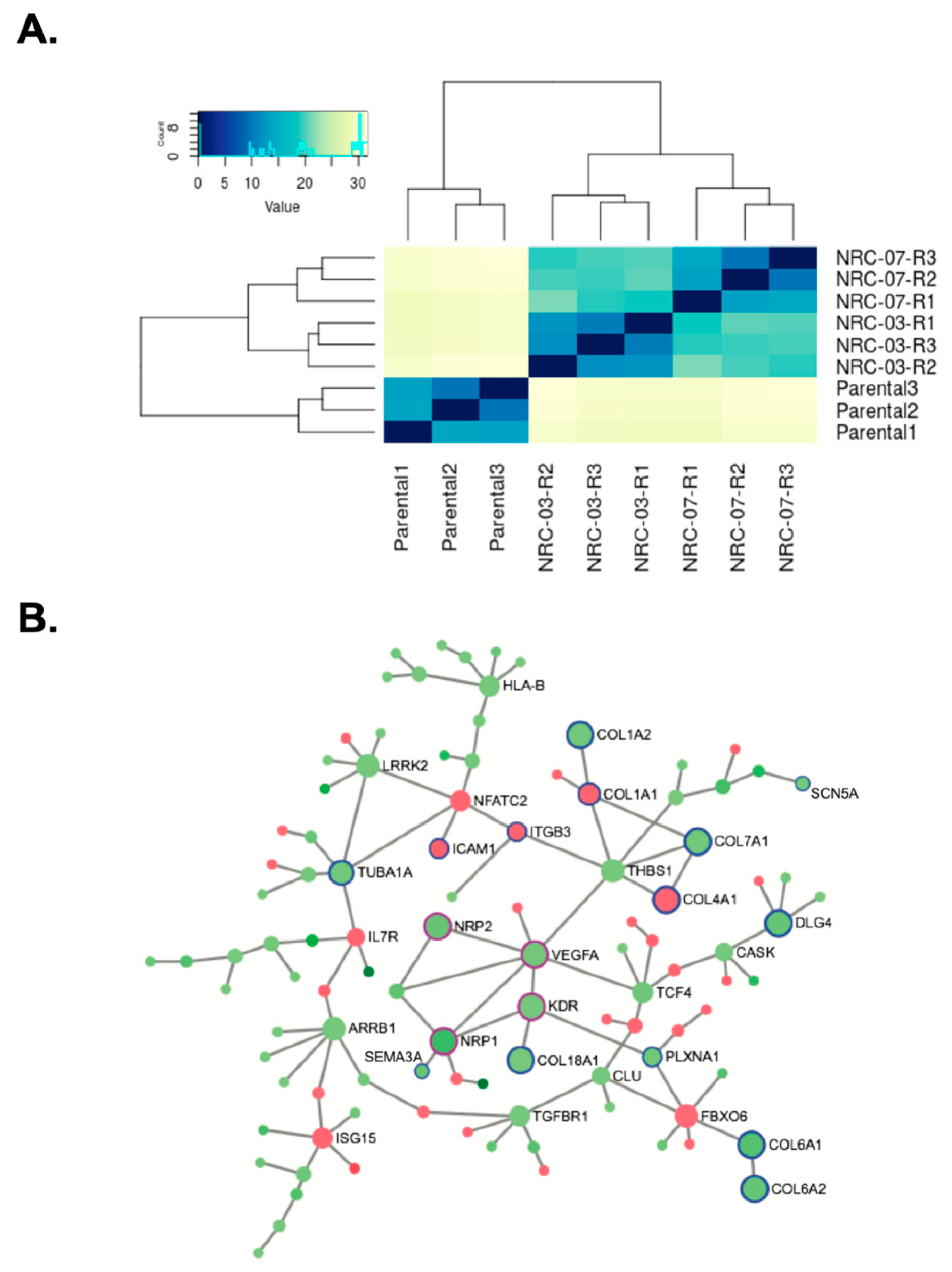
3.5. Resistance to NRC-03 and NRC-07 Is Associated With Altered Expression of Genes Involved in ECM Organization and Angiogenesis
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hilchie, A.L.; Wuerth, K.; Hancock, R.E.W. Immune modulation by multifaceted cationic host defense (antimicrobial) peptides. Nat. Chem. Biol. 2013, 9, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Hoskin, D.W.; Ramamoorthy, A. Studies on anticancer activities of antimicrobial peptides. Biochem. Biophys. Acta 2008, 1778, 357–375. [Google Scholar] [CrossRef] [PubMed]
- Hilchie, A.L.; Power-Coombs, M.R.; Hoskin, D.W. Obstacles and solutions to the use of antimicrobial peptides in the treatment of cancer. In Small Wonders: Peptides for Disease Control; Rajasekaran, K., Cary, J.W., Jaynes, J.M., Montesinos, E., Eds.; American Chemical Society: Washington DC, USA, 2010; pp. 61–78. [Google Scholar]
- Hilchie, A.L.; Hoskin, D.W. The application of cationic antimicrobial peptides in cancer treatment: Laboratory investigations and clinical potential. In Emerging Cancer Therapy; Fialho, A., Chakrabarty, A., Eds.; John Wiley & Sons, Inc.: Hoboken NJ, USA, 2010; pp. 309–332. [Google Scholar]
- Hilchie, A.L.; Doucette, C.D.; Pinto, D.M.; Patrzykat, A.; Douglas, S.; Hoskin, D.W. Pleurocidin-family cationic antimicrobial peptides are cytolytic for breast carcinoma cells and prevent growth of tumor xenografts. Breast Cancer Res. 2011, 13, 102. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, S.S.; Bang, Y.J.; Kim, S.J.; Lee, B.J. In vitro activities of native and designed peptide antibiotics against drug sensitive and resistant tumor cell lines. Peptides 2003, 24, 945–953. [Google Scholar] [CrossRef]
- Johnstone, S.A.; Gelmon, K.; Mayer, L.D.; Hancock, R.E.W.; Bally, M.B. In vitro characterization of the anticancer activity of membrane-active cationic peptides. I. Peptide-mediated cytotoxicity and peptide-enhanced cytotoxic activity of doxorubicin against wild-type and p-glycoprotein over-expressing tumor cell lines. Anticancer Drug Des. 2000, 15, 151–160. [Google Scholar]
- Hilchie, A.L.; Sharon, A.J.; Haney, E.F.; Hoskin, D.W.; Bally, M.B.; Franco, O.L.; Corcoran, J.A.; Hancock, R.E.W. Mastoparan is a membranolytic anti-cancer peptide that works synergistically with gemcitabine in a mouse model of mammary carcinoma. Biochim. Biophys. Acta 2016, 1858, 3195–3204. [Google Scholar] [CrossRef]
- Hansel, W.; Enright, F.; Leuschner, C. Destruction of breast cancers and their metastases by lytic peptide cnjugates in vitro and in vivo. Mol. Cell. Endocrinol. 2007, 260, 183–189. [Google Scholar] [CrossRef]
- Berge, G.; Eliassen, L.T.; Camillio, K.A.; Bartnes, K.; Sveinbjørnsson, B.; Rekdal, O. Therapeutic vaccination against a murine lymphoma by intratumoral injection of a cationic anticancer peptide. Cancer Immunol. Immunother. 2010, 59, 1285–1294. [Google Scholar] [CrossRef]
- Camilio, K.A.; Berge, G.; Ravuri, C.S.; Rekdal, O.; Sveinbjørnsson, B. Complete regression and systemic protective immune responses obtained in B16 melanomas after treatment with LTX-315. Cancer Immunol. Immunother. 2014, 63, 601–613. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, 36. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genoma Project Data Processing Subgroup, The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Lynn, D.J.; Windsor, G.L.; Chan, C.; Richar, N.; Laird, M.R.; Barsky, A.; Gardy, J.L.; Roche, F.M.; Chan, T.H.; Shah, N.; et al. InnateDB: Facilitating systems-level analyses of the mammalian innate immune response. Mol. Syst. Biol. 2008, 4, 218. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Gill, E.E.; Hancock, R.E.W. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat. Protoc. 2015, 10, 823–844. [Google Scholar] [CrossRef]
- Moreno, M.; Giralt, E. Three valuable peptides from bee and wasp venoms for therapeutic and biotechnological use: Melittin, apamin and mastoparan. Toxins 2015, 7, 1126–1150. [Google Scholar] [CrossRef]
- Liu, C.Y.; Hung, M.H.; Wang, D.S.; Chu, P.Y.; Su, J.C.; Teng, T.H.; Huang, C.T.; Chao, T.T.; Wang, C.Y.; Shiau, C.W.; et al. Tamoxifen induces apoptosis through cancerous inhibitor of protein phosphatase 2A-dependent phospho-Akt inactivation in estrogen receptor-negative human breast cancer cells. Breast Cancer Res. 2014, 16, 431–446. [Google Scholar] [CrossRef]
- Schröder-Borm, H.; Bakalova, R.; Andrä, J. The NK-lysin derived peptide NK-2 preferentially kills cancer cells with increased surface levels of negatively charged phosphatidylserine. FEBS Lett. 2005, 579, 6128–6134. [Google Scholar] [CrossRef]
- Iwasaki, T.; Ishibashi, J.; Tanaka, H.; Sato, M.; Asaoka, A.; Taylor, D.; Yamakawa, M. Selective cancer cell cytotoxicity of enantiomeric 9-mer peptides derived from beetle defensins depends on negatively charged phosphatidylserine on the cell surface. Peptides 2009, 30, 660–668. [Google Scholar] [CrossRef]
- Fadnes, B.; Uhlin-Hansen, L.; Lindin, I.; Rekdal, O. Small lytic peptides escape the inhibitory effect of heparan sulfate on the surface of cancer cells. BMC Cancer 2011, 11, 1–11. [Google Scholar] [CrossRef]
- Hilchie, A.L.; Haney, E.F.; Pinto, D.M.; Hancock, R.E.W.; Hoskin, D.W. Enhanced killing of breast cancer cells by a d-amino acid analog of the winter flounder-derived pleurocidin NRC-03. Exp. Mol. Pathol. 2015, 99, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Mader, J.; Smyth, D.; Marshall, J.; Hoskin, D.W. Bovine lactoferricin inhibits basic fibroblast growth factor- and vascular endothelial growth factor165-induced angiogenesis by competing for heparin-like binding sites on endothelial cells. Am. J. Pathol. 2006, 169, 1753–1766. [Google Scholar] [CrossRef] [PubMed]
| Pathway | Adjusted p-Value | DE Genes in Pathway (Weighted) | Pathway Size | Genes |
|---|---|---|---|---|
| Molecules associated with elastic fibres | p < 0.001 | 23 | 728 | ITGB3;FN1;LTBP3;EFEMP2;FBN1;SERPINH1;PLOD2;MATN3;ADAM12;IL18;OLR1;MMP19;MMP15;COL12A; ICAM1;CDC14B;CXADR;IL7R;SDC4;LUM;COL5A2;COL4A4;COL4A2;COL4A3 |
| Cell-ECM interactions | p < 0.001 | 10 | 72 | LIMS2;FERMT2;FNLC;FBLIM1;LIMS1 |
| VEGF receptor 2-mediated cell proliferation | p < 0.001 | 15 | 365 | PRKZ2;SPHK1;DUSP1;SPTB;RASGRP3;RASA4;FN1;KBTBD7;ITPR2;STPBN2;IL6R |
| Gap junction trafficking and regulation | p < 0.001 | 10 | 211 | TUBA4A;TUBB6;SRF;TTC21B;DIAPH3;FMNL2;RHOD;DAAM1;TTBK2;BORA;IQCB1;PROS1;RHOB;RAB3IP; FN1;ITGB3 |
| Downstream signaling of activated FGF receptor 1 | p < 0.001 | 14 | 570 | PEA15;CNKSR1;CSF2RA;TNFC6C;RASA4;FN1;ITPR2;FOXO1;DUSP8;ITGB3;PRKCZ;SPHK1;IL6R |
| Signalling to RAS | p < 0.001 | 13 | 597 | FN1;ITGB3;DUSP8;RASA4;KBTBD7;PRKCZ;SPHK1;DUSP16;PROS1;COL5A2 |
| Signaling by Interleukins | p < 0.001 | 14 | 620 | IL18;IL7R;IL1A;IL6R;RAPGEF1;GAB2;SPTB;RASA4;DUSP16;DUSP1 |
| SHC1 events in EGF receptor signaling | p < 0.001 | 8 | 222 | CSF2RA;CNKSR1;DUSP8;DUSP16;SPTBN2;RAPGEF1;RIT1 |
| Signalling to p38 via RIT and RIN | p < 0.001 | 6 | 86 | RIT1;SPTBN2;DUSP16;RAPGEF1;RASGRP3;PEA15;DUSP1;KBTBD7 |
| Assembly of collagen fibrils and other multimeric structures | p < 0.001 | 8 | 165 | LOX;LOXL3;COL5A2;COL4A4;COL4A3;COL1A1;COL4A2;COL4A1 |
| RHO GTPases activate formins | p < 0.001 | 12 | 683 | SRF;DIAPH3;FMNL2;RHOD;DAAM1RHOB |
| Collagen degradation | p = 0.002 | 15 | 1545 | COL12A1;MMP19;MMP15;COL5A2;COL4A4;COL4A3;COL4A2;COL4A1;COL1A1 |
| ARMS-mediated activation | p = 0.003 | 8 | 398 | SPTB;RASA4;FN1;ITGB3RAPGEF1;RIT1 |
| RHO GTPase effectors | p = 0.003 | 21 | 3014 | RHPN2;CTTN;PKN3;AB12;BAIAP2;SRF;DIAPH3;FMNL2RHOD;MYL9;MYL12B;TUBB6;RHOB |
| NOD1/2 signaling pathway | p = 0.006 | 4 | 17 | CASP4;CASP2;CYLD;TNFAIP3;BIRC3 |
| Signaling by FGFR1 | p = 0.008 | 17 | 2231 | ITGB3;FOXO1;CSF2RA;SPTB;RASA4;KBTBD7;SPTBN2;DUSP16;DUSP1;DNAL4;ABI2;BAIAP2;IL6R;STAT1; CDC14B;MATN3;ICAM1;FBNM1;NCOA3 |
| Downstream signaling of activated FGF receptor 4 | p = 0.026 | 13 | 1508 | FN1;CNKSR1;PEA15;NR4A1;TNRC6C;RASA4;DUSP!;ITPR2;FOXO1;DNAL4;MATN3;ABI2;BAIAP2;IL6R;STAT1; ICAM1;MMP19;RIT1;MMP15 |
| Effects of PIP2 hydrolysis | p = 0.036 | 11 | 1129 | DGKA;DGKH;MGLL;ITPR2;PRKCz;ADAM12;PROS1;RHOB;GAB2;PLCG2;TUBA4A;SPTB;RASA4;DUSP16; DUSP1;KBTBD7;RASGRP3;SPTBN2;DUSP8 |
| Pathway | Adjusted p-Value | DE Genes in Pathway (Weighted) | Pathway Size | Genes |
|---|---|---|---|---|
| ECM organization | p < 0.001 | 78 | 3507 | MUSK;PTPRS;LRP4;NTN4;LTBP1;LTBP4;LTBP2;BMP4;GDF5;ADAM15;ADAM8;COL27A1;MMP16;CASK; ITGA3;MMP14;KDR;ITGA7;ADAMTS9;CTSD;ITGB8;TIMP1;TNC;THBS1;COL8A1;COL7A1;COL18A1;HSPG2; COL6A1;COL6A2;COL5A1;COL4A5;COL1A2 |
| L1CAM interactions | p < 0.001 | 39 | 991 | CNTNAP1;NRP2;MSN;ANK1;NRCAM;ANK2;SCN9A;SCN5A;EPHB2;DPYSL2;KIF4A;DLG3;DLG1;NRP1; RPS6KA5;TUBA1A;DLG4;KCNQ3 |
| Asymmetric localization of PCP proteins | p < 0.001 | 15 | 93 | SMURF2;PRICKLE1;FZD7;FZD1;FZD8;FZD2 |
| Signaling by VEGF | p < 0.001 | 25 | 406 | PGF;NRP2;VEGFC;VEGFA;NRP1;AXL;CYFIP2;KDR;WASF3;CYBA;RASGRF1;DUSP4;EREG;DUSP6;DUSP10; IL17RD;RASA1;CNKSR2;ANGPT1;NRG1;IRS1;ARRB1;EGF;DLG4 |
| Rho GTPase cycle | p < 0.001 | 82 | 4631 | OPHN1;ARHGAP5;CHN2;RHOU;RHOJ;ARHGAP22;STARD8;ARHGAP29;ARHGAP24;FAM13A;ARHGAP26; FGD1;ARHGEF4;NET1;ARHGEF6 |
| Downregulation of SMAD2/3:SMAD4 transcriptional activity | p < 0.001 | 24 | 425 | NEDD4L;SMURF2;HDAC1;SMAD3;TGIF1;UBE2A;ASB9;DET1;SPSB1;RNF182;PJA1;ASB13;RNF43;ANO2; ANO8;NALCN;CLCN3;SCNN1A;CLCN2;SCNN1D;ANO5;TPCN1;DZIP1;WWTR1;RCHY1;SOX4;SOX9;GPR161; CTNNBIP1;PRKG1;UBE2L6;NGFRAP1;DNER;CDON;CAV1;SLC25A5;GNAO1;FGD1;ARHGEF4;TLE3;NET1; XIAP;ARHGEF6;FZD1;FZD8;FZD2;NGFR;AMER1;CDC20;H2AFZ;ARRB1 |
| O-glycosylation of TSR domain-containing proteins | p < 0.001 | 26 | 618 | SEMA5A;ADAMTS12;SPON2;ADAMTS9;ADAMTS15;ADAMTSL1;THBS2;THBS1 |
| O-linked glycosylation | p < 0.001 | 31 | 909 | GALNT18;GALNT12;GALNT5;GALNT14;C1GALT1C1;B3GNT5;ADAMTS12;SPON2;ADAMTS15;ADAMTSL1; THBS2;SEMA5A;ADAMTS9;B3GNT7;CFP;THBS1;ST6GAL1;ST3GAL4 |
| Signaling by platelet derived growth factor | p < 0.001 | 22 | 499 | PDGFD;PLAT;THBS2;THBS1;PDE1C;CAMK4;COL6A1;COL6A2;COL5A1;COL4A5;RASGRF1;DUSP4;EREG; DLG4;DUSP6;DUSP10;IL17RD;RASA1;CNKSR2;ANGPT1;NRG1;IRS1;ARRB1;EGF |
| Collagen degradation | p < 0.001 | 37 | 1545 | CTSD;COL6A1;COL8A1;COL13A1;COL6A2;COL5A1;MMP14;COL18A1;COL7A1;COL4A5;COL1A2 |
| Degradation of the extracellular matrix | p < 0.001 | 32 | 1242 | ADAM15;ADAM8;MMP16;ADAMTS9;TIMP1;CTSS;CTSD;COL13A1;HSPG2;MMP14;COL8A1;COL7A1; COL6A1;COL6A2;COL5A1;COL4A5;COL1A2;COL18A1 |
| Downstream signaling of activated FGF receptor 1 | p < 0.001 | 21 | 570 | PRKAR2B;PIK3R3;CAMK4;DUSP6;IRS1;DUSP4;EREG;DLG4;DUSP10;IL17RD;RASA1;CNKSR2;PDE1C;NRG1; NRG2;GNAO1;KDR;SYNJ2;PIK3CG;AJUBA;NPHP4;SPRY2;PI4K2B;DLG3;GRIK4;GRIK2;DLG1;GNB4 |
| Interaction between L1 and ankyrins | p < 0.001 | 13 | 219 | ANK1;NRCAM;ANK2;SCN9A;SCN5A;KCNQ3 |
| Glycosaminoglycan metabolism | p < 0.001 | 29 | 1239 | HAS2;PAPSS1;HS3ST1;HS3ST3B1;NDST3;B3GNT7;NAGLU;UST;CHST11;GXYLT2;CHST15;EXT1;ST3GAL4; IDS;IDUA;CSPG4;HSPG2 |
| Chondroitin sulfate/dermatan sulfate metabolism | p < 0.001 | 17 | 450 | HSPG2;UST;GXYLT2;CHST11;CHST15;IDS;IDUA;CSPG4;HS3ST1;HS3ST3B1;NDST3;NAGLU;EXT1 |
| Hexose transport | p < 0.001 | 7 | 62 | SLC2A1;PGLS;SORD;SLC4A7;SLC29A1;SLC9A5;SLC29A3;SLCO4A1;GALK1;SLC16A10;PFKFB4;KHK;PFKL; G6PD;HAS2;HS3ST1;ENO1;UST;CHST11;CHST15;SLC7A5;PAPSS1;SLC1A1;NAGLU;B3GNT7;IDS;GYG2; SLC2A1;PGLS;SORD;SLC4A7;SLC29A1;SLC9A5;SLC29A3;SLCO4A1;GALK1;SLC16A10;PFKFB4;KHK;PFKL;G6PD; HAS2;HS3ST1;ENO1;UST;CHST11;CHST15;SLC7A5;PAPSS1;SLC1A1;NAGLU;B3GNT7;IDS;GYG2;ST3GAL4; HSPG2;PRKAA2ST3GAL4;HSPG2;PRKAA2 |
| Signaling by EGF receptor | p < 0.001 | 16 | 428 | LRIG1;SPRY1;SH3KBP1;PDE1C;CAMK4;RASGRF1;DUSP4;DLG4;ARRB1;DUSP6;DUSP10;IL17RD;RASA1; CNKSR2;ANGPT1;EREG;NRG1;IRS1;SPRY2;EGF |
| O-linked glycosylation of mucins | p < 0.001 | 26 | 1231 | GALNT18;GALNT12;GALNT5;GALNT14;C1GALT1C1;B3GNT5;B3GNT7;ST6GAL1;ST3GAL4 |
| Collagen formation | p = 0.001 | 28 | 1336 | CTSS;COLGALT2;LOXL4;COL13A1;MUSK;PTPRS;LRP4;NTN4;LTBP1;LTBP4;LTBP2;BMP4;GDF5;ADAM15; ADAM8;ITGA7;CASK;ITGA3;TNC;MMP16;CD74;CTSH;CTSC;CTSF;KDR;MMP14;ADAMTS9;ITGB8;THBS1; TIMP1;CTSD;HSPG2;UBE2A;ASB9;DET1;ERAP1;SPSB1;RNF182;PJA1;ASB13;KIF4A;KIF3C;RCHY1;DYNC2H1; SIGIRR;IRAK3;NEDD4L;CENPE;SMURF2;PELI2;MEF2C;RPS6KA5;CDC20;ELK1;TAB3;DUSP4;DUSP6;TUBA1A |
| VEGF receptor 2-mediated cell proliferation | p = 0.001 | 14 | 365 | ITPR1;KDR;IL17RD;ANGPT1;DLG4;NRG2;RASGRF1;DUSP4;ARRB1;EGF;GNAO1;PPP3CA;SPTBN5;SPRY2; PDE3B;PLA2G4A |
| The activation of arylsulfatases | p = 0.006 | 6 | 66 | ARSD;ARSE;ARSJ;ARSI |
| HS-GAG biosynthesis | p = 0.006 | 10 | 236 | HS3ST1;HS3ST3B1;NDST3;EXT1;HSPG2 |
| MyD88:Mal cascade initiated on plasma membrane | p = 0.006 | 8 | 101 | IRAK3;SIGIRR;SAA1;MEF2C;RPS6KA5;ELK1;TAB3;DUSP4 |
| Post-translational protein modification | p = 0.012 | 106 | 9940 | PIGA;MAN1A1;ARSD;ARSE;ARSJ;ARSI;PHC1;PHC2;FUT8;GAS6;ALG6;ALG13;ST3GAL5;ST6GALNAC5;MGAT4A; GALNT18;GALNT12;GALNT5;ADAMTS12;GALNT14;SPON2;ADAMTS15;C1GALT1C1;B3GNT5;ADAMTSL1;MPI; STAG2;ST8SIA4;THBS2;SRD5A3;AAAS;SEC31A;SEMA5A;ADAMTS9;CALR;B3GNT7;CFP;THBS1;ST6GAL1;ST3GAL4 |
| Heparan sulfate/heparin (HS-GAG) metabolism | p = 0.013 | 26 | 1440 | HS3ST3B1;NDST3;EXT1;GXYLT2;IDUA;CSPG4;PGLS;SORD;PFKFB4;PFKL;G6PD;KHK;ENO1;GALK1;SLC25A10; SLC2A1;GYG2;AAAS;ALG6;ALG13;MPI;SRD5A3;HLCS;CYP2U1;FDXR;TBXAS1;ACACB |
| Ethanol oxidation | p = 0.024 | 8 | 169 | ALDH2;ACSS1;CYP2U1;PTGS1;GLUL;ALDH5A1;CYP2J2;FDXR;TBXAS1;BCHE;CYP39A1;DLG3;GRIK4;GRIK2; CACNB3;PPFIA4;CASK;DLG1;RIMS1;GNB4;CAMK4;DLG4 |
| Constitutive Signaling by EGF receptor vIII | p = 0.031 | 4 | 27 | EGF;LAMP2;CLU;KDR;VEGFC;TIMP1;GAS6;VEGFA;THBS1;SPRY2 |
| Glycerophospholipid biosynthesis | p = 0.042 | 29 | 1731 | PNPLA3;CDS1;PEMT;DGAT2;LPCAT2;MBOAT1;SLC44A5;PLB1;BCHE;GPAT3;GPD1L;PLA2G12A;PLA2G4A |
| Integrin cell surface interactions | p = 0.046 | 16 | 712 | ITGB8;KDR;ITGA3;THBS1;TNC;COL13A1;ITGA7;COL8A1;COL6A1;COL6A2;COL7A1;COL18A1;COL5A1; COL1A2;COL4A5;HSPG2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hilchie, A.L.; Gill, E.E.; Coombs, M.R.P.; Falsafi, R.; Hancock, R.E.W.; Hoskin, D.W. MDA-MB-231 Breast Cancer Cells Resistant to Pleurocidin-Family Lytic Peptides Are Chemosensitive and Exhibit Reduced Tumor-Forming Capacity. Biomolecules 2020, 10, 1220. https://doi.org/10.3390/biom10091220
Hilchie AL, Gill EE, Coombs MRP, Falsafi R, Hancock REW, Hoskin DW. MDA-MB-231 Breast Cancer Cells Resistant to Pleurocidin-Family Lytic Peptides Are Chemosensitive and Exhibit Reduced Tumor-Forming Capacity. Biomolecules. 2020; 10(9):1220. https://doi.org/10.3390/biom10091220
Chicago/Turabian StyleHilchie, Ashley L., Erin E. Gill, Melanie R. Power Coombs, Reza Falsafi, Robert E. W. Hancock, and David W. Hoskin. 2020. "MDA-MB-231 Breast Cancer Cells Resistant to Pleurocidin-Family Lytic Peptides Are Chemosensitive and Exhibit Reduced Tumor-Forming Capacity" Biomolecules 10, no. 9: 1220. https://doi.org/10.3390/biom10091220
APA StyleHilchie, A. L., Gill, E. E., Coombs, M. R. P., Falsafi, R., Hancock, R. E. W., & Hoskin, D. W. (2020). MDA-MB-231 Breast Cancer Cells Resistant to Pleurocidin-Family Lytic Peptides Are Chemosensitive and Exhibit Reduced Tumor-Forming Capacity. Biomolecules, 10(9), 1220. https://doi.org/10.3390/biom10091220







