Biotechnological Valorization of Food Marine Wastes: Microbial Productions on Peptones Obtained from Aquaculture By-Products
Abstract
1. Introduction
2. Materials and Methods
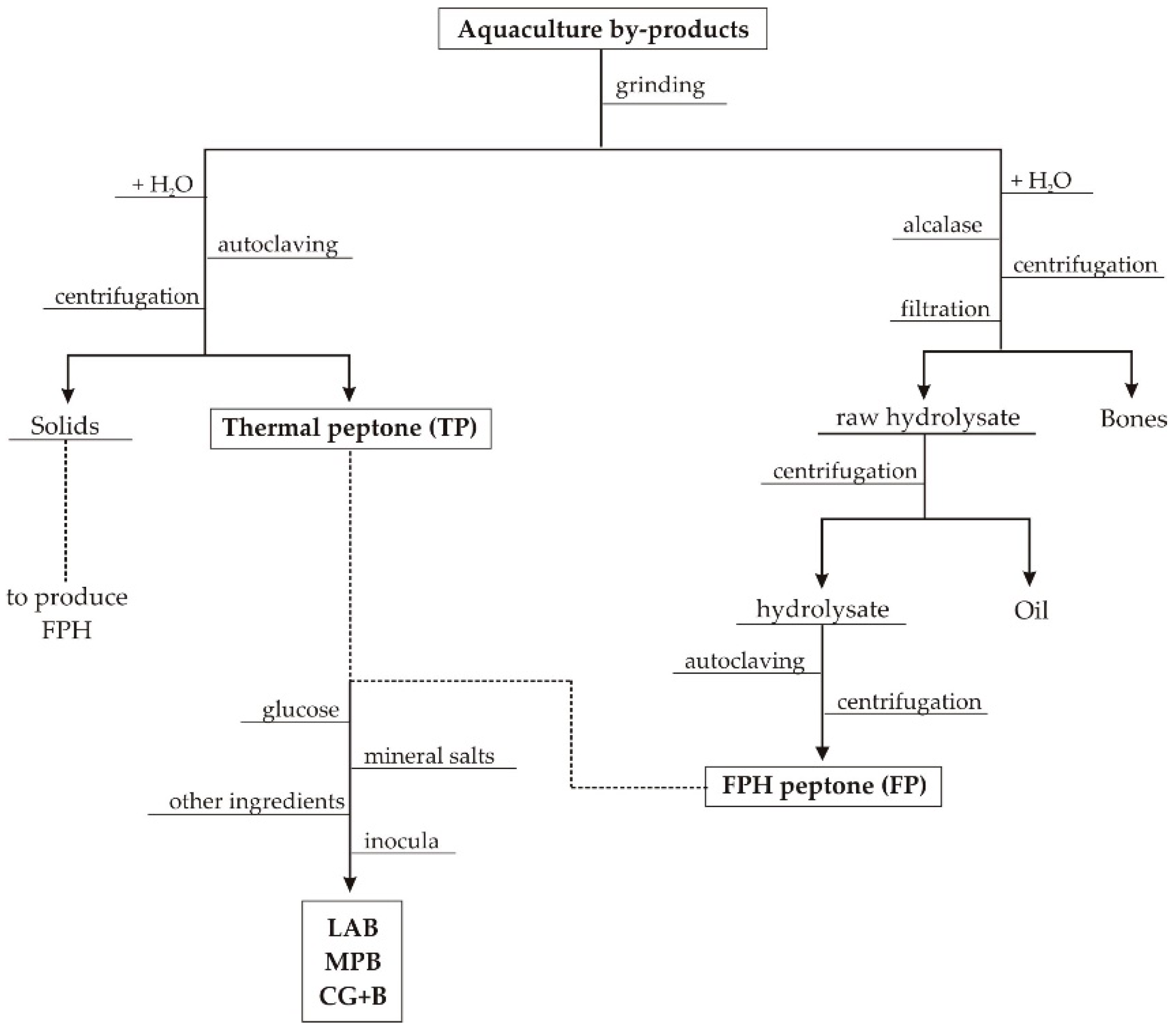
2.1. Wastes from Aquaculture Food Processing
2.2. Microbiological Methods and Culture Media
2.3. Sampling and Determinations of Growth and Metabolites
2.4. Mathematical Modelling of Bacterial Kinetics
2.5. Economical Evaluation of Aquaculture Peptones for Bioproductions
2.6. Numerical Fittings and Statistical Analyses
3. Results and Discussion
3.1. Production and Characteristics of Aquaculture Peptones
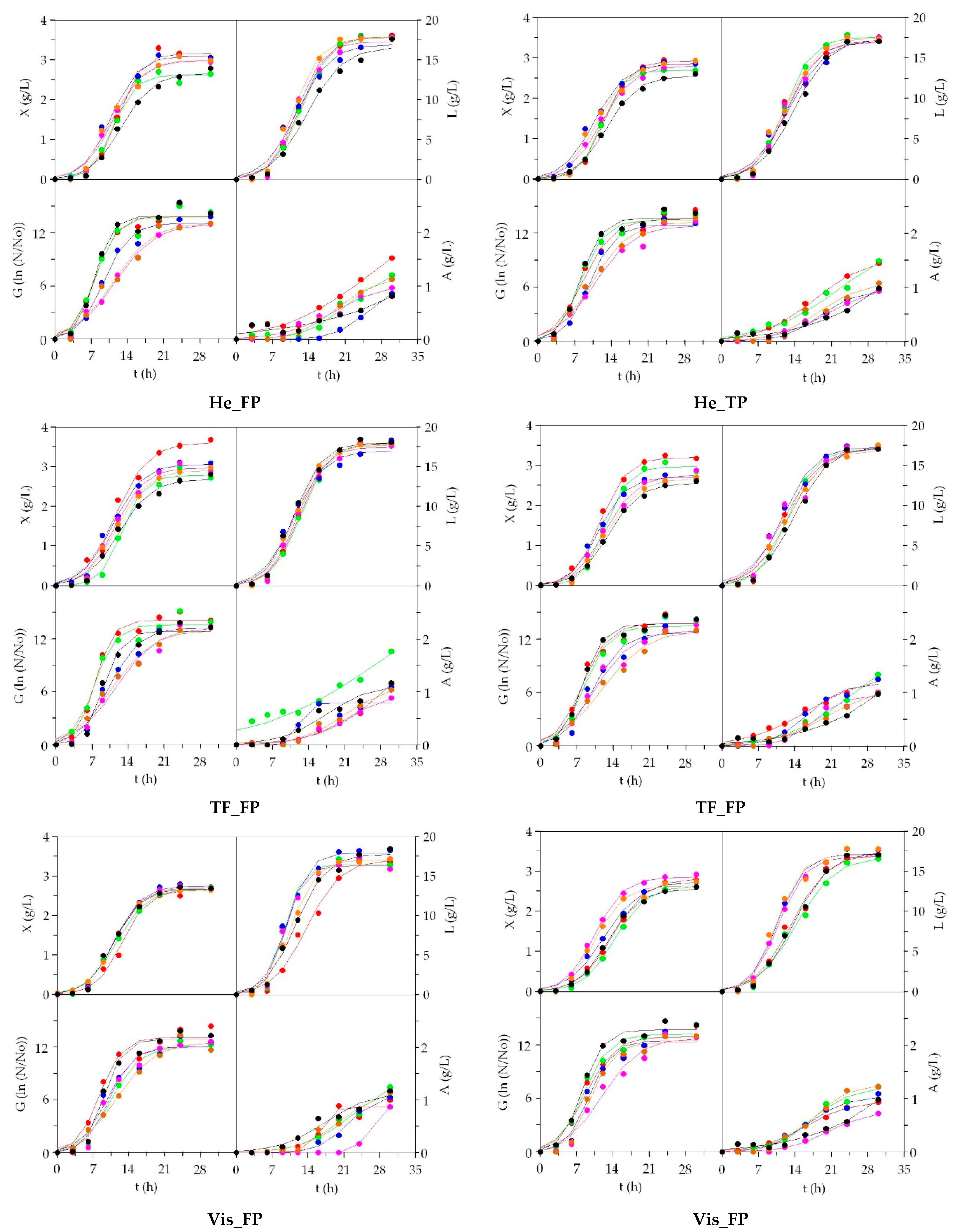
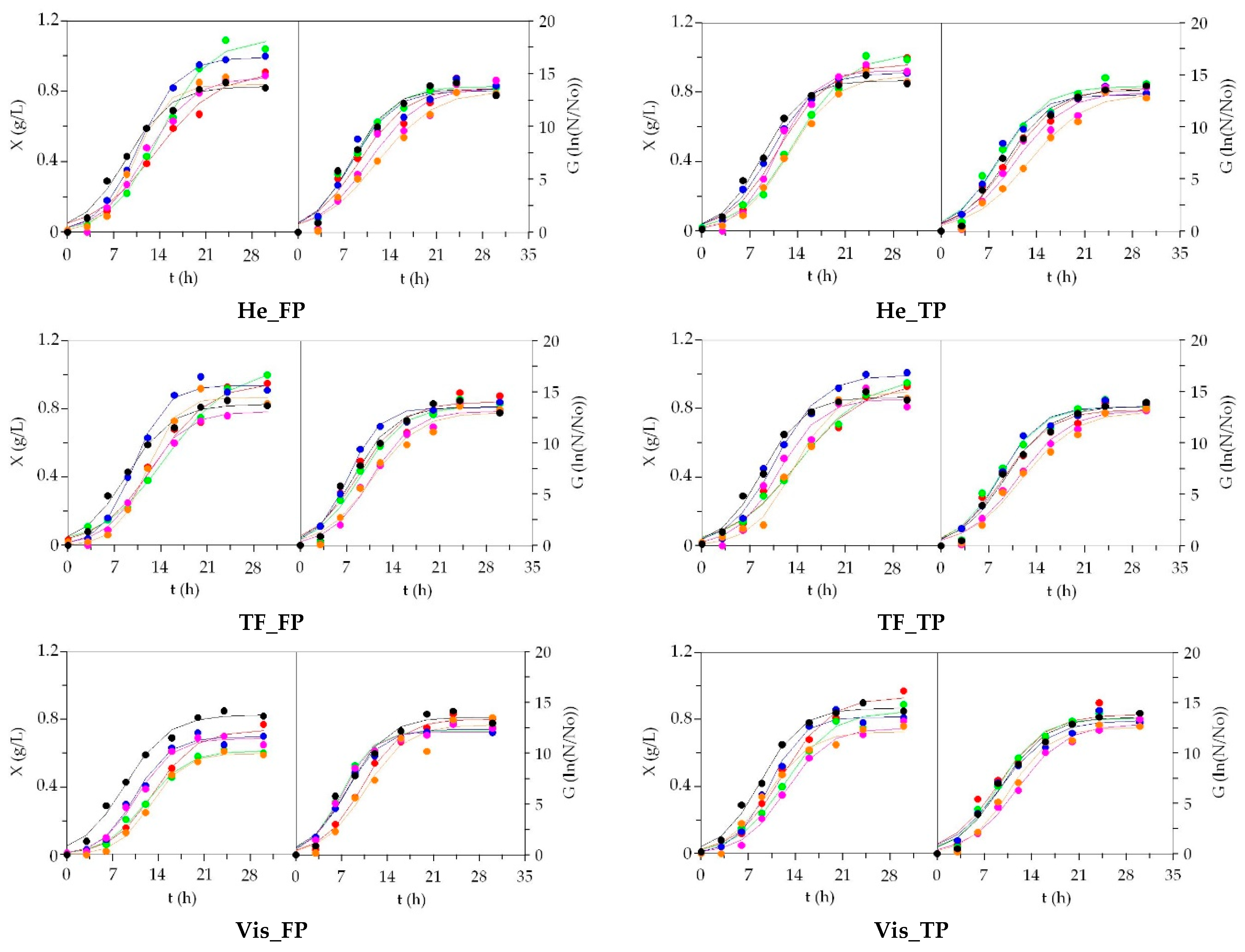
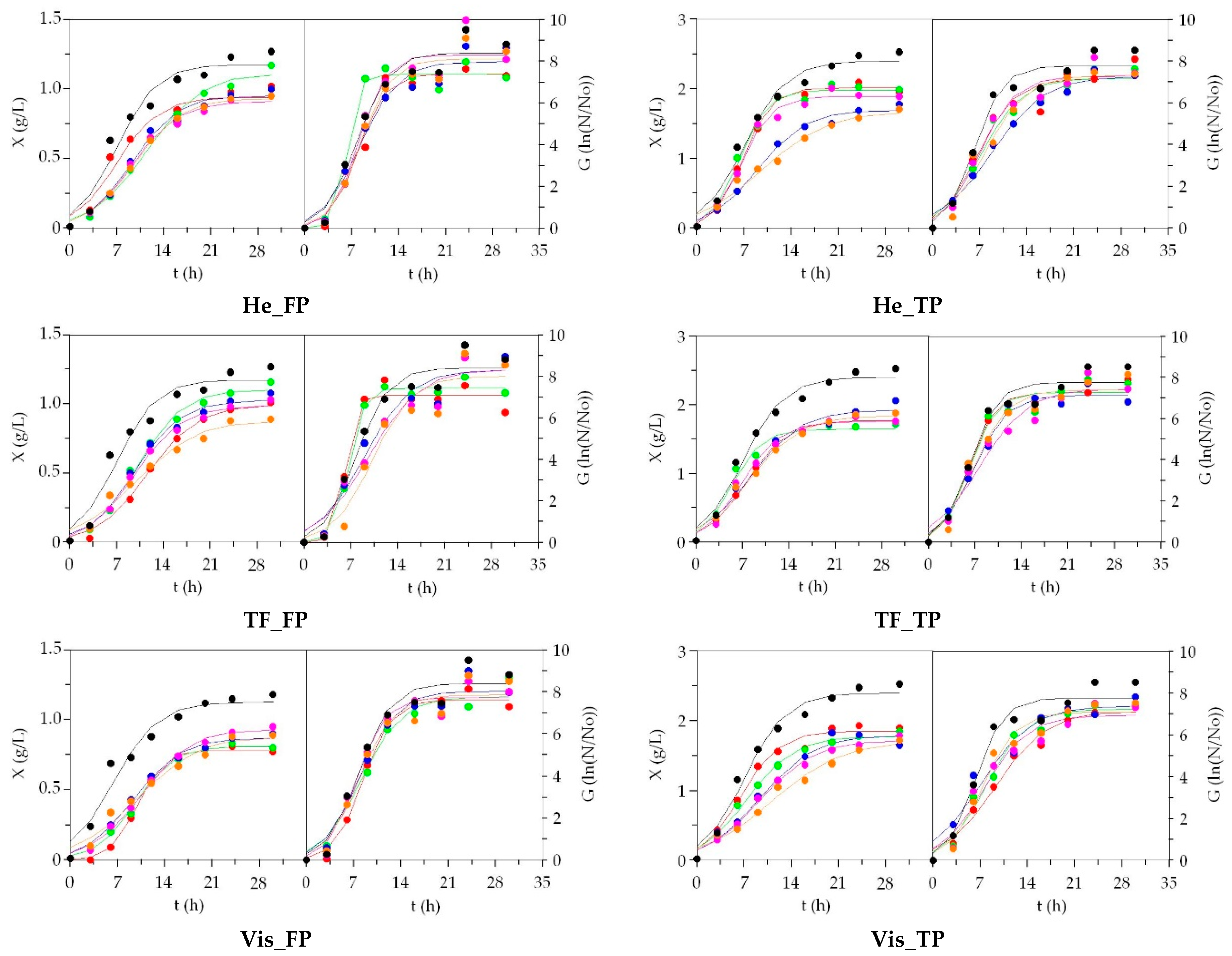
3.2. LAB Bioproductions on Marine Peptones from Aquaculture Wastes
3.3. Growth of Marine Probiotic Bacteria on Media Formulated with Peptones from Aquaculture Wastes
3.4. Growth of Aerobic Gram+ Bacteria Using Aquaculture Peptones
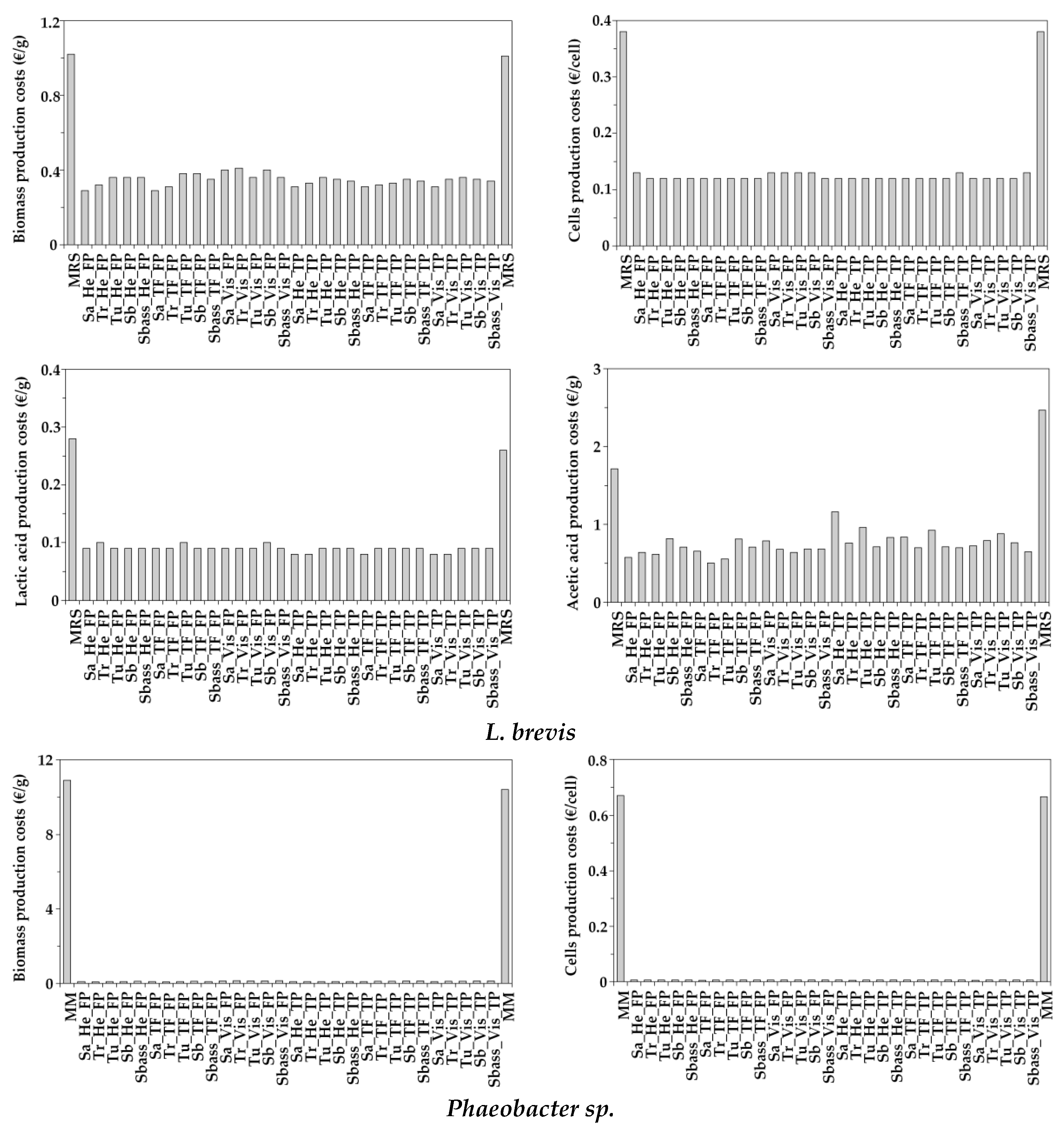
3.5. Simple Economical Evaluation of Aquaculture Peptones for Bacterial Bioproductions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAO. The State of World Fisheries and Aquaculture. 2020. Available online: http://www.fao.org/state-of-fisheries-aquaculture (accessed on 20 May 2020).
- Apromar. La Aquicultura en España. 2018. Available online: http://www.apromar.es (accessed on 23 May 2020).
- Bush, S.R.; Belton, B.; Little, D.C.; Islam, S. Emerging trends in aquaculture value chain research. Aquaculture 2019, 498, 428–434. [Google Scholar] [CrossRef]
- Little, D.C.; Young, J.A.; Zhang, W.; Newton, R.; Al Mamun, A.; Murray, F.J. Sustainable intensification of aquaculture value chains between Asia and Europe: A framework for understanding impacts and challenges. Aquaculture 2018, 493, 338–354. [Google Scholar] [CrossRef]
- Stevens, J.R.; Newton, R.; Tlusty, M.; Little, D.C. The rise of aquaculture by-products: Increasing food production, value, and sustainability through strategic utilisation. Mar. Policy 2018, 90, 115–124. [Google Scholar] [CrossRef]
- Vázquez, J.A.; Amado, I.R.; Sotelo, C.G.; Sanz, N.; Pérez-Martín, R.I.; Valcarcel, J. Production, Characterization, and Bioactivity of Fish Protein Hydrolysates from Aquaculture Turbot (Scophthalmus maximus) Wastes. Biomolecules 2020, 10, 310. [Google Scholar] [CrossRef]
- Liaset, B.; Nortvedt, R.; Lied, E.; Espe, M. Studies on the nitrogen recovery in enzymic hydrolysis of Atlantic salmon (Salmo salar, L.) frames by Protamex™ protease. Process. Biochem. 2002, 37, 1263–1269. [Google Scholar] [CrossRef]
- Mock, T.S.; Francis, D.S.; Jago, M.K.; Glencross, B.D.; Smullen, R.P.; Keast, R.S.; Turchini, G.M. The impact of dietary protein: Lipid ratio on growth performance, fatty acid metabolism, product quality and waste output in Atlantic salmon (Salmo salar). Aquaculture 2019, 501, 191–201. [Google Scholar] [CrossRef]
- He, S.; Franco, C.; Zhang, W.; Franco, C. Process optimisation and physicochemical characterisation of enzymatic hydrolysates of proteins from co-products of Atlantic Salmon (Salmo salar) and Yellowtail Kingfish (Seriola lalandi). Int. J. Food Sci. Technol. 2012, 47, 2397–2404. [Google Scholar] [CrossRef]
- Munekata, P.E.; Pateiro, M.; Domínguez, R.; Zhou, J.; Barba, F.J.; Lorenzo, J.M. Nutritional Characterization of Sea Bass Processing By-Products. Biomolecules 2020, 10, 232. [Google Scholar] [CrossRef]
- Pateiro, M.; Munekata, P.E.; Domínguez, R.; Wang, M.; Barba, F.J.; Bermúdez, R.; Lorenzo, J.M. Nutritional Profiling and the Value of Processing By-Products from Gilthead Sea Bream (Sparus aurata). Mar. Drugs 2020, 18, 101. [Google Scholar] [CrossRef]
- Blanco, M.; Vázquez, J.A.; Pérez-Martín, R.I.; Sotelo, C.G. Hydrolysates of Fish Skin Collagen: An Opportunity for Valorizing Fish Industry Byproducts. Mar. Drugs 2017, 15, 131. [Google Scholar] [CrossRef]
- Sinthusamran, S.; Idowu, A.T.; Benjakul, S.; Prodpran, T.; Yeşilsu, A.F.; Kishimura, H. Effect of proteases and alcohols used for debittering on characteristics and antioxidative activity of protein hydrolysate from salmon frames. J. Food Sci. Technol. 2019, 57, 473–483. [Google Scholar] [CrossRef]
- Franco, D.; Munekata, P.E.S.; Agregán, R.; Bermúdez, R.; López-Pedrouso, M.; Pateiro, M.; Lorenzo, J.M. Application of Pulsed Electric Fields for Obtaining Antioxidant Extracts from Fish Residues. Antioxidants 2020, 9, 90. [Google Scholar] [CrossRef]
- Harnedy, P.A.; Fitzgerald, R.J. Bioactive peptides from marine processing waste and shellfish: A review. J. Funct. Foods 2012, 4, 6–24. [Google Scholar] [CrossRef]
- Halim, N.; Yusof, H.; Sarbon, N.M. Functional and bioactive properties of fish protein hydolysates and peptides: A comprehensive review. Trends Food Sci. Technol. 2016, 51, 24–33. [Google Scholar] [CrossRef]
- Fang, B.; Sun, J.; Dong, P.; Xue, C.; Mao, X. Conversion of turbot skin wastes into valuable functional substances with an eco-friendly fermentation technology. J. Clean. Prod. 2017, 156, 367–377. [Google Scholar] [CrossRef]
- Vázquez, J.A.; Durán, A.I.; Menduíña, A.; Nogueira, M.; Fraguas, J.; Mirón, J.; Valcárcel, J. Tailor-made process to recover high added value compounds from fishery by-products (Chapter 4). In Green Extraction and Valorization of By-Products from Food Processing; Francisco, J.B., Elena, R.S., Mladen, B., Jose, M.L.R., Eds.; CRC Press: Boca Raton, FL, USA; Taylor & Francis Group: Abingdon, UK, 2020; pp. 91–141. ISBN 978-0-429-32500-7. [Google Scholar] [CrossRef]
- Ramkumar, A.; Sivakumar, N.; Victor, R. Fish Waste-Potential Low Cost Substrate for Bacterial Protease Production: A Brief Review. Open Biotechnol. J. 2016, 10, 335–341. [Google Scholar] [CrossRef]
- Deraz, S.F.; El-Fawal, G.F.; Abd-Ellatif, S.A.; Khalil, A.A. Autohydrolysed Tilapia nilotica Fish Viscera as a Peptone Source in Bacteriocin Production. Indian J. Microbiol. 2011, 51, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Djellouli, M.; Martínez-Álvarez, Ó.; Arancibia, M.; Florez-Cuadrado, D.; Ugarte-Ruiz, M.; Domínguez, L.; Zadi-Karam, H.; Karam, N.; Roudj, S.; López-Caballero, M.E. Effect of seafood peptones on biomass and metabolic activity by Enterococcus faecalis DM19. LWT 2017, 81, 94–100. [Google Scholar] [CrossRef]
- Shi, S.; Li, J.; Blersch, D.M. Utilization of solid catfish manure waste as carbon and nutrient source for lactic acid production. Appl. Microbiol. Biotechnol. 2018, 102, 4765–4772. [Google Scholar] [CrossRef] [PubMed]
- Tahiri, I.; Desbiens, M.; Lacroix, C.; Kheadr, E.; Fliss, I. Growth of Carnobacterium divergens M35 and production of Divergicin M35 in snow crab by-product, a natural-grade medium. LWT 2009, 42, 624–632. [Google Scholar] [CrossRef]
- Biji, G.D.; Arun, A.; Muthulakshmi, E.; Vijayaraghavan, P.; Arasu, M.V.; Al-Dhabi, N.A. Bio-prospecting of cuttle fish waste and cow dung for the production of fibrinolytic enzyme from Bacillus cereus IND5 in solid state fermentation. 3 Biotech. 2016, 6, 231–244. [Google Scholar] [CrossRef] [PubMed]
- Vázquez, J.A.; González, M.; Murado, M. Preliminary tests on nisin and pediocin production using waste protein sources. Bioresour. Technol. 2006, 97, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Vázquez, J.A.; Sotelo, C.G.; Sanz, N.; Pérez-Martín, R.I.; Amado, I.R.; Valcarcel, J. Valorization of Aquaculture By-Products of Salmonids to Produce Enzymatic Hydrolysates: Process Optimization, Chemical Characterization and Evaluation of Bioactives. Mar. Drugs 2019, 17, 676. [Google Scholar] [CrossRef] [PubMed]
- Bernfeld, P. Enzymes of Starch Degradation and Synthesis. Adv. Enzymol. Relat. Areas Mol. Biol. 2006, 12, 379–428. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Boil. Chem. 1951, 193, 265–275. [Google Scholar]
- Havilah, E.J.; Wallis, D.M.; Morris, R.; Woolnough, J.A. A microcolorimetric method for determination of ammonia in Kjeldahl digests with a manual spectrophotometer. Lab. Prac. 1977, 26, 545–547. [Google Scholar]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric Method for Determination of Sugars and Related Substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Moore, S.; Spackman, D.H.; Stein, W.H. Chromatography of Amino Acids on Sulfonated Polystyrene Resins. An Improved System. Anal. Chem. 1958, 30, 1185–1190. [Google Scholar] [CrossRef]
- Vázquez, J.A.; Nogueira, M.; Durán, A.; Prieto, M.A.; Amado, I.R.; Rial, D.; González, M.; Murado, M. Preparation of marine silage of swordfish, ray and shark visceral waste by lactic acid bacteria. J. Food Eng. 2011, 103, 442–448. [Google Scholar] [CrossRef]
- Vázquez, J.A.; Murado, M. Mathematical tools for objective comparison of microbial cultures. Application to evaluation of 15 peptones for lactic acid bacteria productions. Biochem. Eng. J. 2008, 39, 276–287. [Google Scholar] [CrossRef]
- García-Santiago, X.; Franco-Uría, A.; Antelo, L.T.; Vázquez, J.A.; Pérez-Martín, R.; Moreira, M.T.; Feijoo, G. Eco-efficiency of a marine biorefinery for valorisation of cartilaginous fish biomass. J. Ind. Ecol. accepted in press. [CrossRef]
- Gbogouri, G.; Linder, M.; Fanni, J.; Parmentier, M. Influence of Hydrolysis Degree on the Functional Properties of Salmon Byproducts Hydrolysates. J. Food Sci. 2004, 69, 615–622. [Google Scholar] [CrossRef]
- Safari, R.; Motamedzadegan, A.; Ovissipour, M.; Regenstein, J.M.; Gildberg, A.; Rasco, B. Use of Hydrolysates from Yellowfin Tuna (Thunnus albacares) Heads as a Complex Nitrogen Source for Lactic Acid Bacteria. Food Bioprocess Technol. 2009, 5, 73–79. [Google Scholar] [CrossRef]
- Aspmo, S.I.; Horn, S.J.; Eijsink, V.G.H. Enzymatic hydrolysis of Atlantic cod (Gadus morhua L.) viscera. Process. Biochem. 2005, 40, 1957–1966. [Google Scholar] [CrossRef]
- Vázquez, J.A.; Blanco, M.; Fraguas, J.; Pastrana, L.; Pérez-Martín, R.I. Optimisation of the extraction and purification of chondroitin sulphate from head by-products of Prionace glauca by environmental friendly processes. Food Chem. 2016, 198, 28–35. [Google Scholar] [CrossRef]
- Vázquez, J.A.; Ramos, P.; Valcarcel, J.; Antelo, L.T.; Novoa-Carballal, R.; Reis, R.L.; Pérez-Martín, R.I. An integral and sustainable valorisation strategy of squid pen by-products. J. Clean. Prod. 2018, 201, 207–218. [Google Scholar] [CrossRef]
- Jensen, P.R.; Hammer, K. Minimal Requirements for Exponential Growth of Lactococcus lactis. Appl. Environ. Microbiol. 1993, 59, 4363–4366. [Google Scholar] [CrossRef]
- Kowalczyk, M.; Mayo, B.; Fernández, M.; Aleksandrzak-Piekarczyk, T. Updates on Metabolism in Lactic Acid Bacteria in Light of “Omic” Technologies. Biotechnol. Lact. Acid Bact. 2015, 30, 1–24. [Google Scholar] [CrossRef]
- Laranjo, M.; Potes, M.E.; Elias, M. Role of Starter Cultures on the Safety of Fermented Meat Products. Front. Microbiol. 2019, 10, 853. [Google Scholar] [CrossRef]
- Martins, E.M.F.; Ramos, A.M.; Lago-Vanzela, E.S.; Stringheta, P.C.; Pinto, C.L.D.O.; Martins, J.M.; Martins, J.M. Products of vegetable origin: A new alternative for the consumption of probiotic bacteria. Food Res. Int. 2013, 51, 764–770. [Google Scholar] [CrossRef]
- Linares, D.M.; Gómez, C.; Renes, E.; Fresno, J.M.; Tornadijo, M.E.; Ross, R.P.; Stanton, C. Lactic Acid Bacteria and Bifidobacteria with Potential to Design Natural Biofunctional Health-Promoting Dairy Foods. Front. Microbiol. 2017, 8, 846–853. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.; Samak, G. Protection and Restitution of Gut Barrier by Probiotics: Nutritional and Clinical Implications. Curr. Nutr. Food Sci. 2013, 9, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Atienza, E.; Araújo, C.; Magadan, S.; Hernandez, P.E.; Herranz, C.; Santos, Y.; Cintas, L.M. In vitro and in vivo evaluation of lactic acid bacteria of aquatic origin as probiotics for turbot (Scophthalmus maximus L.) farming. Fish Shellfish. Immunol. 2014, 41, 570–580. [Google Scholar] [CrossRef]
- Shin, Y.; Kim, T.; Kim, K. Probiotics Used for Respiratory Diseases. J. Bacteriol. Virol. 2015, 45, 79–92. [Google Scholar] [CrossRef][Green Version]
- Vázquez, J.A.; Caprioni, R.; Nogueira, M.; Menduiña, A.; Ramos, P.; Pérez-Martín, R.I. Valorisation of effluents obtained from chemical and enzymatic chitin production of Illex argentinus pen by-products as nutrient supplements for various bacterial fermentations. Biochem. Eng. J. 2016, 116, 34–44. [Google Scholar] [CrossRef]
- Vázquez, J.A.; Docasal, S.; Prieto, M.A.; Gonzalez, M.; Murado, M. Growth and metabolic features of lactic acid bacteria in media with hydrolysed fish viscera. An approach to bio-silage of fishing by-products. Bioresour. Technol. 2008, 99, 6246–6257. [Google Scholar] [CrossRef] [PubMed]
- Prol-García, M.J.; Gómez, M.; Sánchez, L.; Pintado, J. Phaeobacter grown in biofilter: A new strategy for the control of Vibrionaceae in aquaculture. Aquacult. Res. 2014, 45, 1012–1025. [Google Scholar] [CrossRef]
- D’Alvise, P.W.; Lillebø, S.; Wergeland, H.I.; Gram, L.; Bergh, Ø. Protection of cod larvae from vibriosis by Phaeobacter spp.: A comparison of strains and introduction times. Aquaculture 2013, 384, 82–86. [Google Scholar] [CrossRef]
- Prol-García, M.J.; Pintado, J. Effectiveness of Probiotic Phaeobacter Bacteria Grown in Biofilters Against Vibrio anguillarum Infections in the Rearing of Turbot (Psetta maxima) Larvae. Mar. Biotechnol. 2013, 15, 726–738. [Google Scholar] [CrossRef]
- Vázquez, J.A.; González, M.; Murado, M. A new marine medium: Use of different fish peptones and comparative study of the growth of selected species of marine bacteria. Enzym. Microb. Technol. 2004, 35, 385–392. [Google Scholar] [CrossRef]
- Martone, C. Fishery by-product as a nutrient source for bacteria and archaea growth media. Bioresour. Technol. 2005, 96, 383–387. [Google Scholar] [CrossRef] [PubMed]
- Esakkiraj, P.; Dhas, G.A.J.; Palavesam, A.; Immanuel, G. Media Preparation Using Tuna-Processing Wastes for Improved Lipase Production by Shrimp Gut Isolate Staphylococcus epidermidis CMST Pi 2. Appl. Biochem. Biotechnol. 2009, 160, 1254–1265. [Google Scholar] [CrossRef]
- Taskin, M.; Kurbanoglu, E. Evaluation of waste chicken feathers as peptone source for bacterial growth. J. Appl. Microbiol. 2011, 111, 826–834. [Google Scholar] [CrossRef]
- Poernomo, A.; Buckle, K. Crude peptones from cowtail ray (Trygon sephen) viscera as microbial growth media. World J. Microbiol. Biotechnol. 2002, 18, 337–344. [Google Scholar] [CrossRef]
- Ellouz, Y. Production of protease by Bacillus subtilis grown on sardinelle heads and viscera flour. Bioresour. Technol. 2001, 80, 49–51. [Google Scholar] [CrossRef]
- Sanghi, A.; Garg, N.; Sharma, J.; Kuhar, K.; Kuhad, R.C.; Gupta, V.K. Optimization of xylanase production using inexpensive agro-residues by alkalophilic Bacillus subtilis ASH in solid-state fermentation. World J. Microbiol. Biotechnol. 2007, 24, 633–640. [Google Scholar] [CrossRef]
- Kumar, R.; Balaji, S.; Uma, T.S.; Mandal, A.B.; Sehgal, P. Optimization of Influential Parameters for Extracellular Keratinase Production by Bacillus subtilis (MTCC9102) in Solid State Fermentation Using Horn Meal—A Biowaste Management. Appl. Biochem. Biotechnol. 2008, 160, 30–39. [Google Scholar] [CrossRef]
- Pleissner, D.; Qi, Q.; Gao, C.; Rivero, C.P.; Webb, C.; Lin, C.S.K.; Venus, J. Valorization of organic residues for the production of added value chemicals: A contribution to the bio-based economy. Biochem. Eng. J. 2016, 116, 3–16. [Google Scholar] [CrossRef]
- Pleissner, D.; Dietz, D.; Van Duuren, J.B.J.H.; Wittmann, C.; Yang, X.; Lin, C.S.K.; Venus, J. Biotechnological Production of Organic Acids from Renewable Resources. Adv. Biochem. Eng. Biotechnol. 2017, 43, 373–410. [Google Scholar] [CrossRef]
- Lübeck, M.; Lübeck, P.S. Application of lactic acid bacteria in green biorefineries. FEMS Microbiol. Lett. 2019, 366, 60–67. [Google Scholar] [CrossRef]
| FP Peptones | Pr (g/L) | N (g/L) | RS (g/L) | TS (g/L) | TP Peptones | Pr (g/L) | N (g/L) | RS (g/L) | TS (g/L) |
|---|---|---|---|---|---|---|---|---|---|
| Sa_He | 61.0 ± 1.3 | 14.6 ± 2.1 | 0.19 ± 0.04 | 1.29 ± 0.09 | Sa_He | 47.9 ± 1.2 | 11.3 ± 0.3 | 0.10 ± 0.01 | 0.60 ± 0.03 |
| Sa_TF | 69.7 ± 2.1 | 15.9 ± 1.4 | 0.27 ± 0.10 | 1.50 ± 0.10 | Sa_TF | 36.8 ± 0.7 | 8.6 ± 0.4 | 0.12 ± 0.04 | 0.80 ± 0.10 |
| Sa_Vis | 42.7 ± 2.2 | 10.0 ± 0.8 | 0.44 ± 0.18 | 1.37 ± 0.14 | Sa_Vis | 30.3 ± 1.4 | 7.2 ± 0.5 | 0.24 ± 0.07 | 1.17 ± 0.09 |
| RT_He | 47.8 ± 4.8 | 14.1 ± 1.1 | 0.21 ± 0.05 | 1.40 ± 0.10 | RT_He | 36.3 ± 1.4 | 10.4 ± 0.7 | 0.13 ± 0.01 | 0.97 ± 0.07 |
| RT_TF | 53.9 ± 5.1 | 14.7 ± 0.6 | 0.20 ± 0.02 | 1.22 ± 0.10 | RT_TF | 19.6 ± 0.3 | 5.4 ± 0.3 | 0.05 ± 0.02 | 0.52 ± 0.04 |
| RT_Vis | 50.6 ± 3.2 | 14.4 ± 0.9 | 0.16 ± 0.06 | 1.09 ± 0.15 | RT_Vis | 34.3 ± 1.6 | 9.5 ± 1.0 | 0.31 ± 0.02 | 1.40 ± 0.06 |
| Tu_He | 73.5 ± 4.9 | 18.4 ± 1.0 | 0.38 ± 0.06 | 1.26 ± 0.14 | Tu_He | 73.1 ± 1.5 | 18.6 ± 0.6 | 0.19 ± 0.04 | 1.29 ± 0.10 |
| Tu_TF | 73.9 ± 3.8 | 18.5 ± 0.7 | 0.36 ± 0.04 | 1.34 ± 0.17 | Tu_TF | 63.6 ± 3.7 | 15.9 ± 0.8 | 0.22 ± 0.05 | 1.36 ± 0.11 |
| Tu_Vis | 61.6 ± 2.8 | 15.4 ± 1.1 | 0.30 ± 0.09 | 1.39 ± 0.25 | Tu_Vis | 36.6 ± 2.3 | 9.2 ± 0.6 | 0.13 ± 0.06 | 1.52 ± 0.03 |
| Sb_He | 61.6 ± 1.6 | 17.6 ± 0.8 | 0.49 ± 0.06 | 1.78 ± 0.12 | Sb_He | 28.2 ± 2.1 | 9.0 ± 0.3 | 0.06 ± 0.00 | 0.72 ± 0.40 |
| Sb_TF | 81.2 ± 3.1 | 23.2 ± 1.3 | 0.32 ± 0.07 | 1.31 ± 0.07 | Sb_TF | 18.8 ± 0.2 | 6.3 ± 0.6 | 0.31 ± 0.09 | 1.24 ± 0.06 |
| Sb_Vis | 37.9 ± 1.7 | 12.1 ± 0.6 | 0.09 ± 0.01 | 0.84 ± 0.09 | Sb_Vis | 28.0 ± 1.4 | 8.9 ± 0.5 | 0.24 ± 0.07 | 1.67 ± 0.09 |
| Sbass_He | 63.3 ± 0.4 | 18.2 ± 0.8 | 0.35 ± 0.05 | 1.42 ± 0.12 | Sbass_He | 30.6 ± 0.2 | 8.5 ± 0.3 | 0.17 ± 0.05 | 0.88 ± 0.10 |
| Sbass_TF | 78.9 ± 5.1 | 20.8 ± 1.3 | 0.43 ± 0.08 | 1.67 ± 0.16 | Sbass_TF | 20.1 ± 2.3 | 5.8 ± 0.6 | 0.10 ± 0.03 | 0.80 ± 0.08 |
| Sbass_Vis | 33.0 ± 1.6 | 9.2 ± 0.5 | 0.08 ± 0.02 | 0.94 ± 0.07 | Sbass_Vis | 18.6 ± 0.6 | 5.6 ± 0.3 | 0.08 ± 0.01 | 0.56 ± 0.11 |
| Amino Acids | Sa_He | Sa_TF | Sa_Vis | RT_He | RT_TF | RT_Vis | Tu_He | Tu_TF | Tu_Vis | Sb_He | Sb_TF | Sb_Vis | Sbass_He | Sbass_TF | Sbass_Vis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Asp | 9.61 ± 0.30 | 10.33 ± 0.06 | 9.89 ± 0.47 | 9.78 ± 0.19 | 10.32 ± 0.20 | 9.54 ± 0.07 | 8.81 ± 0.16 | 9.63 ± 0.10 | 9.26 ± 0.42 | 9.46 ± 0.03 | 10.80 ± 0.01 | 9.52 ± 0.00 | 9.25 ± 0.17 | 9.82 ± 0.03 | 9.69 ± 0.16 |
| Thr | 3.83 ± 0.39 | 2.95 ± 0.04 | 3.49 ± 1.15 | 4.38 ± 0.22 | 4.44 ± 0.15 | 5.30 ± 0.10 | 3.64 ± 0.16 | 3.90 ± 0.06 | 3.85 ± 0.18 | 4.52 ± 0.03 | 4.30 ± 0.07 | 4.63 ± 0.07 | 4.31 ± 0.05 | 4.48 ± 0.04 | 4.80 ± 0.03 |
| Ser | 4.98 ± 0.05 | 4.98 ± 0.11 | 5.02 ± 0.06 | 5.00 ± 0.20 | 4.83 ± 0.06 | 6.09 ± 0.03 | 5.67 ± 0.14 | 5.37 ± 0.18 | 5.39 ± 0.36 | 4.82 ± 0.01 | 4.68 ± 0.04 | 5.07 ± 0.07 | 4.89 ± 0.06 | 4.62 ± 0.02 | 5.60 ± 0.03 |
| Glu | 13.42 ± 0.45 | 13.23 ± 0.08 | 13.41 ± 0.65 | 13.89 ± 0.14 | 14.98 ± 0.34 | 13.05 ± 0.18 | 12.86 ± 0.18 | 13.55 ± 0.08 | 13.12 ± 0.32 | 13.83 ± 0.11 | 15.11 ± 0.11 | 12.91 ± 0.85 | 13.63 ± 0.07 | 14.21 ± 0.05 | 13.04 ± 0.21 |
| Gly | 12.49 ± 1.11 | 11.08 ± 0.27 | 11.66 ± 0.98 | 9.93 ± 1.10 | 8.94 ± 2.94 | 6.93 ± 0.06 | 14.50 ± 0.32 | 12.57 ± 0.15 | 13.00 ± 0.50 | 10.04 ± 0.08 | 8.63 ± 0.17 | 8.57 ± 0.01 | 10.84 ± 0.11 | 9.66 ± 0.01 | 7.75 ± 0.04 |
| Ala | 7.92 ± 0.45 | 8.45 ± 0.03 | 7.86 ± 1.01 | 7.19 ± 0.31 | 6.98 ± 0.22 | 7.17 ± 0.01 | 8.38 ± 0.24 | 8.06 ± 0.05 | 7.83 ± 0.22 | 7.39 ± 0.08 | 7.69 ± 0.15 | 7.23 ± 0.07 | 7.60 ± 0.01 | 7.56 ± 0.04 | 7.54 ± 0.01 |
| Cys | 0.75 ± 0.11 | 0.83 ± 0.03 | 0.99 ± 0.30 | 0.76 ± 0.09 | 0.74 ± 0.05 | 0.70 ± 0.02 | 0.62 ± 0.05 | 0.77 ± 0.10 | 0.71 ± 0.11 | 0.62 ± 0.01 | 0.89 ± 0.01 | 0.97 ± 0.09 | 0.69 ± 0.10 | 0.68 ± 0.05 | 0.96 ± 0.16 |
| Val | 3.39 ± 0.16 | 3.44 ± 0.30 | 3.37 ± 0.27 | 4.35 ± 0.33 | 4.24 ± 0.22 | 5.67 ± 0.14 | 2.96 ± 0.09 | 3.21 ± 0.11 | 3.23 ± 0.12 | 4.36 ± 0.01 | 3.70 ± 0.23 | 4.90 ± 0.02 | 4.23 ± 0.06 | 4.15 ± 0.03 | 5.54 ± 0.11 |
| Met | 3.13 ± 0.28 | 3.82 ± 0.33 | 3.45 ± 0.90 | 3.16 ± 0.10 | 3.33 ± 0.15 | 2.82 ± 0.14 | 2.76 ± 0.15 | 2.90 ± 0.04 | 2.84 ± 0.17 | 2.95 ± 0.14 | 3.33 ± 0.14 | 2.83 ± 0.05 | 2.76 ± 0.04 | 3.04 ± 0.01 | 2.62 ± 0.11 |
| Ile | 2.28 ± 0.22 | 2.02 ± 0.16 | 2.22 ± 0.28 | 3.22 ± 0.29 | 3.21 ± 0.23 | 4.48 ± 0.11 | 1.97 ± 0.11 | 2.19 ± 0.08 | 2.29 ± 0.12 | 3.38 ± 0.17 | 2.58 ± 0.04 | 3.65 ± 0.01 | 3.17 ± 0.04 | 3.40 ± 0.01 | 4.03 ± 0.22 |
| Leu | 6.17 ± 0.33 | 6.36 ± 0.22 | 6.27 ± 0.07 | 7.09 ± 0.33 | 7.19 ± 0.07 | 7.97 ± 0.15 | 5.37 ± 0.11 | 5.93 ± 0.11 | 5.86 ± 0.17 | 6.65 ± 0.14 | 7.07 ± 0.09 | 7.54 ± 0.01 | 6.31 ± 0.06 | 6.72 ± 0.03 | 7.75 ± 0.13 |
| Tyr | 3.37 ± 0.45 | 4.40 ± 0.17 | 3.88 ± 0.97 | 3.36 ± 0.18 | 3.39 ± 0.22 | 3.64 ± 0.26 | 2.90 ± 0.16 | 3.18 ± 0.08 | 3.11 ± 0.20 | 3.42 ± 0.02 | 3.59 ± 0.05 | 3.51 ± 0.08 | 3.34 ± 0.02 | 3.07 ± 0.02 | 3.99 ± 0.21 |
| Phe | 4.93 ± 0.89 | 7.15 ± 1.13 | 7.00 ± 3.41 | 4.38 ± 0.25 | 4.09 ± 0.15 | 4.55 ± 0.00 | 4.16 ± 0.18 | 4.51 ± 0.17 | 4.31 ± 0.37 | 4.03 ± 0.07 | 4.64 ± 0.12 | 4.93 ± 0.23 | 4.14 ± 0.09 | 3.98 ± 0.01 | 4.96 ± 0.15 |
| His | 2.00 ± 0.13 | 2.11 ± 0.17 | 2.10 ± 0.03 | 2.20 ± 0.49 | 2.18 ± 0.02 | 2.50 ± 0.02 | 1.61 ± 0.07 | 1.75 ± 0.04 | 1.85 ± 0.11 | 2.36 ± 0.05 | 2.67 ± 0.02 | 2.26 ± 0.01 | 2.07 ± 0.01 | 2.13 ± 0.01 | 2.08 ± 0.05 |
| Lys | 7.04 ± 0.46 | 7.96 ± 0.37 | 6.38 ± 0.33 | 7.78 ± 0.42 | 8.60 ± 0.13 | 6.97 ± 0.05 | 5.57 ± 0.14 | 6.15 ± 0.11 | 6.18 ± 0.23 | 7.13 ± 0.07 | 7.81 ± 0.12 | 7.62 ± 0.06 | 6.93 ± 0.11 | 8.05 ± 0.07 | 7.55 ± 0.01 |
| Arg | 5.69 ± 0.40 | 4.44 ± 0.03 | 5.18 ± 1.52 | 5.97 ± 0.10 | 5.96 ± 0.14 | 6.03 ± 0.01 | 6.53 ± 0.24 | 6.34 ± 0.08 | 6.59 ± 0.36 | 6.56 ± 0.15 | 5.73 ± 0.05 | 6.37 ± 0.06 | 6.39 ± 0.09 | 6.47 ± 0.02 | 5.28 ± 0.06 |
| OHPro | 2.85 ± 0.62 | 2.00 ± 0.14 | 2.36 ± 0.78 | 2.25 ± 0.37 | 1.86 ± 0.38 | 1.43 ± 0.22 | 4.00 ± 0.19 | 3.14 ± 0.18 | 3.42 ± 0.35 | 2.70 ± 0.05 | 1.92 ± 0.14 | 2.42 ± 0.69 | 3.18 ± 0.13 | 2.31 ± 0.13 | 1.85 ± 0.17 |
| Pro | 6.15 ± 0.83 | 4.45 ± 0.28 | 5.46 ± 1.67 | 5.30 ± 0.34 | 4.72 ± 0.16 | 5.18 ± 0.24 | 7.67 ± 0.25 | 6.86 ± 0.13 | 7.14 ± 0.37 | 5.79 ± 0.16 | 4.84 ± 0.02 | 5.06 ± 0.04 | 6.27 ± 0.10 | 5.66 ± 0.03 | 4.97 ± 0.40 |
| Amino Acids | Sa_He | Sa_TF | Sa_Vis | RT_He | RT_TF | RT_Vis | Tu_He | Tu_TF | Tu_Vis | Sb_He | Sb_TF | Sb_Vis | Sbass_He | Sbass_TF | Sbass_Vis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Asp | 6.68 ± 0.22 | 6.84 ± 0.30 | 6.69 ± 0.24 | 7.68 ± 0.12 | 7.17 ± 0.02 | 9.31 ± 0.17 | 9.51 ± 0.29 | 7.66 ± 0.08 | 8.56 ± 0.11 | 7.06 ± 0.03 | 6.52 ± 0.16 | 9.81 ± 0.24 | 7.67 ± 0.14 | 6.54 ± 0.20 | 8.95 ± 0.03 |
| Thr | 2.68 ± 0.06 | 2.80 ± 0.06 | 2.74 ± 0.16 | 3.45 ± 0.09 | 3.04 ± 0.13 | 5.14 ± 0.14 | 2.94 ± 0.11 | 3.09 ± 0.09 | 3.65 ± 0.07 | 3.20 ± 0.01 | 2.93 ± 0.01 | 5.14 ± 0.09 | 3.31 ± 0.01 | 2.81 ± 0.10 | 4.59 ± 0.20 |
| Ser | 4.21 ± 0.06 | 4.64 ± 0.01 | 4.45 ± 0.54 | 4.97 ± 0.12 | 5.17 ± 0.46 | 6.47 ± 0.09 | 6.16 ± 0.06 | 6.10 ± 0.06 | 6.24 ± 0.14 | 4.33 ± 0.25 | 3.82 ± 0.28 | 5.09 ± 0.25 | 4.59 ± 0.03 | 4.20 ± 0.06 | 5.25 ± 0.19 |
| Glu | 10.74 ± 0.43 | 11.52 ± 0.39 | 11.24 ± 0.57 | 12.35 ± 0.07 | 11.88 ± 1.05 | 13.11 ± 0.09 | 12.11 ± 0.01 | 12.16 ± 0.13 | 13.38 ± 0.44 | 12.28 ± 0.78 | 11.33 ± 0.15 | 13.30 ± 0.59 | 12.67 ± 0.18 | 11.99 ± 0.25 | 13.95 ± 0.04 |
| Gly | 21.42 ± 0.19 | 20.04 ± 0.18 | 20.21 ± 2.57 | 16.87 ± 0.22 | 19.07 ± 0.30 | 9.48 ± 0.05 | 17.59 ± 0.46 | 17.07 ± 0.27 | 14.92 ± 0.13 | 17.18 ± 0.06 | 19.61 ± 0.13 | 7.49 ± 0.20 | 16.25 ± 0.25 | 19.59 ± 0.06 | 9.40 ± 0.27 |
| Ala | 9.03 ± 0.13 | 9.14 ± 0.42 | 9.09 ± 0.01 | 7.99 ± 0.22 | 8.25 ± 0.63 | 6.73 ± 0.06 | 10.05 ± 0.40 | 9.86 ± 0.27 | 9.59 ± 0.09 | 9.07 ± 0.02 | 9.44 ± 0.12 | 6.91 ± 0.01 | 9.35 ± 0.18 | 10.25 ± 0.10 | 7.76 ± 0.02 |
| Cys | 0.85 ± 0.07 | 0.41 ± 0.05 | 0.71 ± 0.35 | 0.52 ± 0.06 | 0.61 ± 0.24 | 0.57 ± 0.04 | 0.39 ± 0.07 | 0.47 ± 0.04 | 0.65 ± 0.04 | 0.86 ± 0.16 | 1.17 ± 0.06 | 1.35 ± 0.30 | 0.85 ± 0.12 | 0.91 ± 0.13 | 1.45 ± 0.02 |
| Val | 2.39 ± 0.06 | 2.53 ± 0.09 | 2.48 ± 0.11 | 3.19 ± 0.05 | 2.96 ± 0.19 | 4.96 ± 0.04 | 2.21 ± 0.05 | 2.33 ± 0.04 | 3.05 ± 0.14 | 2.88 ± 0.28 | 3.24 ± 0.38 | 5.63 ± 0.16 | 2.93 ± 0.15 | 2.25 ± 0.12 | 4.95 ± 0.02 |
| Met | 2.36 ± 0.03 | 2.66 ± 0.19 | 2.56 ± 0.37 | 2.54 ± 0.10 | 2.55 ± 0.17 | 2.79 ± 0.09 | 2.62 ± 0.12 | 2.71 ± 0.11 | 2.26 ± 0.07 | 2.51 ± 0.26 | 2.23 ± 0.18 | 2.58 ± 0.08 | 2.40 ± 0.10 | 2.13 ± 0.01 | 2.53 ± 0.16 |
| Ile | 1.47 ± 0.07 | 1.50 ± 0.15 | 1.52 ± 0.04 | 2.25 ± 0.15 | 1.74 ± 0.11 | 3.89 ± 0.11 | 1.37 ± 0.18 | 1.40 ± 0.08 | 1.66 ± 0.08 | 1.97 ± 0.14 | 1.30 ± 0.01 | 3.85 ± 0.25 | 1.80 ± 0.04 | 1.47 ± 0.20 | 3.03 ± 0.11 |
| Leu | 3.18 ± 0.06 | 3.08 ± 0.02 | 3.17 ± 0.09 | 4.51 ± 0.18 | 3.50 ± 0.28 | 6.61 ± 0.12 | 3.91 ± 0.15 | 3.98 ± 0.03 | 4.80 ± 0.05 | 4.63 ± 0.07 | 3.95 ± 0.14 | 7.32 ± 0.04 | 4.54 ± 0.01 | 3.61 ± 0.13 | 6.45 ± 0.17 |
| Tyr | 1.07 ± 0.06 | 0.97 ± 0.09 | 0.99 ± 0.09 | 1.56 ± 0.10 | 1.35 ± 0.26 | 2.92 ± 0.08 | 1.87 ± 0.12 | 1.92 ± 0.14 | 2.34 ± 0.06 | 1.89 ± 0.33 | 1.46 ± 0.16 | 3.91 ± 0.14 | 2.00 ± 0.13 | 1.24 ± 0.10 | 3.61 ± 0.11 |
| Phe | 3.19 ± 0.13 | 3.23 ± 0.02 | 3.33 ± 0.15 | 3.56 ± 0.05 | 3.47 ± 0.13 | 3.43 ± 0.06 | 3.27 ± 0.20 | 3.44 ± 0.06 | 3.38 ± 0.08 | 3.10 ± 0.47 | 2.73 ± 0.01 | 4.34 ± 0.09 | 3.49 ± 0.36 | 3.08 ± 0.16 | 4.29 ± 0.44 |
| His | 1.49 ± 0.01 | 1.58 ± 0.09 | 1.56 ± 0.16 | 1.89 ± 0.08 | 1.52 ± 0.21 | 2.47 ± 0.13 | 1.25 ± 0.06 | 1.33 ± 0.14 | 1.53 ± 0.06 | 1.69 ± 0.02 | 2.25 ± 0.05 | 2.62 ± 0.04 | 1.57 ± 0.03 | 1.25 ± 0.02 | 2.14 ± 0.04 |
| Lys | 6.39 ± 0.14 | 5.21 ± 2.39 | 6.35 ± 0.23 | 6.09 ± 0.21 | 6.77 ± 0.08 | 7.14 ± 0.08 | 4.47 ± 0.18 | 4.58 ± 0.12 | 5.13 ± 0.10 | 5.17 ± 0.12 | 6.12 ± 0.18 | 8.02 ± 0.09 | 5.68 ± 0.09 | 5.44 ± 0.08 | 7.88 ± 0.07 |
| Arg | 7.09 ± 0.11 | 7.25 ± 0.14 | 7.23 ± 0.16 | 7.00 ± 0.09 | 6.73 ± 0.25 | 6.93 ± 0.38 | 6.64 ± 0.10 | 6.72 ± 0.11 | 6.28 ± 0.07 | 6.79 ± 0.13 | 7.05 ± 0.05 | 6.13 ± 0.01 | 6.60 ± 0.13 | 6.52 ± 0.02 | 5.41 ± 0.04 |
| OHPro | 6.78 ± 0.56 | 7.06 ± 0.11 | 6.68 ± 0.37 | 5.76 ± 0.17 | 6.54 ± 0.06 | 2.31 ± 0.11 | 6.29 ± 0.18 | 6.00 ± 0.11 | 4.69 ± 0.18 | 6.29 ± 0.17 | 6.42 ± 0.43 | 1.58 ± 0.21 | 5.76 ± 0.71 | 7.26 ± 0.83 | 2.32 ± 0.34 |
| Pro | 8.97 ± 0.43 | 9.55 ± 0.37 | 8.99 ± 0.48 | 7.81 ± 0.21 | 7.67 ± 0.21 | 5.72 ± 0.05 | 9.35 ± 0.25 | 9.17 ± 0.10 | 7.89 ± 0.00 | 9.10 ± 0.09 | 8.42 ± 0.50 | 4.92 ± 0.26 | 8.55 ± 0.29 | 9.46 ± 0.41 | 6.03 ± 0.14 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vázquez, J.A.; Durán, A.I.; Menduíña, A.; Nogueira, M. Biotechnological Valorization of Food Marine Wastes: Microbial Productions on Peptones Obtained from Aquaculture By-Products. Biomolecules 2020, 10, 1184. https://doi.org/10.3390/biom10081184
Vázquez JA, Durán AI, Menduíña A, Nogueira M. Biotechnological Valorization of Food Marine Wastes: Microbial Productions on Peptones Obtained from Aquaculture By-Products. Biomolecules. 2020; 10(8):1184. https://doi.org/10.3390/biom10081184
Chicago/Turabian StyleVázquez, José Antonio, Ana I. Durán, Araceli Menduíña, and Margarita Nogueira. 2020. "Biotechnological Valorization of Food Marine Wastes: Microbial Productions on Peptones Obtained from Aquaculture By-Products" Biomolecules 10, no. 8: 1184. https://doi.org/10.3390/biom10081184
APA StyleVázquez, J. A., Durán, A. I., Menduíña, A., & Nogueira, M. (2020). Biotechnological Valorization of Food Marine Wastes: Microbial Productions on Peptones Obtained from Aquaculture By-Products. Biomolecules, 10(8), 1184. https://doi.org/10.3390/biom10081184






