Dietary Supplementation with Sugar Beet Fructooligosaccharides and Garlic Residues Promotes Growth of Beneficial Bacteria and Increases Weight Gain in Neonatal Lambs
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Sampling and Experimental Setup
2.3. Total DNA Extraction, 16S rRNA Gene Amplicon Library Preparation and Sequencing
2.4. Bioinformatics and Data Analysis
3. Results
3.1. Effect of the Diet on Weight Gain and Blood Immunoglobulin Concentration of Lambs
3.2. Sequencing Depth, Alpha Diversity and Lamb Fecal Bacteria Composition
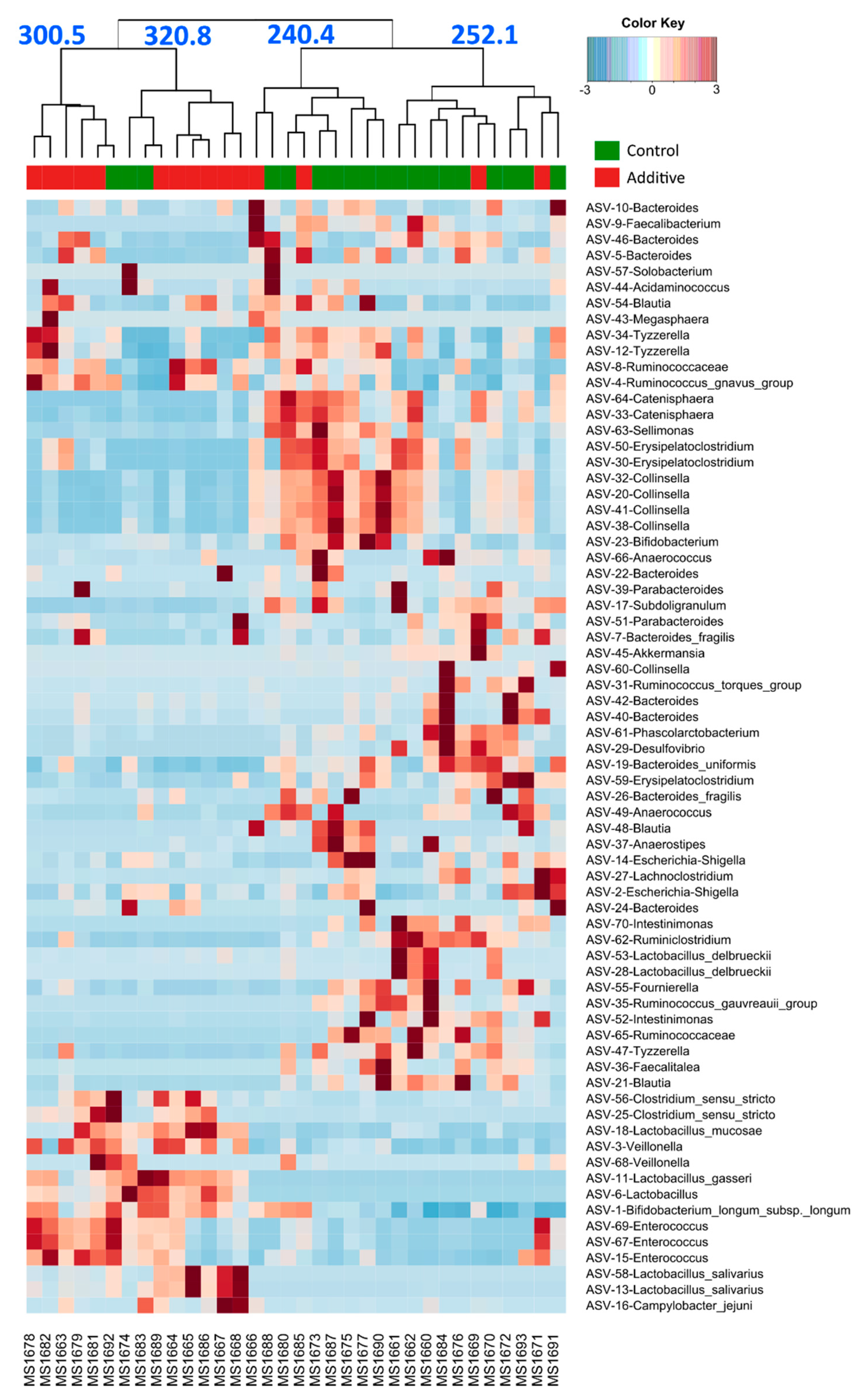
3.3. Evaluation of the Effect of Additive Intake over Lamb Fecal Microbiota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tanca, A.; Fraumene, C.; Manghina, V.; Palomba, A.; Abbondio, M.; Deligios, M.; Pagnozzi, D.; Addis, M.F.; Uzzau, S. Diversity and functions of the sheep faecal microbiota: A multi-omic characterization. Microb. Biotechnol. 2017, 10, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J.; Bayer, E.A.; Rincon, M.T.; Lamed, R.; White, B.A. Polysaccharide utilization by gut bacteria: Potential for new insights from genomic analysis. Nat. Rev. Microbiol. 2008, 6, 121. [Google Scholar] [CrossRef] [PubMed]
- Hess, M.; Sczyrba, A.; Egan, R.; Kim, T.W.; Chokhawala, H.; Schroth, G.; Luo, S.; Clark, D.S.; Chen, F.; Zhang, T.; et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 2011, 331, 463–467. [Google Scholar] [CrossRef] [PubMed]
- Ellison, M.J.; Conant, G.C.; Cockrum, R.R.; Austin, K.J.; Truong, H.; Becchi, M.; Lamberson, W.R.; Cammack, K.M. Diet alters both the structure and taxonomy of the ovine gut microbial ecosystem. DNA Res. 2013, 21, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Malmuthuge, N.; Griebel, P.J.; Guan, L.L. Taxonomic identification of commensal bacteria associated with the mucosa and digesta throughout the gastrointestinal tracts of pre-weaned calves. Appl. Environ. Microbiol. 2014, 80, 2012–2028. [Google Scholar] [CrossRef]
- Mao, S.; Zhang, M.; Liu, J.; Zhu, W. Characterising the bacterial microbiota across the gastrointestinal tracts of dairy cattle: Membership and potential function. Sci. Rep. 2015, 5, 16116. [Google Scholar] [CrossRef]
- Mackie, R.I. Mutualistic fermentative digestion in the gastrointestinal tract: Diversity and evolution. Integr. Comp. Biol. 2002, 42, 319–326. [Google Scholar] [CrossRef]
- Henderson, G.; Cox, F.; Ganesh, S.; Jonker, A.; Young, W.; Global Rumen Census Collaborators; Ariza, C. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 2015, 5, 14567. [Google Scholar] [CrossRef]
- Maynard, C.L.; Elson, C.O.; Hatton, R.D.; Weaver, C.T. Reciprocal interactions of the intestinal microbiota and immune system. Nature 2012, 489, 231–241. [Google Scholar] [CrossRef]
- Wopereis, H.; Oozeer, R.; Knipping, K.; Belzer, C.; Knol, J. The first thousand days-intestinal microbiology of early life: Establishing a symbiosis. Pediat. Allerg. Immunol. UK 2014, 25, 428–438. [Google Scholar]
- Flint, H.J.; Duncan, S.H.; Scott, K.P.; Louis, P. Interactions and competition within the microbial community of the human colon: Links between diet and health. Environ. Microbiol. 2007, 9, 1101–1111. [Google Scholar] [CrossRef] [PubMed]
- Sommer, F.; Backhed, F. The gut microbiota–masters of host development and physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Malmuthuge, N.; Griebel, P.J.; Guan, L.L. The gut microbiome and its potential role in the development and function of newborn calf gastrointestinal tract. Front. Vet. Sci. 2015, 2, 36. [Google Scholar] [CrossRef] [PubMed]
- Jost, T.; Lacroix, C.; Braegger, C.P.; Chassard, C. New insights in gut microbiota establishment in healthy breast-fed neonates. PLoS ONE 2012, 7, e44595. [Google Scholar] [CrossRef] [PubMed]
- Danzeisen, J.L.; Calvert, A.J.; Noll, S.L.; McComb, B.; Sherwood, J.S.; Logue, C.M.; Johnson, T.J. Succession of the turkey gastrointestinal bacterial microbiome related to weight gain. PeerJ 2013, 1, e237. [Google Scholar] [CrossRef] [PubMed]
- Uyeno, Y.; Sekiguchi, Y.; Kamagata, Y. rRNA-based analysis to monitor succession of faecal bacterial communities in Holstein calves. Lett. Appl. Microbiol. 2010, 51, 570–577. [Google Scholar] [CrossRef]
- Jami, E.; Israel, A.; Kotser, A.; Mizrahi, I. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 2013, 7, 1069. [Google Scholar] [CrossRef]
- Oikonomou, G.; Teixeira, A.G.V.; Foditsch, C.; Bicalho, M.L.; Machado, V.S.; Bicalho, R.C. Fecal microbial diversity in pre-weaned dairy calves as described by pyrosequencing of metagenomic 16S rDNA. Associations of Faecalibacterium species with health and growth. PLoS ONE 2013, 8, e63157. [Google Scholar] [CrossRef]
- Klein-Jöbstl, D.; Schornsteiner, E.; Mann, E.; Wagner, M.; Drillich, M.; Schmitz-Esser, S. Pyrosequencing reveals diverse fecal microbiota in Simmental calves during early development. Front. Microbiol. 2014, 5, 622. [Google Scholar]
- Kim, M.; Kim, J.; Kuehn, L.; Bono, J.L.; Berry, E.D.; Kalchayanand, N.; Freetly, H.C.; Benson, A.K.; Wells, J.E. Investigation of bacterial diversity in the feces of cattle fed different diets. J. Anim. Sci. 2014, 92, 683–694. [Google Scholar] [CrossRef]
- Bi, Y.; Cox, M.S.; Zhang, F.; Suen, G.; Zhang, N.; Tu, Y.; Diao, Q. Feeding modes shape the acquisition and structure of the initial gut microbiota in newborn lambs. Environ. Microbiol. 2019, 21, 2333–2346. [Google Scholar] [CrossRef] [PubMed]
- Lyons, T.; Boland, T.; Storey, S.; Doyle, E. Linseed oil supplementation of lambs’ diet in early life leads to persistent changes in rumen microbiome structure. Front. Microbiol. 2017, 8, 1656. [Google Scholar] [CrossRef] [PubMed]
- Yáñez-Ruiz, D.R.; Abecia, L.; Newbold, C.J. Manipulating rumen microbiome and fermentation through interventions during early life: A review. Front. Microbiol. 2015, 6, 1133. [Google Scholar] [CrossRef] [PubMed]
- Abecia, L.; Ramos-Morales, E.; Martinez-Fernandez, G.; Arco, A.; Martin-Garcia, A.I.; Newbold, C.J. Feeding management in early life influences microbial colonisation and fermentation in the rumen of newborn goat kids. Anim. Prod. Sci. 2014, 54, 1449–1454. [Google Scholar] [CrossRef]
- Rey, M.; Enjalbert, F.; Combes, S.; Cauquil, L.; Bouchez, O.; Monteils, V. Establishment of ruminal bacterial community in dairy calves from birth to weaning is sequential. J. Appl. Microbiol. 2014, 116, 245–257. [Google Scholar] [CrossRef]
- Huang, Q.; Holman, D.B.; Alexander, T.; Hu, T.; Jin, L.; Xu, Z.; McAllister, T.A.; Acharya, S.; Zhao, G.; Wang, Y. Fecal microbiota of lambs fed purple prairie clover (Dalea purpurea Vent.) and alfalfa (Medicago sativa). Arch. Microbiol. 2018, 200, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Rautava, S.; Luoto, R.; Salminen, S.; Isolauri, E. Microbial contact during pregnancy, intestinal colonization and human disease. Nat. Rev. Gastro. Hepat. 2012, 9, 565–576. [Google Scholar] [CrossRef]
- Stinson, L.F.; Payne, M.S.; Keelan, J.A. Planting the seed: Origins, composition, and postnatal health significance of the fetal gastrointestinal microbiota. Crit. Rev. Microbiol. 2017, 43, 352–369. [Google Scholar] [CrossRef]
- Christian, M.; Sabrina, D.; Francesca, B.; Eoghan, C.; Francesca, T.; Jennifer, M.; Belzer, C.; Palacio, S.D.; Montes, S.A.; Mancabelli, L.; et al. The first microbial colonizers of the human gut: Composition, activities, and health implications of the infant gut microbiota. Microbiol. Mol. Biol. 2017, 81, e00036-17. [Google Scholar]
- Uyeno, Y.; Shigemori, S.; Shimosato, T. Effect of probiotics/prebiotics on cattle health and productivity. Microbes Env. 2015, 30, 126–132. [Google Scholar] [CrossRef]
- Weimer, P.J. Redundancy, resilience, and host specificity of the ruminal microbiota: Implications for engineering improved ruminal fermentations. Front. Microbiol. 2015, 6, 296. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R.; Probert, H.M.; Loo, J.V.; Rastall, R.A.; Roberfroid, M.B. Dietary modulation of the human colonic microbiota: Updating the concept of prebiotics. Nutr. Res. Rev. 2004, 17, 259–275. [Google Scholar] [CrossRef] [PubMed]
- Quigley, J.D., III; Kost, C.J.; Wolfe, T.A. Effects of spray-dried animal plasma in milk replacers or additives containing serum and oligosaccharides on growth and health of calves. J. Dairy Sci. 2002, 85, 413–421. [Google Scholar] [CrossRef]
- Gaggìa, F.; Mattarelli, P.; Biavati, B. Probiotics and prebiotics in animal feeding for safe food production. Int. J. Food Microbiol. 2010, 141, 15–28. [Google Scholar] [CrossRef]
- Grand, E.; Respondek, F.; Martineau, C.; Detilleux, J.; Bertrand, G. Effects of short-chain fructooligosaccharides on growth performance of pre-ruminant veal calves. J. Dairy Sci. 2013, 96, 1094–1101. [Google Scholar] [CrossRef]
- Ricke, S.C. Potential of fructooligosaccharide prebiotics in alternative and nonconventional poultry production systems. Poult. Sci. 2015, 94, 1411–1418. [Google Scholar] [CrossRef] [PubMed]
- Wanapat, M.; Khejornsart, P.; Pakdee, P.; Wanapat, S. Effect of supplementation of garlic powder on rumen ecology and digestibility of nutrients in ruminants. J. Sci. Food Agric. 2008, 88, 2231–2237. [Google Scholar] [CrossRef]
- Filocamo, A.; Nueno-Palop, C.; Bisignano, C.; Mandalari, G.; Narbad, A. Effect of garlic powder on the growth of commensal bacteria from the gastrointestinal tract. Phytomedicine 2012, 19, 707–711. [Google Scholar] [CrossRef]
- Chen, K.; Xie, K.; Liu, Z.; Nakasone, Y.; Sakao, K.; Hossain, A.; Hou, D.X. Preventive Effects and Mechanisms of Garlic on Dyslipidemia and Gut Microbiome Dysbiosis. Nutrients 2019, 11, 1225. [Google Scholar] [CrossRef] [PubMed]
- Ruddock, P.S.; Liao, M.; Foster, B.C.; Lawson, L.; Arnason, J.T.; Dillon, J.A.R. Garlic natural health products exhibit variable constituent levels and antimicrobial activity against Neisseria gonorrhoeae, Staphylococcus aureus and Enterococcus faecalis. Phytother. Res. 2005, 19, 327–334. [Google Scholar] [PubMed]
- Ruiz, R.; Garci, M.P.; Lara, A.; Rubio, L.A. Garlic derivatives (PTS and PTS-O) differently affect the ecology of swine faecal microbiota In Vitro. Vet. Microbiol. 2010, 144, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Peinado, M.J.; Ruiz, R.; Echavarri, A.; Rubio, L.A. Garlic derivative PTS-O is effective against broiler pathogens In Vivo. Poult. Sci. 2012, 91, 2148–2157. [Google Scholar] [CrossRef] [PubMed]
- Vogt, L.; Meyer, D.; Pullens, G.; Faas, M.; Smelt, M.; Venema, K.; Ramasamy, U.; Schols, H.A.; De Vos, P. Immunological properties of inulin-type fructans. Crit. Rev. Food Sci. Nutr. 2014, 55, 414–436. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glockner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with qiime 2′s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, 60. [Google Scholar] [CrossRef]
- Dehority, B.A. Numbers, factors affecting the population and distribution of rumen bacteria. In Rumen Microbiology; Dehority, B.A., Ed.; Nottingham Univ. Press: Nottingham, UK, 2003; pp. 265–294. [Google Scholar]
- Li, R.W.; Connor, E.E.; Li, C.; Baldwin Vi, R.L.; Sparks, M.E. Characterization of the rumen microbiota of pre-ruminant calves using metagenomic tools. Environ. Microbiol. 2012, 14, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Elekwachi, C.; Jiao, J.; Wang, M.; Tang, S.; Zhou, C.; Tan, Z.; Forster, R.J. Changes in metabolically active bacterial community during rumen development, and their alteration by rhubarb root powder revealed by 16S rRNA amplicon sequencing. Front. Microbiol. 2017, 8, 1–22. [Google Scholar] [CrossRef]
- Mayo, B.; Van Sinderen, D. Bifidobacteria Genomics and Molecular Aspects; Caister Academic Press: Poole, UK, 2010. [Google Scholar]
- Al-Saiady, M.Y. Effect of probiotic bacteria on immunoglobulin G concentration and other blood components of newborn calves. J. Anim. Vet. Adv. 2010, 9, 604–609. [Google Scholar] [CrossRef]
- Douillard, F.P.; de Vos, W.M. Functional genomics of lactic acid bacteria: From food to health. Microb. Cell Fact. 2014, 13 (Suppl. 1), S8. [Google Scholar] [CrossRef] [PubMed]
- Abe, F.; Ishibashi, N.; Shimamura, S. Effect of administration of bifidobacteria and lactic acid bacteria to newborn calves and piglets. J. Dairy Sci. 1995, 73, 2838–2846. [Google Scholar] [CrossRef]
- Rada, V.; Vlkov, E.; Nevoral, J.; Trojanov, I. Comparison of bacterial flora and enzymatic activity in faeces of infants and calves. FEMS Microbiol. Lett. 2006, 258, 25–28. [Google Scholar] [CrossRef]
- Vlkova, E.; Nevoral, J.; Rada, V. Distribution of bifidobacteria in the gastrointestinal tract of calves. Folia Microbiol. 2006, 51, 325–328. [Google Scholar] [CrossRef]
- Li, F.; Guan, L.L. Metatranscriptomic profiling reveals linkages between the active rumen microbiome and feed efficiency in beef cattle. Appl. Env. Microbiol. 2017, 83, e00061-17. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, H.; Hutkins, R.W. Fermentation of fructooligosaccharides by lactic acid bacteria and bifidobacteria. Appl. Environ. Microbiol. 2000, 66, 2682–2684. [Google Scholar] [CrossRef] [PubMed]
- Saulnier, D.M.; Molenaar, D.; de Vos, W.M.; Gibson, G.R.; Kolida, S. Identification of prebiotic fructooligosaccharide metabolism in Lactobacillus plantarum WCFS1 through microarrays. Appl. Environ. Microb. 2007, 73, 1753–1765. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.H.; Macfarlane, G.T. Role of intestinal bacteria in nutrient metabolism. Clin. Nutr. 1997, 16, 3–11. [Google Scholar] [CrossRef]
- Hartemink, R.; Van Laere, K.M.J.; Rombouts, F.M. Growth of enterobacteria on fructo-oligosaccharides. J. Appl. Microbiol. 1997, 83, 367–374. [Google Scholar] [CrossRef]
- Benyacoub, J.F.; Rochat, K.-Y.; Saudan, I.; Rochat, N.; Antille, C.; Cherbut, T.; von der Weid, T.; Schiffrin, E.J.; Blum, S. Feeding a diet containing a fructooligosaccharide mix can enhance Salmonella vaccine efficacy in mice. J. Nutr. 2008, 138, 123–129. [Google Scholar] [CrossRef][Green Version]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639. [Google Scholar] [CrossRef]
- Nocek, J.E.; Kautz, W.P.; Leedle, J.A.; Allman, J.G. Ruminal supplementation of direct-fed microbials on diurnal pH variation and in situ digestion in dairy cattle. J. Dairy Sci. 2002, 85, 429–433. [Google Scholar] [CrossRef]
- Emmanuel, D.G.; Jafari, A.; Beauchemin, K.A.; Leedle, J.A.; Ametaj, B.N. Feeding live cultures of Enterococcus faecium and Saccharomyces cerevisiae induces an inflammatory response in feedlot steers. J. Anim. Sci. 2007, 85, 233–239. [Google Scholar] [CrossRef]
- Nocek, J.E.; Kautz, W.P. Direct-fed microbial supplementation on ruminal digestion, health, and performance of pre- and postpartum dairy cattle. J. Dairy Sci. 2006, 89, 260–266. [Google Scholar] [CrossRef]
- Berry, D.; Schwab, C.; Milinovich, G.; Reichert, J.; Mahfoudh, K.B.; Decker, T.; Engel, M.; Hai, B.; Hainzl, E.; Heider, S.; et al. Phylotype-level 16S rRNA analysis reveals new bacterial indicators of health state in acute murine colitis. ISME J. 2012, 6, 2091. [Google Scholar] [CrossRef] [PubMed]
- Quijada, N.M.; De Filippis, F.; Sanz, J.J.; del Camino García-Fernández, M.; Rodriguez-Lazaro, D.; Ercolini, D.; Hernández, M. Different Lactobacillus populations dominate in “Chorizo de León” manufacturing performed in different production plants. Food Microbiol. 2018, 70, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Ercolini, D. High-Throughput Sequencing and Metagenomics: Moving Forward in the Culture-Independent Analysis of Food Microbial Ecology. Appl. Environ. Microbiol. 2013, 79, 3148–3155. [Google Scholar] [CrossRef]
- Mayo, B.; Rachid, C.T.C.C.; Alegría, Á; Leite, A.M.O.; Peixoto, R.S.; Delgado, S. Impact of Next Generation Sequencing Techniques in Food Microbiology. Curr. Genomics 2014, 15, 293–309. [Google Scholar] [CrossRef] [PubMed]
- Jovel, J.; Patterson, J.; Wang, W.; Hotte, N.; O’Keefe, S.; Mitchel, T.; Perry, T.; Kao, D.; Mason, A.L.; Madsen, K.L.; et al. Characterization of the Gut Microbiome Using 16S or Shotgun Metagenomics. Front. Microbiol. 2016, 7, 459. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Li, C.; Chen, Y.; Liu, J.; Zhang, C.; Irving, B.; Fitzsimmons, C.; Plastow, G.; Guan, L.L. Host genetics influence the rumen microbiota and heritable rumen microbial features associate with feed efficiency in cattle. Microbiome 2019, 7, 92. [Google Scholar] [CrossRef]
- Creevey, C.J.; Kelly, W.J.; Henderson, G.; Leahy, S.C. Determining the culturability of the rumen bacterial microbiome. Microb. Biotechnol. 2014, 7, 467–479. [Google Scholar] [CrossRef]
- Seshadri, R.; Leahy, S.C.; Attwood, G.T.; Teh, K.H.; Lambie, S.C.; Cookson, A.L.; Eloe-Fadrosh, E.A.; Pavlopoulos, G.A.; Hadjithomas, M.; Varghese, N.J.; et al. Cultivation and sequencing of rumen microbiome members from the Hungate1000 Collection. Nat. Biotechnol. 2018, 36, 359. [Google Scholar] [CrossRef]
- Stewart, R.D.; Auffret, M.D.; Warr, A.; Wiser, A.H.; Press, M.O.; Langford, K.W.; Liachko, I.; Snelling, T.J.; Dewhurst, R.J.; Walker, A.W.; et al. Assembly of 913 microbial genomes from metagenomic sequencing of the cow rumen. Nat. Commun. 2018, 9, 870. [Google Scholar] [CrossRef]
- Stewart, R.D.; Auffret, M.D.; Warr, A.; Walker, A.W.; Roehe, R.; Watson, M. Compendium of 4,941 rumen metagenome-assembled genomes for rumen microbiome biology and enzyme discovery. Nat. Biotechnol. 2019, 37, 953–961. [Google Scholar] [CrossRef]
- Klein-Jöbstl, D.; Quijada, N.M.; Dzieciol, M.; Feldbacher, B.; Wagner, M.; Drillich, M.; Schmitz-Esser, S.; Mann, E. Microbiota of newborn calves and their mothers reveals possible transfer routes for newborn calves’ gastrointestinal microbiota. PLoS ONE 2019, 14, e0220554. [Google Scholar] [CrossRef] [PubMed]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quijada, N.M.; Bodas, R.; Lorenzo, J.M.; Schmitz-Esser, S.; Rodríguez-Lázaro, D.; Hernández, M. Dietary Supplementation with Sugar Beet Fructooligosaccharides and Garlic Residues Promotes Growth of Beneficial Bacteria and Increases Weight Gain in Neonatal Lambs. Biomolecules 2020, 10, 1179. https://doi.org/10.3390/biom10081179
Quijada NM, Bodas R, Lorenzo JM, Schmitz-Esser S, Rodríguez-Lázaro D, Hernández M. Dietary Supplementation with Sugar Beet Fructooligosaccharides and Garlic Residues Promotes Growth of Beneficial Bacteria and Increases Weight Gain in Neonatal Lambs. Biomolecules. 2020; 10(8):1179. https://doi.org/10.3390/biom10081179
Chicago/Turabian StyleQuijada, Narciso M., Raúl Bodas, Jose M. Lorenzo, Stephan Schmitz-Esser, David Rodríguez-Lázaro, and Marta Hernández. 2020. "Dietary Supplementation with Sugar Beet Fructooligosaccharides and Garlic Residues Promotes Growth of Beneficial Bacteria and Increases Weight Gain in Neonatal Lambs" Biomolecules 10, no. 8: 1179. https://doi.org/10.3390/biom10081179
APA StyleQuijada, N. M., Bodas, R., Lorenzo, J. M., Schmitz-Esser, S., Rodríguez-Lázaro, D., & Hernández, M. (2020). Dietary Supplementation with Sugar Beet Fructooligosaccharides and Garlic Residues Promotes Growth of Beneficial Bacteria and Increases Weight Gain in Neonatal Lambs. Biomolecules, 10(8), 1179. https://doi.org/10.3390/biom10081179







