Oxidation of Isodrimeninol with PCC Yields Drimane Derivatives with Activity against Candida Yeast by Inhibition of Lanosterol 14-Alpha Demethylase
Abstract
1. Introduction
2. Materials and Methods
2.1. General Information
2.2. Purification of Isodrimeninol from Drimys Winteri
2.3. Oxidation of C1 with Pyridinium Chlorochromate (PCC)
2.4. Structural Identification of Compounds
2.5. Antifungal Assays against Candida Species
2.6. Simulated Systems
2.7. Molecular Docking Calculations
2.8. Molecular Dynamics Simulations
2.9. Trajectory Analysis
3. Results
3.1. Hemisynthetic Compounds by Oxidation of Isodrimeninol
3.2. Anti-Candida Activity Assay
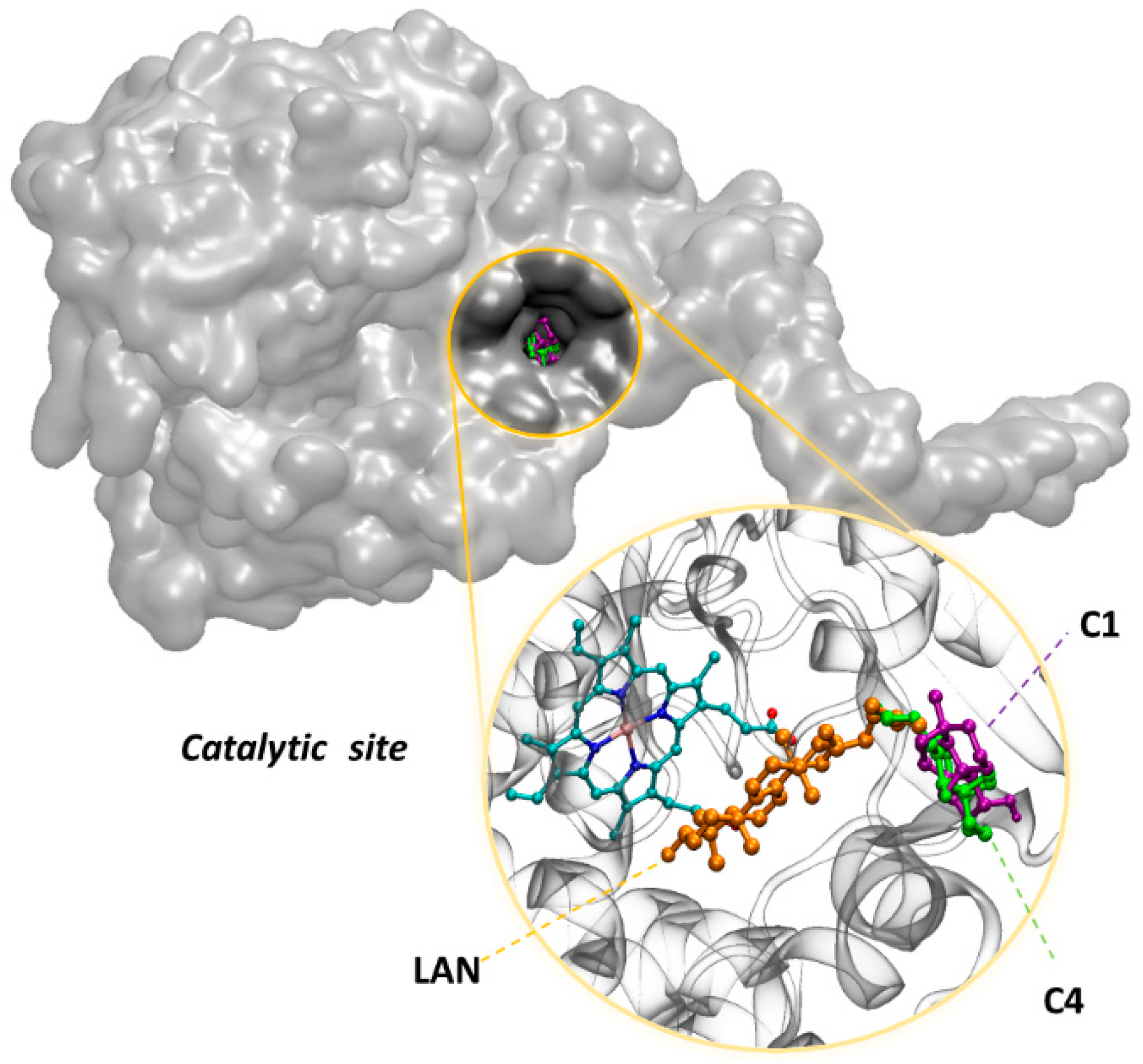
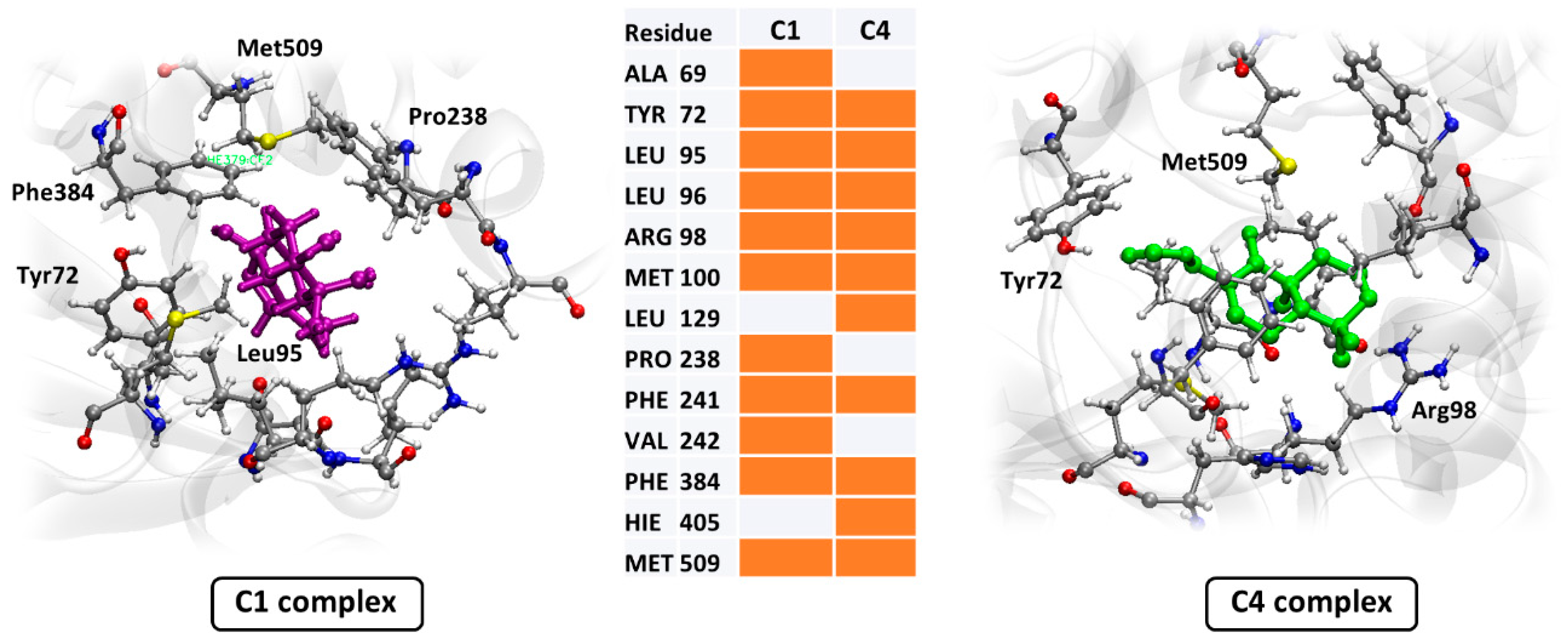
3.3. In Silico Assay
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kullberg, B.J.; Arendrup, M.C. Invasive candidiasis. N. Eng. J. Med. 2015, 373, 1445–1456. [Google Scholar] [CrossRef]
- Silva, S.; Negri, M.; Henriques, M.; Oliveira, R.; Williams, D.W.; Azeredo, J. Candida glabrata, Candida parapsilosis and Candida tropicalis: Biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol. Rev. 2012, 36, 288–305. [Google Scholar] [CrossRef]
- Arendrup, M.C.; Patterson, T.F. Multidrug-resistant Candida: Epidemiology, molecular mechanisms, and treatment. Int. J. Infect. Dis. 2017, 216, S445–S451. [Google Scholar] [CrossRef]
- Spampinato, C.; Leonardi, D. Candida infections, causes, targets, and resistance mechanisms: Traditional and alternative antifungal agents. Biomed. Res. Int. 2013, 2013, 13. [Google Scholar] [CrossRef] [PubMed]
- Whaley, S.G.; Berkow, E.L.; Rybak, J.M.; Nishimoto, A.T.; Barker, K.S.; Rogers, P.D. Azole antifungal resistance in Candida albicans and emerging non-albicans Candida species. Front. Microbiol. 2017, 7, 2173. [Google Scholar] [CrossRef]
- Whaley, S.G.; Rogers, P.D. Azole resistance in Candida glabrata. Curr. Infect. Dis. Rep. 2016, 18, 41. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Lee, J.R.; Lunde, C.S.; Kubo, I. In Vitro Anti-Fungal Susceptibilities of Candida albicans and Other Fungal Pathogens to Polygodial, a Sesquiterpene Dialdehyde. Planta Med. 1999, 65, 204–208. [Google Scholar] [CrossRef] [PubMed]
- Bombaça, A.C.S.; Von Dossow, D.; Barbosa, J.M.C.; Paz, C.; Burgos, V.; Menna-Barreto, R.F.S. TrypanocidalActivity of Natural Sesquiterpenoids Involves Mitochondrial Dysfunction, ROS Production and Autophagic Phenotype in Trypanosoma cruzi. Molecules 2018, 23, 2800. [Google Scholar] [CrossRef]
- Paz, C.; Burgos, V.; Iturra, A.; Rebolledo, R.; Ortiz, L.; Baggio, R.; Cespedes-Acuña, C.L. Assessment of insecticidal responses of extracts and compounds of Drimys winteri, Lobelia tupa, Viola portalesia and Vestia foetida against the granary weevil Sitophilus granarius. Ind. Crops Prod. 2018, 122, 232–238. [Google Scholar] [CrossRef]
- Kubo, I.; Lee, S.H.; Shimizu, K. Combination Effect of Miconazole with Polygodial against Candida albicans. J. Med. Microbiol. 2011, 1, 7–11. [Google Scholar]
- Kubo, I.; Himejima, M. Anethole, a Synergist of Polygodial against Filamentous Microorganisms. J. Agric. Food Chem. 1991, 39, 2290–2292. [Google Scholar] [CrossRef]
- Kubo, I.; Fujita, K.; Lee, S.H. Antifungal Mechanism of Polygodial. J. Agric. Food Chem. 2001, 49, 1607–1611. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, Y.; Tanabe, M.; Kayama, Y.; Abe, M.; Kashio, M.; Koizumi, K.; Okumura, Y.; Morimitsu, Y.; Tominaga, M.; Ozawa, Y.; et al. Miogadial and miogatrial with α,β-unsaturated 1,4-dialdehyde moieties—Novel and potent TRPA1 agonists. Life Sci. 2009, 85, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Arias, H.R.; Feuerbach, D.; Schmidt, B.; Heydenreich, M.; Paz, C.; Ortells, M.O. Drimane sesquiterpenoids noncompetitively inhibit human α4β2 nicotinic acetylcholine receptors with higher potency compared to human α3β4 and α7 subtypes. J. Nat. Prod. 2018, 81, 811–817. [Google Scholar] [CrossRef] [PubMed]
- Carcamo, G.; Silva, M.; Becerra, J.; Urrutia, H.; Sossa, K.; Paz, C. Inhibition of quorum sensing by drimane lactones from Chilean flora. J. Chil. Chem. Soc. 2014, 59, 2622–2624. [Google Scholar] [CrossRef]
- Paz, C.; Cárcamo, G.; Silva, M.; Becerra, J.; Urrutia, H.; Sossa, K. Drimendiol, a drimane sesquiterpene with quorum sensing inhibition activity. Nat. Prod. Commun. 2013, 8, 1934578X1300800201. [Google Scholar] [CrossRef]
- Zárraga, M.; Zárraga, A.M.; Rodríguez, B.; Pérez, C.; Paz, C.; Paz, P.; Sanhueza, C. Synthesis of a new nitrogenated drimane derivative with antifungal activity. Tetrahedron Lett. 2008, 49, 4775–4776. [Google Scholar] [CrossRef]
- CLSI document M27-A3. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standard–third edition; Clinical and Laboratory Standards Institute: Wayne, NJ, USA, 2008. [Google Scholar]
- Anandakrishnan, R.; Aguilar, B.; Onufriev, A.V. H++3.0: Automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res. 2012, 40, W537–W541. [Google Scholar] [CrossRef]
- Gordon, J.C.; Myers, J.B.; Folta, T. H++: A server for estimating pK(a)s and adding missing hydrogens to macromolecules. Nucleic Acids Res. 2005, 33, W368–W371. [Google Scholar] [CrossRef] [PubMed]
- Sterling, T.; Irwin, J.J. ZINC 15 – Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Liu, Y.; Grimm, M.; Dai, W.T. CB-Dock: A web server for cavity detection-guided protein–ligand blind docking. Acta Pharmacol. Sin. 2020, 41, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem 2002, 23, 1624–1641. [Google Scholar] [CrossRef]
- Martínez, L. Automatic Identification of Mobile and Rigid Substructures in Molecular Dynamics Simulations and Fractional Structural Fluctuation Analysis. PLoS ONE 2015, 10, e0119264. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.R.; McGee, T.D.; Swails, J.M. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Fukuyama, Y.; Sato, T.; Asakawa, Y.; Takemoto, T. A potent cytotoxic warburganal and related drimane-type sesquiterpenoids from Polygonum hydropiper. Phytochemistry 1980, 21, 2895–2898. [Google Scholar] [CrossRef]
- Rodríguez, B.; Zapata, N.; Medina, P.; Viñuela, E. A complete 1H and 13C NMR data assignment for four drimane sesquiterpenoids isolated from Drimys winterii. Magn. Res. Chem. 2005, 43, 82–84. [Google Scholar] [CrossRef] [PubMed]
- Marcos, I.S.; Moro, R.F.; Carballares, M.S.; Urones, J.G. An Efficient Total Synthesis of Isodrimeninol from Zamoranic Acid. Synlett 2000, 2000, 541–543. [Google Scholar]
- Nakano, T.; Villamizar, J.; Maillo, M.A. Highly efficient synthesis of optically active drimanic sesquiterpenes, (+)-fuegin, (+)-epifutronolide (7β-hydroxyisodrimenin) and (+)-7-ketoisodrimenin. Tetrahedron 1999, 55, 1561–1568. [Google Scholar] [CrossRef]
- Vlad, P.F.; Ciocarlan, A.; Edu, C.; Aricu, A.; Biriiac, A.; Coltsa, M.; D’Ambrosio, M.; Deleanu, C.; Nicolescu, A.; Shova, S.; et al. Regio- and stereoselective synthesis of (+)-6-ketoeuryfuran, (+)-6-ketowinterin, and (−)-7-ketoeuryfuran from accessible labdane diterpenoids (+)-larixol and (−)-sclareol. Tetrahedron 2013, 69, 918–926. [Google Scholar] [CrossRef]
- Fernandes, R.A. PCC: Novel Oxidation Reactions. Synlett 2003, 2003, 741–742. [Google Scholar] [CrossRef][Green Version]
- Akita, H.; Nozawa, M.; Mitsuda, A.; Ohsawa, H. A convenient synthesis of (+)-albicanol based on enzymatic function: Total syntheses of (+)-albicanyl acetate, (−)-albicanyl 3,4-dihydroxycinnamate, (−)-drimenol, (−)-drimenin and (−)-ambrox. Tetrahedron Asymmetry 2000, 11, 1375–1388. [Google Scholar] [CrossRef]
- Fetizon, M.; Goulaouic, P.; Hanna, I. 1,4-dioxene in organic chemistry. Part VII regiospecific oxidative cleavage of 1,4-dioxenyl carbinols with pyridinium chlorochromate. A new method for the preparation of α-hydroxy acids and α-keto acids. Tetrahedron Lett. 1988, 29, 6261–6264. [Google Scholar] [CrossRef]
- Harrigan, G.G.; Ahmad, A.; Baj, N.; Glass, T.E.; Gunatilaka, A.A.L.; Kingston, D.G.I. Bioactive and Other Sesquiterpenoids from Porella cordeana. J. Nat. Prod. 1993, 56, 921–925. [Google Scholar] [CrossRef] [PubMed]
- Hargrove, T.Y.; Friggeri, L.; Wawrzak, Z.; Qi, A.; Hoekstra, W.J.; Schotzinger, R.J.; York, J.D.; Guengerich, F.P.; Lepesheva, G.I. Structural analyses of Candida albicans sterol 14α-demethylase complexed with azole drugs address the molecular basis of azole-mediated inhibition of fungal sterol biosynthesis. J. Biol. Chem. 2017, 292, 6728–6743. [Google Scholar] [CrossRef]
- Castanheira, M.; Deshpande, L.M.; Messer, S.A.; Rhomberg, P.R.; Pfaller, M.A. Analysis of global antifungal surveillance results reveals predominance of Erg11 Y132F alteration among azole-resistant Candida parapsilosis and Candida tropicalis and country-specific isolate dissemination. Int. J. Antimicrob. Agents 2020, 55, 105799. [Google Scholar] [CrossRef]
- Berkow, E.L.; Lockhart, S.R. Fluconazole resistance in Candida species: A current perspective. Infect. Drug Resist. 2017, 10, 237. [Google Scholar] [CrossRef]


| H | C1 | C2 | C3 | C4 | C5 |
|---|---|---|---|---|---|
| 1 | 1.24 (1H, m) 1.79 (1H, ddd, 13.1, 5.0, 2.8) | 1.56 (1H, m) 2.07 (1H, m) | 1.22 (1H, m) 2.49 (1H, dq, 13.4, 2.7) | 1.36 (1H, m) 1.89 (1H, m) | 1.33 (1H, ddd, 13.5, 13.5, 3.8) 2.65 (1H, m) |
| 2 | 1.46 (1H, m) 1.59 (1H, m) | 1.58 (1H, m) 1.71 (1H, m) | 1.49 (1H, m) 1.59 (1H, dq, 13.7, 3.2) | 1.58 (2H, m) | 1.60 (1H, m) 1.70 (1H, m) |
| 3 | 1.22 (1H, m) 1.46 (1H, m) | 1.25 (1H, m) 1.51 (1H, m) | 1.49 (1H, m) 1.24 (1H, m) | 1.18 (1H, m) 1.45 (1H, m) | 1.25 (1H, dd, 13.3, 4.1) 1.52 (1H, m) |
| 5 | 1.30 (1H, dd, 11.7, 5.4) | 1.78 (1H, dd, 13.4, 3.6) | 1.36 (1H, dd, 11.7,5.3) | 1.63 (1H, dd, 11.4, 4.4) | 1.87 (1H, dd, 14.3, 3.0) |
| 6 | 1.91 (1H, m) 2.14 (1H, m) | 2.49 (1H, dd, 18.0, 13.4) 2.57 (1H, ddd, 18.0, 3.6, 0.5) | 2.21 (1H, m) 1.98 (1H, m) | 2.33 (1H, dddd, 19.6, 11.4, 3.9, 2.0) 2.44 (1H, ddd, 19.6, 5.0, 5.0) | 2.50 (1H, dd, 17.5, 14.3) 2.63 (1H, dd, 17.3, 3.3) |
| 7 | 5.51 (1H, m) | - | 5.74 (1H, br, s) | 6.98 (1H, d, 5.8) | - |
| 9 | 2.20 (1H, m) | - | 2.78 (1H, br, s) | - | - |
| 11 | 5.28 (1H, t, 3.9) 11-OH: 3.29 (1H, d, 4.1) | 7.91 (1H, d, 1.5) | - | 8.08 (1H, s) | - |
| 12 | 4.18 (1H, dddd, 11.3, 3.1, 1.5, 1.5) 4.48 (1H, ddddd, 11.3, 3.0, 3.0, 2.0, 2.0) | 7.17 (1H, d, 1.5) | 4.66 (2H, m) | 4.76 (1H, d, 12.7) 4.83 (1H, d, 12.7) | 4.82 (2H, s) |
| 13 | 0.82 (3H, s) | 1.25 (3H, d, 0.6) | 0.90 (3H, s) | 1.06 (3H, d, 0.6) | 1.28 (3H, s) |
| 14 | 0.92 (3H, s) | 0.96 (3H, s) | 0.92 (3H, s) | 0.99 (3H, s) | 0.96 (3H, s) |
| 15 | 0.88 (3H, s) | 0.92 (3H, s) | 0.88 (3H, s) | 0.91 (3H, s) | 0.92 (3H, s) |
| C | C1 | C2 | C3 | C4 | C5 |
|---|---|---|---|---|---|
| 1 | 40.0 | 38.3 | 38.4 | 33.1 | 33.2b |
| 2 | 18.6 | 18.7 | 18.2 | 18.2 | 18.0 |
| 3 | 42.5 | 41.7 | 42.3 | 41.7 | 41.2 |
| 4 | 33.1 | 33.3a | 33.0 | 33.8 | 33.3b |
| 5 | 50.0 | 51.3 | 49.6 | 49.1 | 52.1 |
| 6 | 23.8 | 37.3 | 23.3 | 24.6 | 36.2 |
| 7 | 117.3 | 196.4 | 121.2 | 147.7 | 196.5 |
| 8 | 136.6 | 123.3 | 129.8 | 131.6 | 149.2 |
| 9 | 61.6 | 138.7 | 53.6 | 204.0 | 152.6 |
| 10 | 33.5 | 33.9 | 34.3 | 45.3 | 36.8 |
| 11 | 99.5 | 144.2 | 175.4 | 160.9 | 171.0 |
| 12 | 69.1 | 136.0 | 69.8 | 61.6 | 67.4 |
| 13 | 14.2 | 23.3 | 33.0 | 17.2 | 18.2 |
| 14 | 21.6 | 21.5 | 21.4 | 22.4 | 21.1 |
| 15 | 33.3 | 33.1a | 13.9 | 32.4 | 32.9 |
| Compound | C. albicans ATCC 90028 | C. glabrata ATCC 90030 | C. krusei ATCC 6258 |
|---|---|---|---|
| C1 | 125 | 125 | 125 |
| C2 | >200 | >200 | >200 |
| C3 | >200 | >200 | >200 |
| C4 | 75 | 75 | 75 |
| C5 | >200 | >200 | >200 |
| Fluconazole | 8 | 50 | 75 |
| Ligand | Binding Free Energy Terms (kcal mol−1) | ||||
|---|---|---|---|---|---|
| EVdW | Eel | EGB | Esurf | ∆Gbind | |
| LAN | −60.8 ± 0.2 | −7.0 ± 0.2 | 20.0 ± 0.1 | −7.3 ± 0.1 | −55.1 ± 0.2 |
| C1 | −30.9 ± 0.1 | −1.9 ± 0.2 | 11.1 ± 0.1 | −3.9 ± 0.1 | −25.7 ± 0.2 |
| C4 | −36.1 ± 0.1 | −3.4 ± 0.2 | 13.7 ± 0.1 | −4.9 ± 0.1 | −30.7 ± 0.2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marin, V.; Iturra, A.; Opazo, A.; Schmidt, B.; Heydenreich, M.; Ortiz, L.; Jiménez, V.A.; Paz, C. Oxidation of Isodrimeninol with PCC Yields Drimane Derivatives with Activity against Candida Yeast by Inhibition of Lanosterol 14-Alpha Demethylase. Biomolecules 2020, 10, 1101. https://doi.org/10.3390/biom10081101
Marin V, Iturra A, Opazo A, Schmidt B, Heydenreich M, Ortiz L, Jiménez VA, Paz C. Oxidation of Isodrimeninol with PCC Yields Drimane Derivatives with Activity against Candida Yeast by Inhibition of Lanosterol 14-Alpha Demethylase. Biomolecules. 2020; 10(8):1101. https://doi.org/10.3390/biom10081101
Chicago/Turabian StyleMarin, Victor, Andres Iturra, Andres Opazo, Bernd Schmidt, Matthias Heydenreich, Leandro Ortiz, Verónica A. Jiménez, and Cristian Paz. 2020. "Oxidation of Isodrimeninol with PCC Yields Drimane Derivatives with Activity against Candida Yeast by Inhibition of Lanosterol 14-Alpha Demethylase" Biomolecules 10, no. 8: 1101. https://doi.org/10.3390/biom10081101
APA StyleMarin, V., Iturra, A., Opazo, A., Schmidt, B., Heydenreich, M., Ortiz, L., Jiménez, V. A., & Paz, C. (2020). Oxidation of Isodrimeninol with PCC Yields Drimane Derivatives with Activity against Candida Yeast by Inhibition of Lanosterol 14-Alpha Demethylase. Biomolecules, 10(8), 1101. https://doi.org/10.3390/biom10081101






