Folding of Truncated Granulin Peptides
Abstract
:1. Introduction
2. Materials and Methods
2.1. Peptide Synthesis
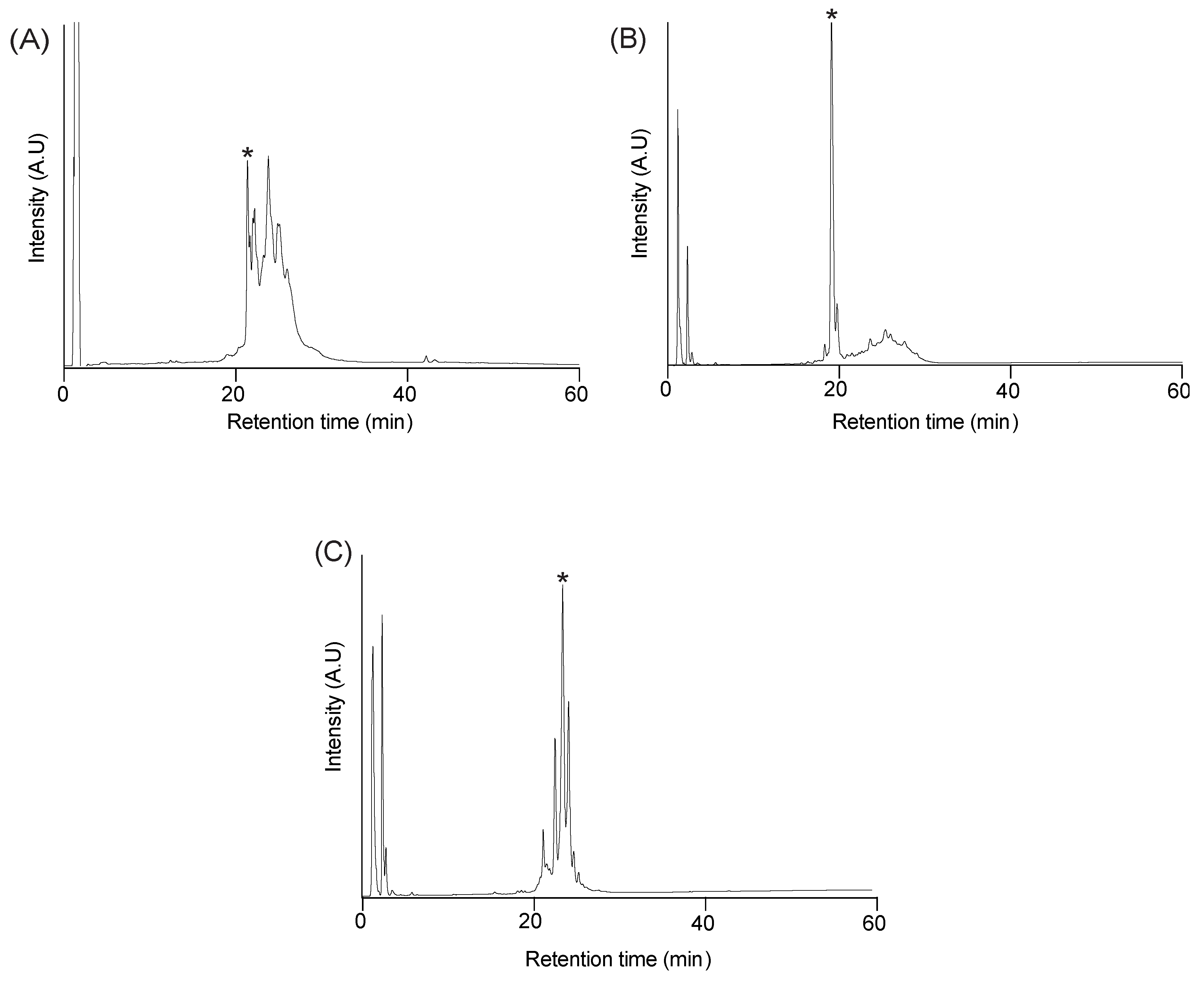
2.2. Purification
2.3. Disulfide Bond Formation
2.4. NMR Spectroscopy
2.5. Structure Calculations
2.6. Mammalian Cell Culture
2.7. Cell Proliferation Monitoring in Real Time Using xCELLigence
3. Results
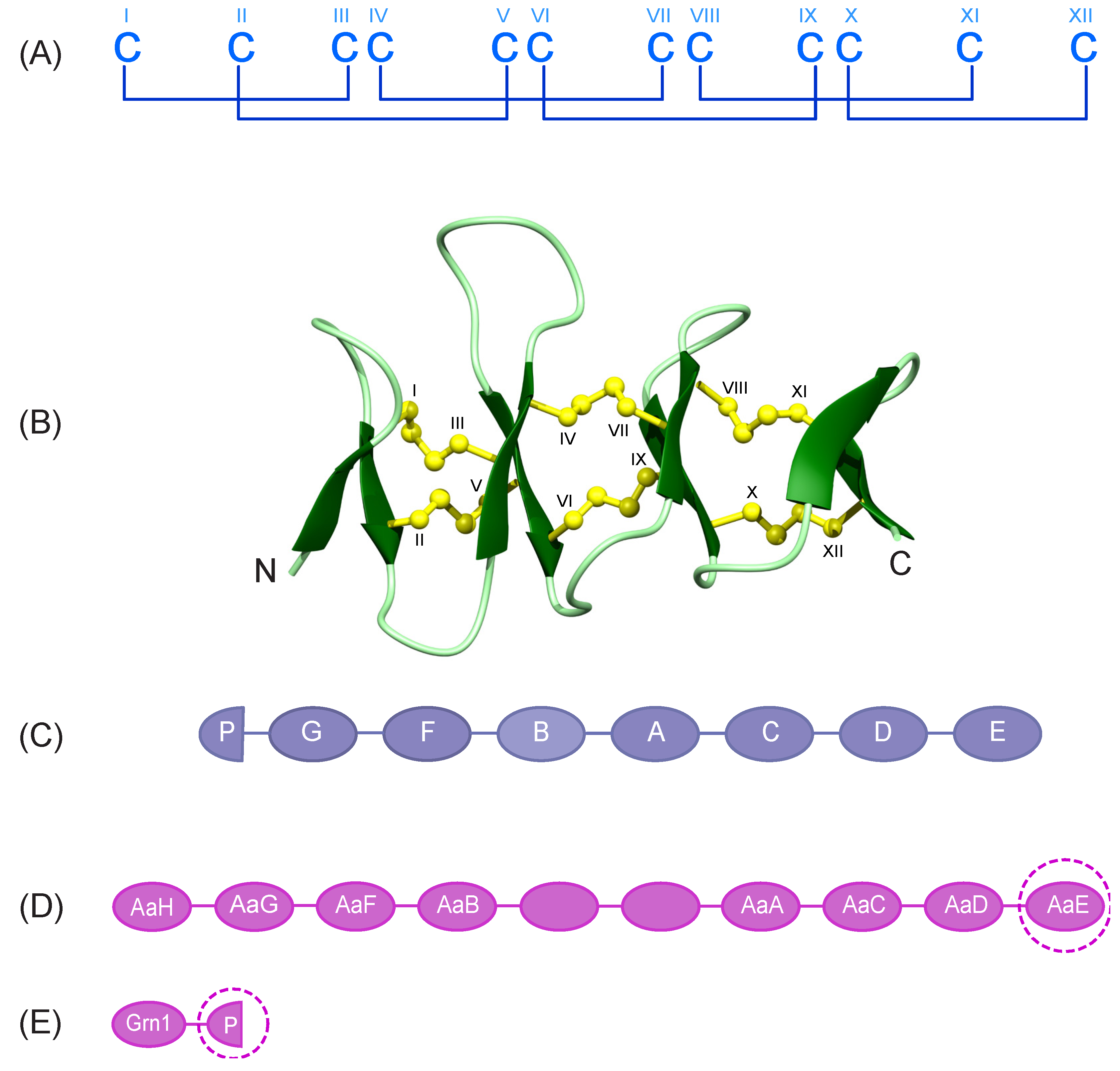
3.1. Design and Synthesis of Zebrafish Granulin Peptides
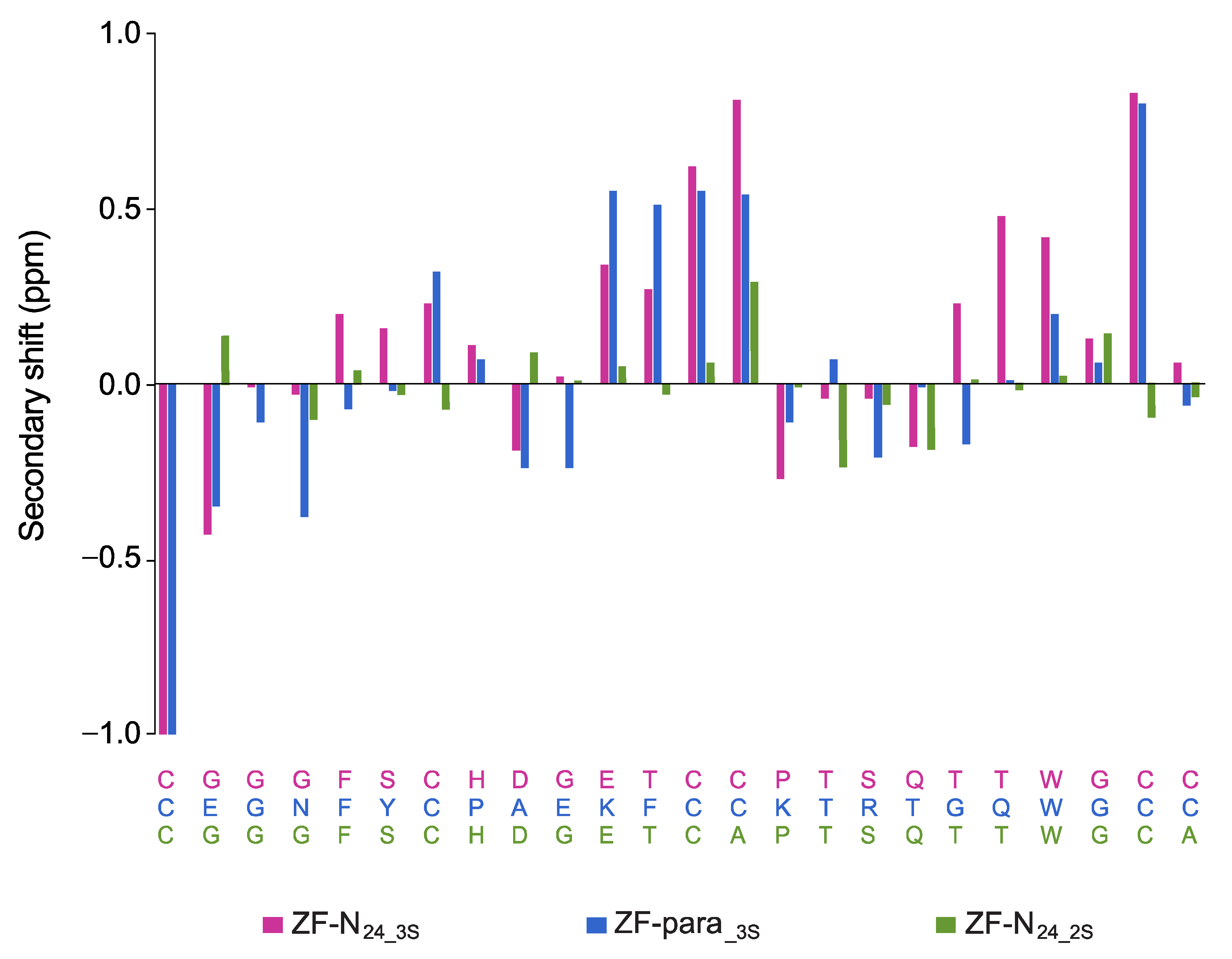
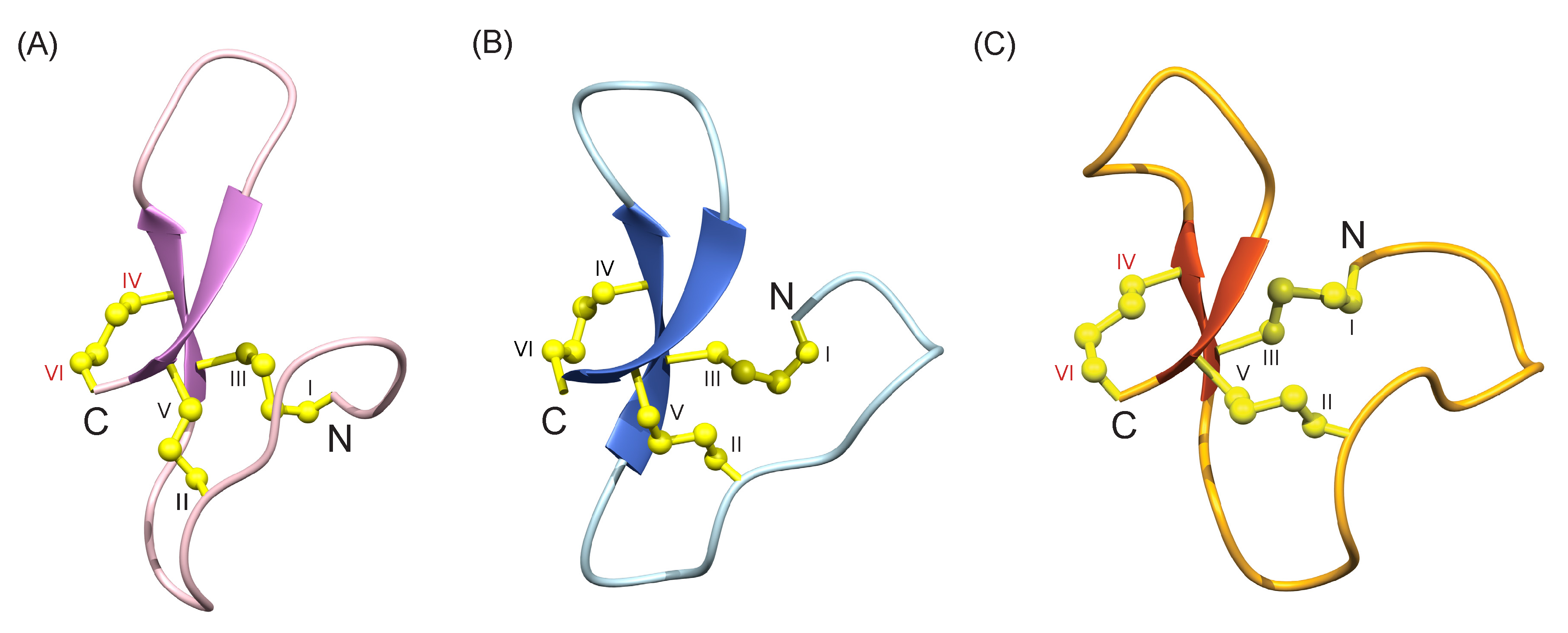
3.2. Structural Analysis with NMR Spectroscopy
3.3. Cell Proliferation Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Palfree, R.G.; Bennett, H.P.; Bateman, A. The evolution of the secreted regulatory protein progranulin. PLoS ONE 2015, 10, e0133749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bansal, P.S.; Smout, M.J.; Wilson, D.; Caceres, C.C.; Dastpeyman, M.; Sotillo, J.; Seifert, J.; Brindley, P.J.; Loukas, A.; Daly, N.L. Development of a potent wound healing agent based on the liver fluke granulin structural fold. J. Med. Chem. 2017, 60, 4258–4266. [Google Scholar] [CrossRef] [PubMed]
- Dastpeyman, M.; Bansal, P.S.; Wilson, D.; Sotillo, J.; Brindley, P.J.; Loukas, A.; Smout, M.J.; Daly, N.L. Structural variants of a liver fluke derived granulin peptide potently stimulate wound healing. J. Med. Chem. 2018, 61, 8746–8753. [Google Scholar] [CrossRef] [PubMed]
- Smout, M.J.; Sotillo, J.; Laha, T.; Papatpremsiri, A.; Rinaldi, G.; Pimenta, R.N.; Chan, L.Y.; Johnson, M.S.; Turnbull, L.; Whitchurch, C.B.; et al. Carcinogenic parasite secretes growth factor that accelerates wound healing and potentially promotes neoplasia. PLoS Pathog. 2015, 11, e1005209. [Google Scholar] [CrossRef]
- Botelho, M.C.; Alves, H.; Richter, J. Wound healing and cancer progression in Opisthorchis viverrini associated cholangiocarcinoma. Parasitol. Res. 2016, 115, 2913–2914. [Google Scholar] [CrossRef]
- Ding, H.; Wei, J.; Zhao, Y.; Liu, Y.; Liu, L.; Cheng, L. Progranulin derived engineered protein Atsttrin suppresses TNF-α-mediated inflammation in intervertebral disc degenerative disease. Oncotarget 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Qiao, G.; Xu, H.L.; Li, C.; Li, X.; Farooqi, A.A.; Zhao, Y.M.; Liu, X.H.; Liu, M.; Stagos, D.; Lin, X.K. Granulin A synergizes with cisplatin to inhibit the growth of human hepatocellular carcinoma. Int. J. Mol. Sci. 2018, 19, 3060. [Google Scholar] [CrossRef] [Green Version]
- Chitramuthu, B.P.; Bennett, H.P.J.; Bateman, A. Progranulin: A new avenue towards the understanding and treatment of neurodegenerative disease. Brain 2017, 140, 3081–3104. [Google Scholar] [CrossRef]
- Jian, J.; Tian, Q.-Y.; Hettinghouse, A.; Zhao, S.; Liu, H.; Liu, C.-J.; Wei, J.; Grunig, G.; Zhang, W.; Setchell, K.D.R.; et al. Progranulin recruits HSP70 to β-glucocerebrosidase and is therapeutic against Gaucher disease. EBioMedicine 2016, 13, 212–224. [Google Scholar] [CrossRef] [Green Version]
- Abella, V.; Pino, J.; Scotece, M.; Conde, J.; Lagoa, F.; Gonzalez-Gay, M.A.; Mera, A.; Gomez, R.; Mobasheri, A.; Gualillol, O. Progranulin as a biomaker and potential therapeutic agent. Drug Discov. Today 2017, 22, 1557–1564. [Google Scholar] [CrossRef]
- Pogonowska, M.; Poniatowski, Ł.A.; Wawrzyniak, A.; Królikowska, K.; Kalicki, B. The role of progranulin (PGRN) in the modulation of anti-inflammatory response in asthma. Cent. Eur. J. Immunol. 2019, 44, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Dastpeyman, M.; Smout, M.J.; Wilson, D.; Loukas, A.; Daly, N.L. Folding of granulin domains. Pept. Sci. 2018, 110. [Google Scholar] [CrossRef]
- Ong, C.H.P.; Bateman, A. Progranulin (granulin-epithelin precursor, PC-cell derived growth factor, acrogranin) in proliferation and tumorigenesis. Histol. Histopathol. 2003, 18, 1275–1288. [Google Scholar] [PubMed]
- Hrabal, R.; Chen, Z.; James, S.; Bennett, H.P.J.; Ni, F. The hairpin stack fold, a novel protein architecture for a new family of protein growth factors. Nat. Struct. Biol. 1996, 3, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Chitramuthu, B.; Bateman, A.; Bennett, H.P.J.; Xu, P.; Ni, F. Structure dissection of zebrafish progranulins identifies a well-folded granulin/epithelin module protein with pro-cell survival activities: Dynamic structures of zebrafish granulin modules. Protein Sci. 2018, 27, 1476–1490. [Google Scholar] [CrossRef] [Green Version]
- Tolkatchev, D.; Malik, S.; Vinogradova, A.; Wang, P.; Chen, Z.; Xu, P.; Bennett, H.P.J.; Bateman, A.; Ni, F. Structure dissection of human progranulin identifies well-folded granulin/epithelin modules with unique functional activities. Protein Sci. 2008, 17, 711–724. [Google Scholar] [CrossRef] [Green Version]
- Tolkatchev, D.; Ng, A.; Vranken, W.; Ni, F. Design and solution structure of a well-folded stack of two β-hairpins based on the amino-terminal fragment of human granulin A. Biochemistry 2000, 39, 2878–2886. [Google Scholar] [CrossRef]
- Vranken, W.F.; Chen, Z.G.; Xu, P.; James, S.; Bennett, H.P.J.; Ni, F. A 30-residue fragment of the carp granulin-1 protein folds into a stack of two β-hairpins similar to that found in the native protein. J. Pept. Res. 1999, 53, 590–597. [Google Scholar] [CrossRef]
- Vranken, W.F.; James, S.; Bennett, H.P.J.; Ni, F. Solution structures of a 30-residue amino-terminal domain of the carp granulin-1 protein and its amino-terminally truncated 3-30 subfragment: Implications for the conformational stability of the stack of two β-hairpins. Proteins 2002, 47, 14–24. [Google Scholar] [CrossRef]
- Tolkatchev, D.; Xu, P.; Ni, F. A peptide derived from the C-terminal part of a plant cysteine protease folds into a stack of two β-hairpins, a scaffold present in the emerging family of granulin-like growth factors. J. Pept. Res. 2001, 57, 227–233. [Google Scholar] [CrossRef]
- Niezgoda, J.A.; Van Gils, C.C.; Frykberg, R.G.; Hodde, J.P. Randomized clinical trial comparing OASIS Wound Matrix to Regranex Gel for diabetic ulcers. Adv. Skin Wound Care 2005, 18, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Chan, R.K.; Liu, P.H.; Pietramaggiori, G.; Ibrahim, S.I.; Hechtman, H.B.; Orgill, D.P. Effect of recombinant platelet-derived growth factor (Regranex®) on wound closure in genetically diabetic mice. J. Burn Care Res. 2006, 27, 202–205. [Google Scholar] [CrossRef] [PubMed]
- Georgoulia, P.S.; Glykos, N.M. Molecular simulation of peptides coming of age: Accurate prediction of folding, dynamics and structures. Arch. Biochem. Biophys. 2019, 664, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.F. Structure Genomics of Zebrafish Granulins; McGill University: Montreal, QC, Canada, 2004. [Google Scholar]
- Wüthrich, K. NMR studies of structure and function of biological macromolecules (Nobel Lecture). J. Biomol. NMR 2003, 27, 13–39. [Google Scholar] [CrossRef]
- Vranken, W.F.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, M.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 2005, 59, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Cierpicki, T.; Otlewski, J. Amide proton temperature coefficients as hydrogen bond indicators in proteins. J. Biomol. NMR 2001, 21, 249–261. [Google Scholar] [CrossRef]
- Wishart, D.S.; Bigam, C.G.; Holm, A.; Hodges, R.S.; Sykes, B.D. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J. Biomol. NMR 1995, 5, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Koradi, R.; Billeter, M.; Wüthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996, 14, 51–55. [Google Scholar] [CrossRef]
- Güntert, P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 2004, 278, 353–378. [Google Scholar]
- Shen, Y.; Bax, A. Protein structural information derived from NMR chemical shift with the neural network program TALOS-N. Methods Mol. Biol. 2015, 1260, 17–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhandari, V.; Roger, G.E.P.; Bateman, A. Isolation and sequence of the granulin precursor cDNA from human bone marrow reveals tandem cysteine-rich granulin domains. Proc. Natl. Acad. Sci. USA 1992, 89, 1715–1719. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; He, B.-J.; Jang, R.; Zhang, Y.; Shen, H.-B. Accurate disulfide-bonding network predictions improve ab initio structure prediction of cysteine-rich proteins. Bioinformatics 2015, 31, 3773–3781. [Google Scholar] [CrossRef] [PubMed]
- Paul George, A.A.; Heimer, P.; Maaß, A.; Hamaekers, J.; Hofmann-Apitius, M.; Biswas, A.; Imhof, D. Insights into the folding of disulfide-rich μ-conotoxins. ACS Omega 2018, 3, 12330–12340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewandowska, A.; Ołdziej, S.; Liwo, A.; Scheraga, H.A. β-hairpin-forming peptides; models of early stages of protein folding. Biophys. Chem. 2010, 151, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, L.D.; Foged, M.M.; Albert, A.; Bertelsen, A.B.; Søltoft, C.L.; Robinson, S.D.; Petersen, S.V.; Purcell, A.W.; Olivera, B.M.; Norton, R.S.; et al. The three-dimensional structure of an H-superfamily conotoxin reveals a granulin fold arising from a common ICK cysteine framework. J. Biol. Chem. 2019, 294, 8745–8759. [Google Scholar] [CrossRef] [PubMed]
- Jin, A.H.; Dekan, Z.; Smout, M.J.; Wilson, D.; Dutertre, S.; Vetter, I.; Lewis, R.J.; Loukas, A.; Daly, N.L.; Alewood, P.F. Conotoxin ϕ-MiXXVIIA from the superfamily G2 employs a novel cysteine framework that mimics granulin and displays anti-apoptotic activity. Angew. Chem. Int. Ed. 2017, 56, 14973–14976. [Google Scholar] [CrossRef]
- Uversky, V.N. A decade and a half of protein intrinsic disorder: Biology still waits for physics. Protein Sci. 2013, 22, 693–724. [Google Scholar] [CrossRef] [Green Version]
- Ghag, G.; Wolf, L.M.; Reed, R.G.; Van Der Munnik, N.P.; Mundoma, C.; Moss, M.A.; Rangachari, V. Fully reduced granulin-B is intrinsically disordered and displays concentration-dependent dynamics. Protein Eng. Des. Sel. 2016, 29, 177–186. [Google Scholar] [CrossRef] [Green Version]
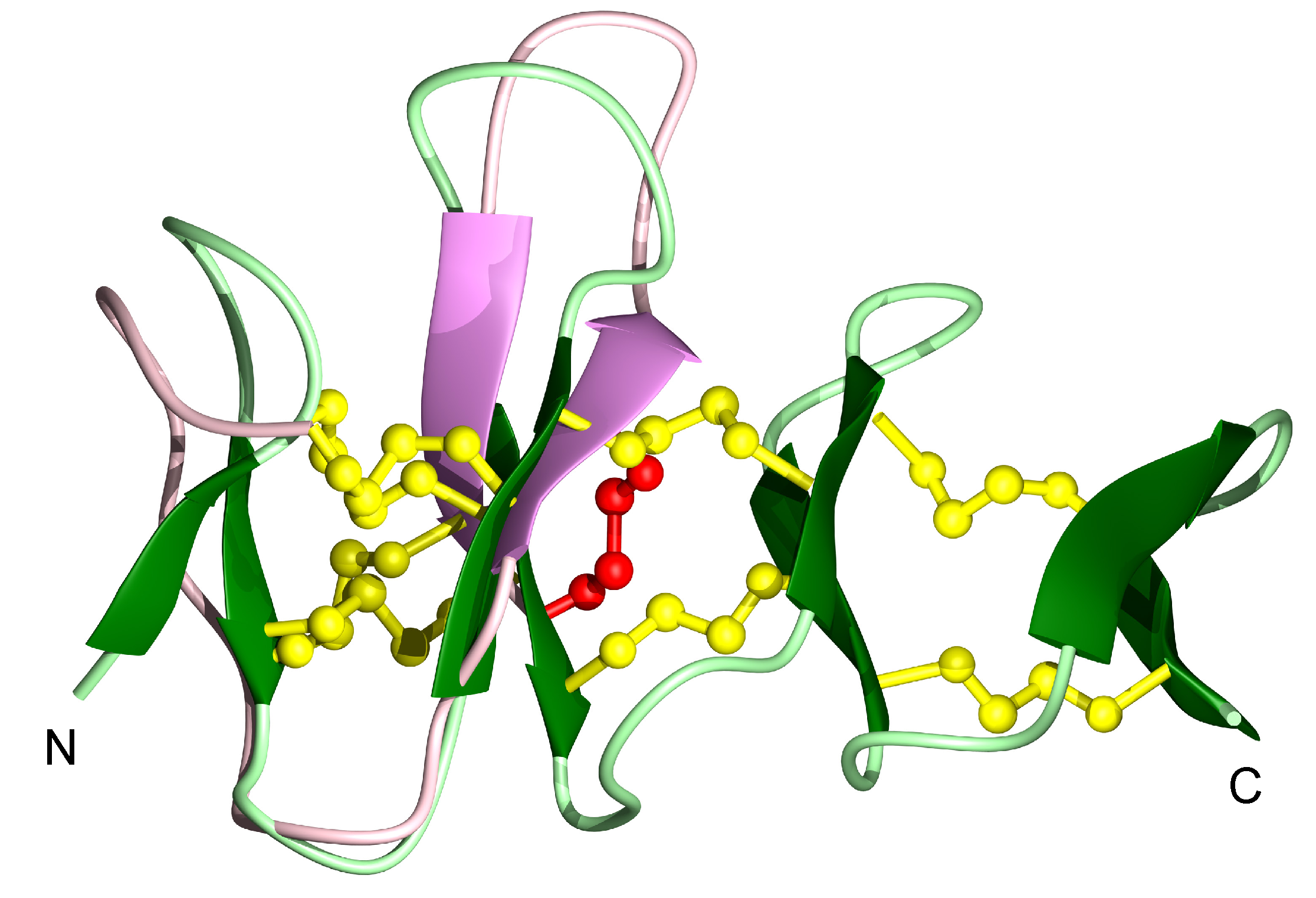
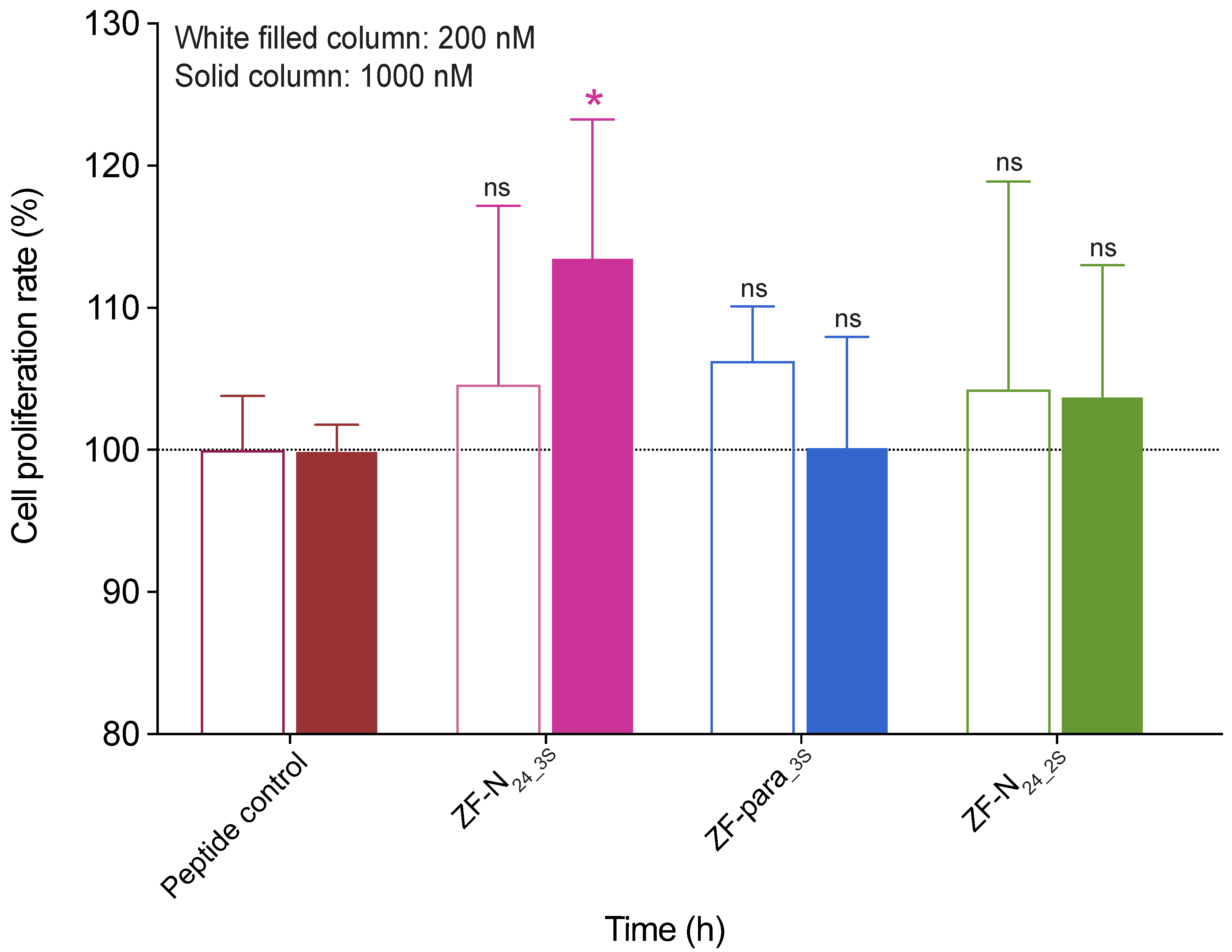
| Peptide | Source | Sequence |
|---|---|---|
| ZF-N24_3s | Zebrafish granulin AaE | CGGGF-SCHDGETCCPTSQTTWGCC |
| ZF-N24_2s | Zebrafish granulin AaE | CGGGF-SCHDGETCAPTSQTTWGCA |
| ZF-para_3s | Zebrafish paragranulin | CEGNFY-CPAEKFCCKTRTGQWGCC |
| Ov-GRN12-35_3s# | Ov-GRN-1 | CPDPVYTCRPGQTCCRGLHG-YGCC |
| Experimental Restraints | ZF-N24_3s | ZF-para_3s |
|---|---|---|
| Interproton distance restraints | ||
| Intra-residue, |i − j| = 0 | 41 | 72 |
| Sequential, |i − j| = 1 | 65 | 88 |
| Medium range, 1 < |i − j| < 5 | 12 | 36 |
| Long range, |i − j| > = 5 | 31 | 64 |
| Disulfide-bond restraints (3 restraints per bond) | 9 | 9 |
| Dihedral-angle restraints | 27 | 32 |
| Hydrogen bond restraints (2 restraints per bond) | 4 | 8 |
| Root Mean Square Deviations from Mean Coordinate Structure (Å) | ||
| Backbone atoms | 1.29 ± 0.38 | 0.63 ± 0.20 |
| All heavy atoms | 1.81 ± 0.41 | 1.31 ± 0.35 |
| Ramachandran Statistics | ||
| % in most favoured region | 80.6 | 96.4 |
| % Residues in additionally allowed regions | 19.4 | 3.6 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takjoo, R.; Wilson, D.; Bansal, P.S.; Loukas, A.; Smout, M.J.; Daly, N.L. Folding of Truncated Granulin Peptides. Biomolecules 2020, 10, 1152. https://doi.org/10.3390/biom10081152
Takjoo R, Wilson D, Bansal PS, Loukas A, Smout MJ, Daly NL. Folding of Truncated Granulin Peptides. Biomolecules. 2020; 10(8):1152. https://doi.org/10.3390/biom10081152
Chicago/Turabian StyleTakjoo, Rozita, David Wilson, Paramjit S. Bansal, Alex Loukas, Michael J. Smout, and Norelle L. Daly. 2020. "Folding of Truncated Granulin Peptides" Biomolecules 10, no. 8: 1152. https://doi.org/10.3390/biom10081152
APA StyleTakjoo, R., Wilson, D., Bansal, P. S., Loukas, A., Smout, M. J., & Daly, N. L. (2020). Folding of Truncated Granulin Peptides. Biomolecules, 10(8), 1152. https://doi.org/10.3390/biom10081152






