Survey and Validation of tRNA Modifications and Their Corresponding Genes in Bacillus subtilis sp Subtilis Strain 168
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatic Analyses
2.2. Strains and Media and Bulk tRNA Preparation
2.3. Mass Spectrometry Analyses
2.3.1. LC-MS of Nucleosides
2.3.2. LC-MS Analysis of Oligonucleotides
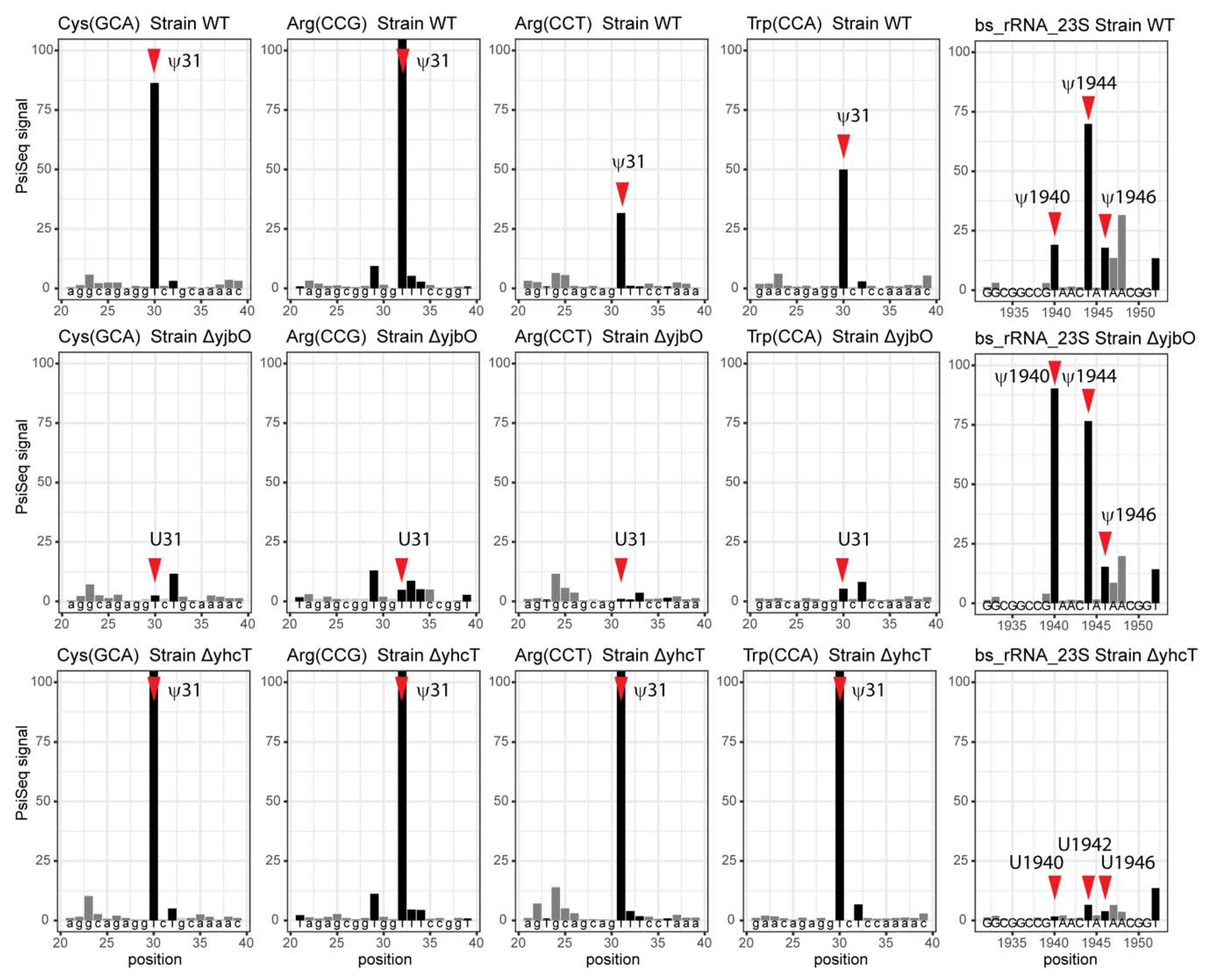
2.4. Identification of Modification by Next Generation Sequencing Methods
3. Results and Discussion
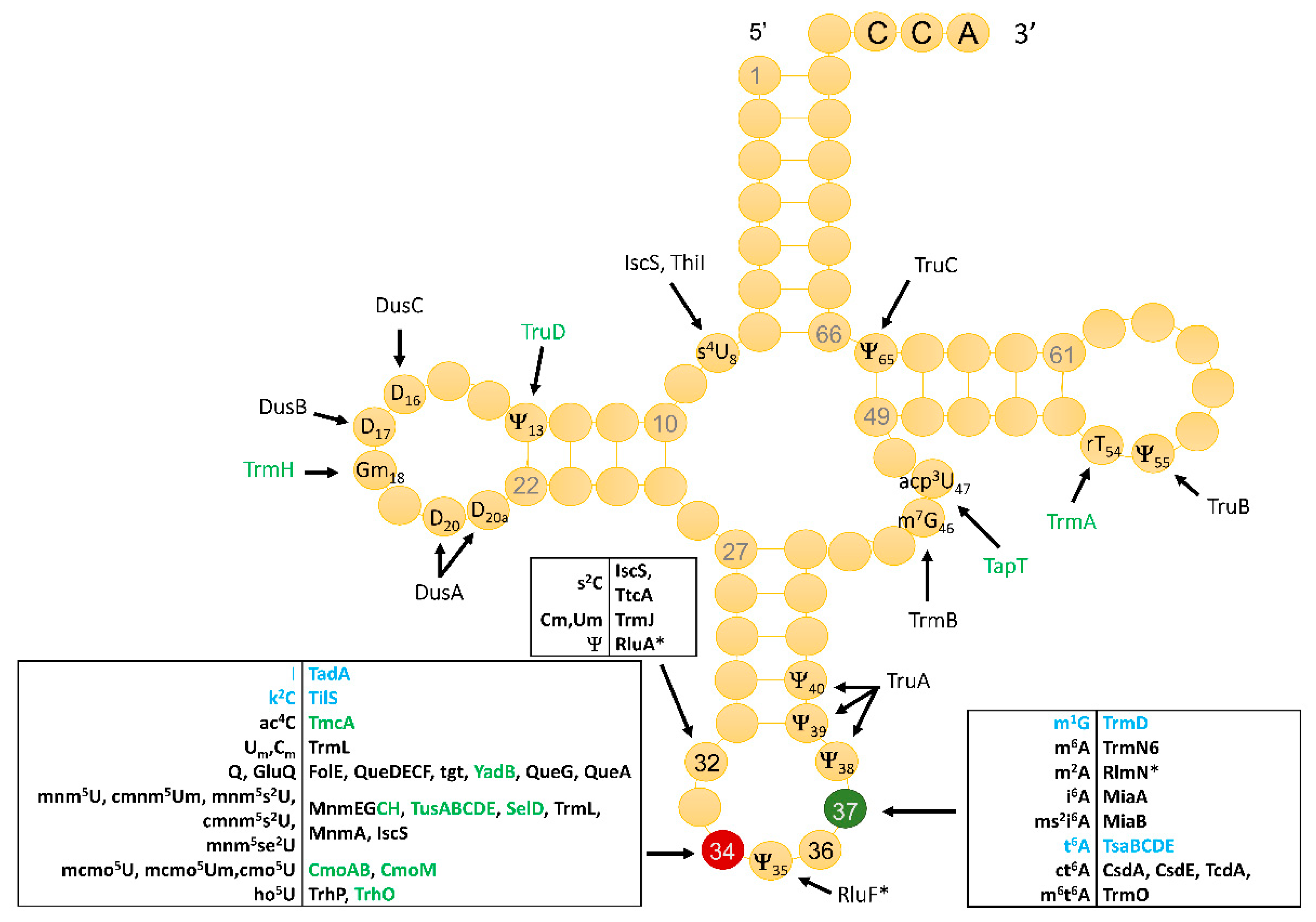
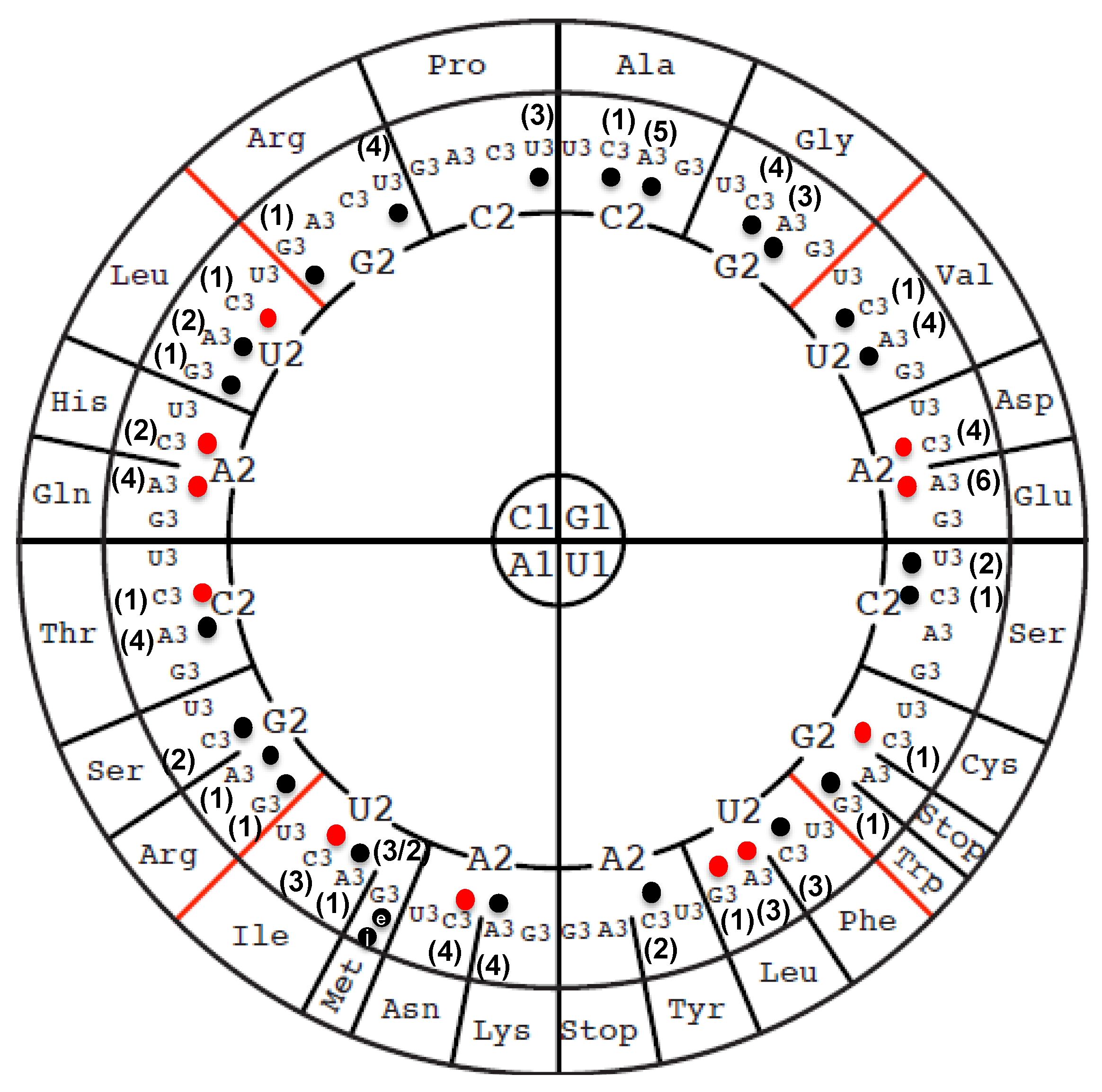
3.1. Identity and Position of tRNA Modifications in the Model Gram-Positive B. subtilis 168
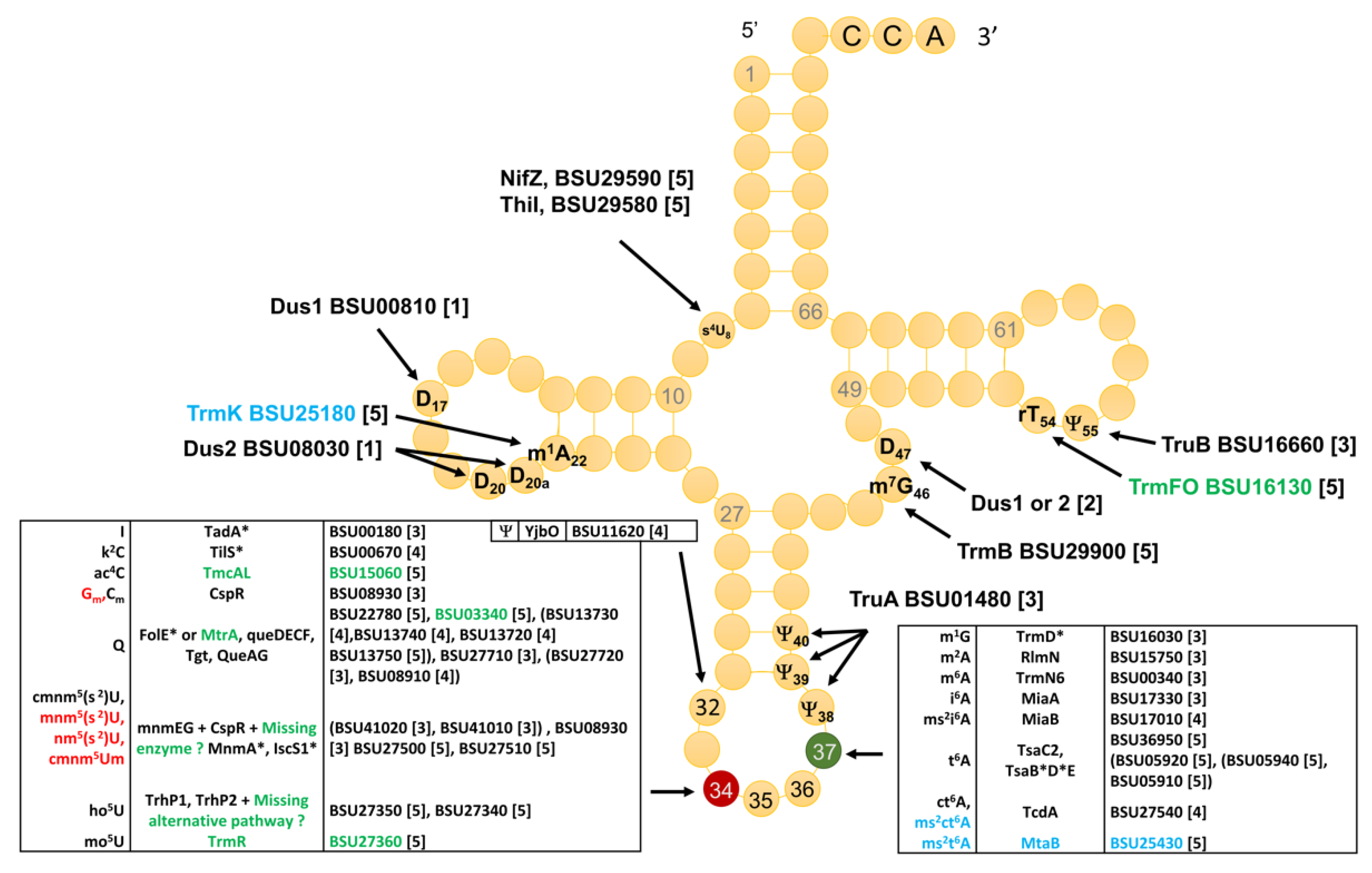
3.2. Compilation of Predicted and Validated B. subtilis tRNA Modification Genes
3.3. The Majority of tRNA Modifications Are Introduced by Orthologs in E. coli and B. subtilis with Some Differences Observed in the Complex Pathways
3.4. Modifications and Enzymes Specific to Bacillus subtilis
3.5. Paralog Issues Make Direct Annotation Transfers Problematic for a Few Modifications
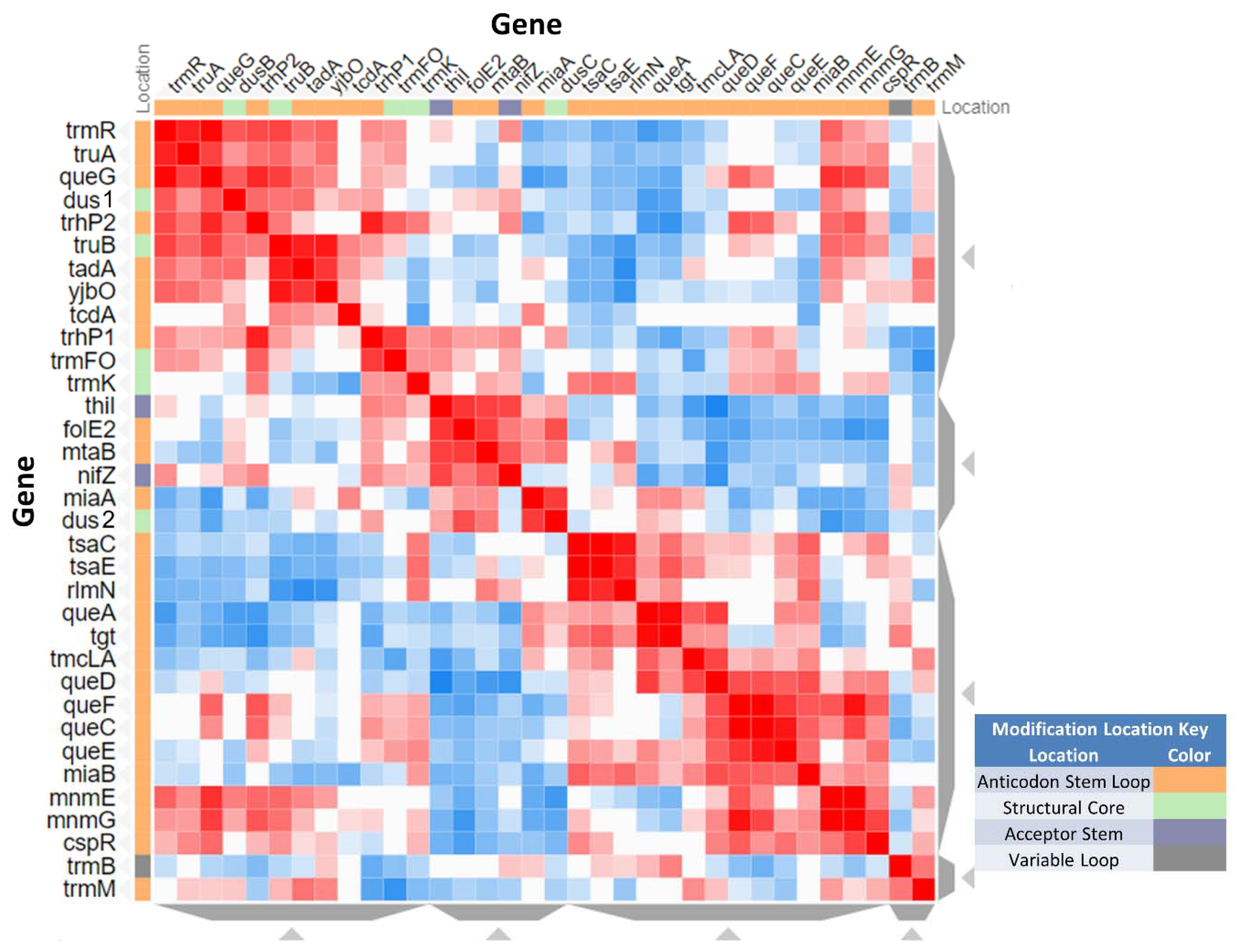
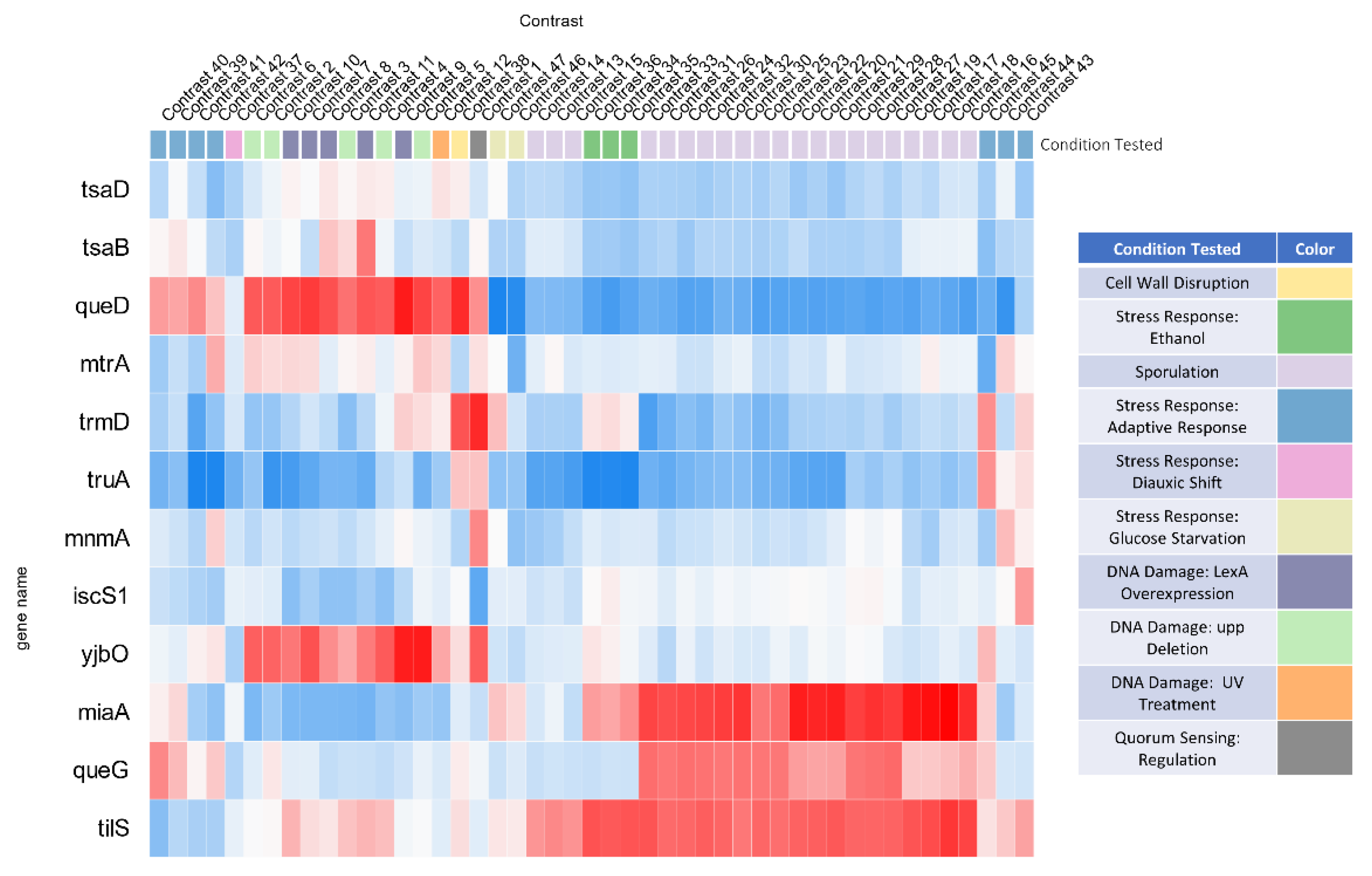
3.6. System Analysis of tRNA Modification Genes in B. subtilis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- El Yacoubi, B.; Bailly, M.; de Crécy-Lagard, V. Biosynthesis and function of posttranscriptional modifications of Transfer RNAs. Annu. Rev. Genet. 2012, 46, 69–95. [Google Scholar] [CrossRef] [PubMed]
- Agris, P.F.; Eruysal, E.R.; Narendran, A.; Väre, V.Y.P.; Vangaveti, S.; Ranganathan, S.V. Celebrating wobble decoding: Half a century and still much is new. RNA Biol. 2018, 15, 537–553. [Google Scholar] [CrossRef]
- Kimura, S.; Waldor, M.K. The RNA degradosome promotes tRNA quality control through clearance of hypomodified tRNA. Proc. Natl. Acad. Sci. USA 2019, 116, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, A.; Chernyakov, I.; Gu, W.; Hiley, S.L.; Hughes, T.R.; Grayhack, E.J.; Phizicky, E.M. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 2006, 21, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.S.; Sarin, L.P. Transfer RNA modification and infection-Implications for pathogenicity and host responses. Biochim. Biophys. Acta-Gene Regul. Mech. 2018, 1861, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, C.; Lünse, C.; Mörl, M. tRNA modifications: Impact on structure and thermal adaptation. Biomolecules 2017, 7, 35. [Google Scholar] [CrossRef]
- Thiaville, P.C.; El Yacoubi, B.; Köhrer, C.; Thiaville, J.J.; Deutsch, C.; Iwata-Reuyl, D.; Bacusmo, J.M.; Armengaud, J.; Bessho, Y.; Wetzel, C.; et al. Essentiality of threonylcarbamoyladenosine (t6A), a universal tRNA modification, in bacteria. Mol. Microbiol. 2015, 98, 1199–1221. [Google Scholar] [CrossRef]
- Soma, A.; Ikeuchi, Y.; Kanemasa, S.; Kobayashi, K.; Ogasawara, N.; Ote, T.; Kato, J.; Watanabe, K.; Sekine, Y.; Suzuki, T.; et al. An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol. Cell. 2003, 12, 689–698. [Google Scholar] [CrossRef]
- Ikeuchi, Y.; Soma, A.; Ote, T.; Kato, J.; Sekine, Y.; Suzuki, T. Molecular mechanism of lysidine synthesis that determines tRNA identity and codon recognition. Mol. Cell 2005, 19, 235–246. [Google Scholar] [CrossRef]
- Fabret, C.; Dervyn, E.; Dalmais, B.; Guillot, A.; Marck, C.; Grosjean, H.; Noirot, P. Life without the essential bacterial tRNAIle2–lysidine synthetase TilS: A case of tRNA gene recruitment in Bacillus subtilis. Mol. Microbiol. 2011, 80, 1062–1074. [Google Scholar] [CrossRef]
- Björk, G.R.; Hagervall, T.G. Transfer RNA modification: Presence, synthesis, and function. EcoSal Plus 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Persson, B.C. Modification of tRNA as a regulatory device. Mol. Microbiol. 1993, 8, 1011–1016. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Begley, T.J.; Dedon, P.C. tRNA modifications regulate translation during cellular stress. FEBS Lett. 2014, 588, 4287–4296. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.; Pham, P.; Dedon, P.C.; Begley, T.J. Lifestyle modifications: Coordinating the tRNA epitranscriptome with codon bias to adapt translation during stress responses. Genome Biol. 2018, 19, 228. [Google Scholar] [CrossRef] [PubMed]
- Pollo-Oliveira, L.; de Crécy-Lagard, V. Can protein expression be regulated by modulation of tRNA Modification profiles? Biochemistry 2019, 58, 355–362. [Google Scholar] [CrossRef]
- Huber, S.M.; Leonardi, A.; Dedon, P.C.; Begley, T.J. The versatile roles of the tRNA epitranscriptome during cellular responses to toxic exposures and environmental stress. Toxics 2019, 7, 17. [Google Scholar] [CrossRef]
- Raina, M.; Ibba, M. tRNAs as regulators of biological processes. Front. Genet. 2014, 5, 171. [Google Scholar] [CrossRef]
- De Crécy-Lagard, V.; Marck, C.; Brochier-Armanet, C.; Grosjean, H. Comparative RNomics and Modomics in Mollicutes: Prediction of gene function and evolutionary implications. IUBMB Life 2007, 59, 634–658. [Google Scholar] [CrossRef]
- Kimura, S.; Dedon, P.C.; Waldor, M.K. Comparative tRNA sequencing and RNA mass spectrometry for surveying tRNA modifications. Nat. Chem. Biol. 2020, in press. [Google Scholar] [CrossRef]
- Puri, P.; Wetzel, C.; Saffert, P.; Gaston, K.W.; Russell, S.P.; Cordero Varela, J.A.; van der Vlies, P.; Zhang, G.; Limbach, P.A.; Ignatova, Z.; et al. Systematic identification of tRNAome and its dynamics in Lactococcus lactis. Mol. Microbiol. 2014, 93, 944–956. [Google Scholar] [CrossRef]
- Cao, X.; Limbach, P.A. Enhanced detection of post-transcriptional modifications using a mass-exclusion list strategy for RNA modification mapping by LC-MS/MS. Anal. Chem. 2015, 87, 8433–8440. [Google Scholar] [CrossRef] [PubMed]
- Chionh, Y.H.; McBee, M.; Babu, I.R.; Hia, F.; Lin, W.; Zhao, W.; Cao, J.; Dziergowska, A.; Malkiewicz, A.; Begley, T.J.; et al. tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nat. Commun. 2016, 7, 13302. [Google Scholar] [CrossRef] [PubMed]
- Kunst, F.; Ogasawara, N.; Moszer, I.; Albertini, A.M.; Alloni, G.; Azevedo, V.; Bertero, M.G.; Bessieres, P.; Bolotin, A.; Borchert, S.; et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 1997, 390, 249–256. [Google Scholar] [CrossRef]
- Flórez, L.A.; Roppel, S.F.; Schmeisky, A.G.; Lammers, C.R.; Stülke, J. A community-curated consensual annotation that is continuously updated: The Bacillus subtilis centered wiki SubtiWiki. Database 2009, 2009, bap012. [Google Scholar] [CrossRef] [PubMed]
- Belda, E.; Sekowska, A.; Le Fèvre, F.; Morgat, A.; Mornico, D.; Ouzounis, C.; Vallenet, D.; Médigue, C.; Danchin, A. An updated metabolic view of the Bacillus subtilis 168 genome. Microbiology 2013, 159, 757–770. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lowe, T.M. GtRNAdb 2.0: An expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016, 44, 184–189. [Google Scholar] [CrossRef]
- Boccaletto, P.; MacHnicka, M.A.; Purta, E.; Pitkowski, P.; Baginski, B.; Wirecki, T.K.; de Crécy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018, 46, 303–307. [Google Scholar] [CrossRef]
- Jühling, F.; Mörl, M.; Hartmann, R.K.; Sprinzl, M.; Stadler, P.F.; Pütz, J. tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009, 37, 159–162. [Google Scholar] [CrossRef]
- Sajek, M.P.; Woźniak, T.; Sprinzl, M.; Jaruzelska, J.; Barciszewski, J. T-psi-C: User friendly database of tRNA sequences and structures. Nucleic Acids Res. 2020, 48, 256–260. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, 158–169. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.H.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. FASEB J. 1997, 25, 1326. [Google Scholar] [CrossRef] [PubMed]
- NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 46, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 2017, 45, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Fernandez, N.F.; Gundersen, G.W.; Rahman, A.; Grimes, M.L.; Rikova, K.; Hornbeck, P.; Ma’ayan, A. Clustergrammer, a web-based heatmap visualization and analysis tool for high-dimensional biological data. Sci. Data 2017, 4, 170151. [Google Scholar] [CrossRef]
- Moretto, M.; Sonego, P.; Dierckxsens, N.; Brilli, M.; Bianco, L.; Ledezma-Tejeida, D.; Gama-Castro, S.; Galardini, M.; Romualdi, C.; Laukens, K.; et al. COLOMBOS v3.0: Leveraging gene expression compendia for cross-species analyses. Nucleic Acids Res. 2016, 44, 620–623. [Google Scholar] [CrossRef]
- Koo, B.-M.; Kritikos, G.; Farelli, J.D.; Todor, H.; Tong, K.; Kimsey, H.; Wapinski, I.; Galardini, M.; Cabal, A.; Peters, J.M.; et al. Construction and analysis of two genome-scale deletion libraries for Bacillus subtilis. Cell Syst. 2017, 4, 291–305. [Google Scholar] [CrossRef]
- Ross, R.; Cao, X.; Yu, N.; Limbach, P.A. Sequence mapping of transfer RNA chemical modifications by liquid chromatography tandem mass spectrometry. Methods 2016, 107, 73–78. [Google Scholar] [CrossRef]
- Yu, N.; Lobue, P.A.; Cao, X.; Limbach, P.A. RNAModMapper: RNA modification mapping software for analysis of liquid chromatography tandem mass spectrometry data. Anal. Chem. 2017, 89, 10744–10752. [Google Scholar] [CrossRef]
- Marchand, V.; Blanloeil-Oillo, F.; Helm, M.; Motorin, Y. Illumina-based RiboMethSeq approach for mapping of 22032-O-Me residues in RNA. Nucleic Acids Res. 2016, 44, e135. [Google Scholar] [CrossRef]
- Marchand, V.; Pichot, F.; Thüring, K.; Ayadi, L.; Freund, I.; Dalpke, A.; Helm, M.; Motorin, Y. Next-generation sequencing-based ribomethseq protocol for analysis of tRNA 2′-O-methylation. Biomolecules 2017, 7, 13. [Google Scholar] [CrossRef]
- Werner, S.; Schmidt, L.; Marchand, V.; Kemmer, T.; Falschlunger, C.; Sednev, M.V.; Bec, G.; Ennifar, E.; Höbartner, C.; Micura, R.; et al. Machine learning of reverse transcription signatures of variegated polymerases allows mapping and discrimination of methylated purines in limited transcriptomes. Nucleic Acids Res. 2020, 48, 3734–3746. [Google Scholar] [CrossRef] [PubMed]
- Motorin, Y.; Marchand, V. Detection and analysis of RNA ribose 2’-O-methylations: Challenges and solutions. Genes 2018, 9, 642. [Google Scholar] [CrossRef] [PubMed]
- Motorin, Y.; Muller, S.; Behm-Ansmant, I.; Branlant, C. Identification of modified residues in RNAs by reverse transcription-based methods. Methods Enzymol. 2007, 425, 21–53. [Google Scholar] [PubMed]
- Behm-Ansmant, I.; Helm, M.; Motorin, Y. Use of specific chemical reagents for detection of modified nucleotides in RNA. J. Nucleic Acids 2011, 2011, 408053. [Google Scholar] [CrossRef] [PubMed]
- De Brouwer, A.P.M.; Abou Jamra, R.; Körtel, N.; Soyris, C.; Polla, D.L.; Safra, M.; Zisso, A.; Powell, C.A.; Rebelo-Guiomar, P.; Dinges, N.; et al. Variants in PUS7 cause intellectual disability with speech delay, microcephaly, short stature, and aggressive behavior. Am. J. Hum. Genet. 2018, 103, 1045–1052. [Google Scholar] [CrossRef] [PubMed]
- Grosjean, H.; Westhof, E. An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. 2016, 44, 8020–8040. [Google Scholar] [CrossRef]
- Moukadiri, I.; Villarroya, M.; Benítez-Páez, A.; Armengod, M.-E. Bacillus subtilis exhibits MnmC-like tRNA modification activities. RNA Biol. 2018, 15, 1167–1173. [Google Scholar] [CrossRef]
- Taniguchi, T.; Miyauchi, K.; Sakaguchi, Y.; Yamashita, S.; Soma, A.; Tomita, K.; Suzuki, T. Acetate-dependent tRNA acetylation required for decoding fidelity in protein synthesis. Nat. Chem. Biol. 2018, 14, 1010–1020. [Google Scholar] [CrossRef]
- Miles, Z.D.; McCarty, R.M.; Molnar, G.; Bandarian, V. Discovery of epoxyqueuosine (oQ) reductase reveals parallels between halorespiration and tRNA modification. Proc. Natl. Acad. Sci. USA 2011, 108, 7368–7372. [Google Scholar] [CrossRef] [PubMed]
- Moukadiri, I.; Prado, S.; Piera, J.; Velázquez-Campoy, A.; Björk, G.R.; Armengod, M.E. Evolutionarily conserved proteins MnmE and GidA catalyze the formation of two methyluridine derivatives at tRNA wobble positions. Nucleic Acids Res. 2009, 37, 7177–7193. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, K.; Kimura, S.; Suzuki, T. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat. Chem. Biol. 2013, 9, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Suzuki, T. Fine-tuning of the ribosomal decoding center by conserved methyl-modifications in the Escherichia coli 16S rRNA. Nucleic Acids Res. 2010, 38, 1341–1352. [Google Scholar] [CrossRef] [PubMed]
- Emmerechts, G.; Barbe, S.; Herdewijn, P.; Anne, J.; Rozenski, J. Post-transcriptional modification mapping in the Clostridium acetobutylicum 16S rRNA by mass spectrometry and reverse transcriptase assays. Nucleic Acids Res. 2007, 35, 3494–3503. [Google Scholar] [CrossRef]
- Desai, P.M.; Culver, G.M.; Rife, J.P. Site-directed mutants of 16S rRNA reveal important RNA domains for KsgA function and 30S subunit assembly. Biochemistry 2011, 50, 854–863. [Google Scholar] [CrossRef]
- Poldermans, B.; Bakker, H.; Van Knippenberg, P.H. Studies on the function of two adjacent N6, N6-dimethyladenosines near the 3’ end of 16 S ribosomal RNA of Escherichia coli. IV. The effect of the methylgroups on ribosomal subunit interaction. Nucleic Acids Res. 1980, 8, 143–152. [Google Scholar] [CrossRef]
- Grosjean, H.; Edqvist, J.; Straby, K.B.; Gieg’e, R. Enzymatic formation of modified nucleosides in tRNA: Dependence on tRNA architecture. J. Mol. Biol. 1996, 255, 67–85. [Google Scholar] [CrossRef]
- Armengaud, J.; Urbonavicius, J.; Fernandez, B.; Chaussinand, G.; Bujnicki, J.M.; Grosjean, H. N2-methylation of guanosine at position 10 in tRNA is catalyzed by a THUMP domain-containing, S-Adenosylmethionine-dependent methyltransferase, conserved in Archaea and Eukaryota. J. Biol. Chem. 2004, 279, 37142–37152. [Google Scholar] [CrossRef]
- Pallan, P.S.; Kreutz, C.; Bosio, S.; Micura, R.; Egli, M. Effects of N(2),N(2)-dimethylguanosine on RNA structure and stability: Crystal structure of an RNA duplex with tandem m(2) (2)G:A pairs. RNA 2008, 14, 2452–2453. [Google Scholar] [CrossRef]
- Golovina, A.Y.; Dzama, M.M.; Osterman, I.A.; Sergiev, P.V.; Serebryakova, M.V.; Bogdanov, A.A.; Dontsova, O.A. The last rRNA methyltransferase of E. coli revealed: The yhiR gene encodes adenine-N6 methyltransferase specific for modification of A2030 of 23S ribosomal RNA. RNA 2012, 18, 1725–1734. [Google Scholar] [CrossRef] [PubMed]
- Grosjean, H.; Constantinesco, F.; Foiret, D.; Benachenhou, N. A novel enzymatic pathway leading to 1-methylinosine modification in Haloferax volcanii tRNA. Nucleic Acids Res. 1995, 23, 4312–4319. [Google Scholar] [CrossRef] [PubMed]
- Jora, M.; Lobue, P.A.; Ross, R.L.; Williams, B.; Addepalli, B. Detection of ribonucleoside modifications by liquid chromatography coupled with mass spectrometry. Biochim. Biophys. Acta-Gene Regul. Mech. 2019, 1862, 280–290. [Google Scholar] [CrossRef]
- Singh, R.; Farmer, P.B. Liquid chromatography-electrospray ionization-mass spectrometry: The future of DNA adduct detection. Carcinogenesis 2005, 27, 178–196. [Google Scholar] [CrossRef]
- Arnold, H.H.; Keith, G. The nucleotide sequence of phenylalanine tRNA from Bacillus subtilis. Nucleic Acids Res. 1977, 4, 2821–2829. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Matsumoto, T.; Ohta, T.; Kumagai, I.; Oshima, T.; Murao, K.; Hasegawa, T.; Ishikura, H.; Watanabe, K. A thermostable Gm-methylase recognizes the tertiary structure of tRNA 1. J. Biochem. 1987, 101, 1191–1198. [Google Scholar] [CrossRef]
- Vold, B.S.; Keith, D.E., Jr.; Buck, M.; McCloskey, J.A.; Pang, H. Lysine tRNAs from Bacillus subtilis 168: Structural analysis. Nucleic Acids Res. 1982, 10, 3125–3132. [Google Scholar] [CrossRef]
- Kang, B.-I.; Miyauchi, K.; Matuszewski, M.; D’Almeida, G.S.; Rubio, M.A.T.; Alfonzo, J.D.; Inoue, K.; Sakaguchi, Y.; Suzuki, T.; Sochacka, E.; et al. Identification of 2-methylthio cyclic N6-threonylcarbamoyladenosine (ms2ct6A) as a novel RNA modification at position 37 of tRNAs. Nucleic Acids Res. 2017, 45, 2124–2136. [Google Scholar] [CrossRef]
- Sankaran, B.; Bonnett, S.A.; Shah, K.; Gabriel, S.; Reddy, R.; Schimmel, P.; Rodionov, D.A.; de Crécy-Lagard, V.; Helmann, J.D.; Iwata-Reuyl, D.; et al. Zinc-independent folate biosynthesis: Genetic, biochemical, and structural investigations reveal new metal dependence for GTP cyclohydrolase IB. J. Bacteriol. 2009, 191, 6936–6949. [Google Scholar] [CrossRef]
- Dubois, D.Y.; Blaise, M.; Becker, H.D.; Campanacci, V.; Keith, G.; Giegé, R.; Cambillau, C.; Lapointe, J.; Kern, D. An aminoacyl-tRNA synthetase-like protein encoded by the Escherichia coli yadB gene glutamylates specifically tRNAAsp. Proc. Natl. Acad. Sci. USA 2004, 101, 7530–7535. [Google Scholar] [CrossRef]
- El Yacoubi, B.; Lyons, B.; Cruz, Y.; Reddy, R.; Nordin, B.; Agnelli, F.; Williamson, J.R.; Schimmel, P.; Swairjo, M.A.; de Crécy-Lagard, V. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res. 2009, 37, 2894–2909. [Google Scholar] [CrossRef]
- Deutsch, C.; El Yacoubi, B.; De Crécy-Lagard, V.; Iwata-Reuyl, D. Biosynthesis of threonylcarbamoyl adenosine (t6A), a universal tRNA nucleoside. J. Biol. Chem. 2012, 287, 13666–13673. [Google Scholar] [CrossRef]
- Lauhon, C.T. Mechanism of N6-threonylcarbamoyladenonsine (t6A) biosynthesis: Isolation and characterization of the Intermediate threonylcarbamoyl-AMP. Biochemistry 2012, 51, 8950–8963. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Miyauchi, K.; Ikeuchi, Y.; Thiaville, P.C.; de Crécy-Lagard, V.; Suzuki, T. Discovery of the beta-barrel-type RNA methyltransferase. Nucleic Acids Res. 2014, 42, 9350–9365. [Google Scholar] [CrossRef] [PubMed]
- Anton, B.P.; Russell, S.P.; Vertrees, J.; Kasif, S.; Raleigh, E.A.; Limbach, P.A.; Roberts, R.J. Functional characterization of the YmcB and YqeV tRNA methylthiotransferases of Bacillus subtilis. Nucleic Acids Res. 2010, 38, 6195–6205. [Google Scholar] [CrossRef] [PubMed]
- Arragain, S.; Handelman, S.K.; Forouhar, F.; Wei, F.Y.; Tomizawa, K.; Hunt, J.F.; Douki, T.; Fontecave, M.; Mulliez, E.; Atta, M. Identification of eukaryotic and prokaryotic methylthiotransferase for biosynthesis of 2-methylthio-N6-threonylcarbamoyladenosine in tRNA. J. Biol. Chem. 2010, 285, 28425–28433. [Google Scholar] [CrossRef] [PubMed]
- Ikeuchi, Y.; Shigi, N.; Kato, J.; Nishimura, A.; Suzuki, T. Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol. Cell. 2006, 21, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Black, K.A.; Dos Santos, P.C. Diverse mechanisms of sulfur decoration in bacterial tRNA and their cellular functions. Biomolecules 2017, 7, 33. [Google Scholar] [CrossRef]
- Black, K.A.; dos Santos, P.C. Abbreviated pathway for biosynthesis of 2-thiouridine in Bacillus subtilis. J. Bacteriol. 2015, 196, 1952–1962. [Google Scholar] [CrossRef]
- Roovers, M.; Kaminska, K.H.; Tkaczuk, K.L.; Gigot, D.; Droogmans, L.; Bujnicki, J.M. The YqfN protein of Bacillus subtilis is the tRNA: m1A22 methyltransferase (TrmK). Nucleic Acids Res. 2008, 36, 3252–3262. [Google Scholar] [CrossRef]
- Guy, M.P.; Podyma, B.M.; Preston, M.A.; Shaheen, H.H.; Krivos, K.L.; Limbach, P.A.; Hopper, A.K.; Phizicky, E.M. Yeast Trm7 interacts with distinct proteins for critical modifications of the tRNAPhe anticodon loop. RNA 2012, 18, 1921–1933. [Google Scholar] [CrossRef] [PubMed]
- Guy, M.P.; Phizicky, E.M. Conservation of an intricate circuit for crucial modifications of the tRNAPhe anticodon loop in eukaryotes. RNA 2015, 21, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Angelova, M.T.; Dimitrova, D.G.; Da Silva, B.; Marchand, V.; Jacquier, C.; Achour, C.; Brazane, M.; Goyenvalle, C.; Bourguignon-Igel, V.; Shehzada, S.; et al. tRNA 2’-O-methylation by a duo of TRM7/FTSJ1 proteins modulates small RNA silencing in Drosophila. Nucleic Acids Res. 2020, 48, 2050–2072. [Google Scholar] [CrossRef]
- Urbonavicius, J.; Skouloubris, S.; Myllykallio, H.; Grosjean, H. Identification of a novel gene encoding a flavin-dependent tRNA: m5U methyltransferase in bacteria--evolutionary implications. Nucleic Acids Res. 2005, 33, 3955–3964. [Google Scholar] [CrossRef]
- Ikeuchi, Y.; Kitachara, K.; Suzuki, T. The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon. EMBO J. 2008, 27, 2194–2203. [Google Scholar] [CrossRef] [PubMed]
- Rajakovich, L.J.; Tomlinson, J.; Dos Santos, P.C. Functional analysis of Bacillus subtilis genes involved in the biosynthesis of 4-thiouridine in tRNA. J. Bacteriol. 2012, 194, 4933–4940. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Xiao, H.; Bonanno, J.B.; Kalyanaraman, C.; Brown, S.; Tang, X.; Al-Obaidi, N.F.; Patskovsky, Y.; Babbitt, P.C.; Jacobson, M.P.; et al. Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function. Nature 2013, 498, 123–126. [Google Scholar] [CrossRef]
- Sakai, Y.; Miyauchi, K.; Kimura, S.; Suzuki, T. Biogenesis and growth phase-dependent alteration of 5-methoxycarbonylmethoxyuridine in tRNA anticodons. Nucleic Acids Res. 2016, 44, 509–523. [Google Scholar] [CrossRef]
- Ryu, H.; Grove, T.L.; Almo, S.C.; Kim, J. Identification of a novel tRNA wobble uridine modifying activity in the biosynthesis of 5-methoxyuridine. Nucleic Acids Res. 2018, 46, 9160–9169. [Google Scholar] [CrossRef]
- Sakai, Y.; Kimura, S.; Suzuki, T. Dual pathways of tRNA hydroxylation ensure efficient translation by expanding decoding capability. Nat. Commun. 2019, 10, 2858. [Google Scholar] [CrossRef] [PubMed]
- Lauhon, C.T. Identification and characterization of genes required for 5-hydroxyuridine synthesis in Bacillus subtilis and Escherichia coli. J. Bacteriol. 2019, 201, e00433-19. [Google Scholar] [CrossRef] [PubMed]
- Bou-Nader, C.; Montémont, H.; Guérineau, V.; Jean-Jean, O.; Brégeon, D.; Hamdane, D. Unveiling structural and functional divergences of bacterial tRNA dihydrouridine synthases: Perspectives on the evolution scenario. Nucleic Acids Res. 2018, 46, 1386–1394. [Google Scholar] [CrossRef] [PubMed]
- Ofengand, J.; Bakin, A. Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria and chloroplasts. J. Mol. Biol. 1997, 266, 246–268. [Google Scholar] [CrossRef] [PubMed]
- Del Campo, M.; Kaya, Y.; Ofengand, J. Identification and site of action of the remaining four putative pseudouridine synthases in Escherichia coli. RNA 2001, 7, 1603–1615. [Google Scholar] [PubMed]
- Niu, L.; Ofengand, J. Cloning and characterization of the 23S RNA pseudouridine 2633 synthase from Bacillus subtilis. Biochemistry 1999, 38, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Addepalli, B.; Limbach, P.A. Pseudouridine in the anticodon of Escherichia coli tRNATyr(QΨA) Is catalyzed by the dual specificity enzyme RluF. J. Biol. Chem. 2016, 291, 22327–22337. [Google Scholar] [CrossRef]
- Taniguchi, T.; Miyauchi, K.; Nakane, D.; Miyata, M.; Muto, A.; Nishimura, S.; Suzuki, T. Decoding system for the AUA codon by tRNAIle with the UAU anticodon in Mycoplasma mobile. Nucleic Acids Res. 2013, 41, 2621–2631. [Google Scholar] [CrossRef]
- Hill, P.J.; Abibi, A.; Albert, R.; Andrews, B.; Gagnon, M.M.; Gao, N.; Grebe, T.; Hajec, L.I.; Huang, J.; Livchak, S.; et al. Selective inhibitors of bacterial t-RNA-(N1G37) methyltransferase (TrmD) that demonstrate novel ordering of the lid domain. J. Med. Chem. 2013, 56, 7278–7288. [Google Scholar] [CrossRef]
- Kambampati, R.; Lauhon, C.T. MnmA and IscS are required for in vitro 2-thiouridine biosynthesis in Escherichia coli. Biochemistry 2003, 42, 1109–1117. [Google Scholar] [CrossRef]
- Meeske, A.J.; Rodrigues, C.D.A.; Brady, J.; Lim, H.C.; Bernhardt, T.G.; Rudner, D.Z. High-throughput genetic screens identify a large and diverse collection of new sporulation genes in Bacillus subtilis. PLoS Biol. 2016, 14, e1002341. [Google Scholar] [CrossRef]
- Sayer, C.V.; Barat, B.; Popham, D.L. Identification of L-Valine-initiated-germination-active genes in Bacillus subtilis using Tn-seq. PLoS ONE 2019, 14, e0218220. [Google Scholar] [CrossRef] [PubMed]
| Detected in Bulk Nucleoside Analysis and Mapped by RiboMethSeq | Previously Mapped | Mapped by MS | Mapped by NGS | Pathway Intermediate |
|---|---|---|---|---|
| tRNA modifications | ||||
| 1-methyladenosine, m1A | M% | Glu22 UUC1 Tyr22 GUA1 | Cys22 GCA Gln22 UUG Glu22 UUC Leu22 CAA Leu22 UAA Leu22 GAG Tyr22 GUA Ser22 GGA Ser22 GCT Ser22 UGA | |
| 1-methylguanosine, m1G | M | Pro37 UGG Leu37 CAG | ||
| 2-methyladenosine, m2A | Glu37 UUC | |||
| 2-methylthio-N6-isopentenyladenosine, ms2i6A | M | Leu37 UAA | ||
| 2-methylthio-N6-threonylcarbamoyladenosine, ms2t6A | M2 | Ser37 UGA | ||
| 2’-O-methylcytidine, Cm | Leu34 CAA | |||
| 2’-O-methylguanosine, Gm | M | Phe34 GAA | ||
| Modified U* with 2’-O-methyluridine, U*m | Lys34 UUU | |||
| Modified U* with 2’-O-methyluridine, U*m | Gln34 UUG | |||
| 4-thiouridine, s4U | M | Gln8 UUG | Leu8 UAA Gln8 UUG Tyr8 GUA | |
| 5-aminomethyluridine, nm5U | P [49] | |||
| 5-carboxymethylaminomethyl-2′-O-methyluridine, cmnm5Um | P& [49] | Leu34 UAA | Leu34 UAA | |
| 5-carboxymethylaminomethyl-2-thiouridine, cmnm5s2U | M/[49] | Gln34 UUG | ||
| 5-methoxyuridine, mo5U | M | Leu34 UAG | ||
| 5-methylaminomethyl-2-thiouridine, mnm5s2U | [49] | Glu34 UUC Gln34 UUG | ||
| 5-methylaminomethyluridine, mnm5U | Gln34 UUG | [49] | ||
| 5-methyluridine, m5U | M | |||
| 7-methylguanosine, m7G | M | |||
| cyclic N6-threonylcarbamoyladenosine, ct6A | M2 | Leu37 UAA Ile37 GAU Thr37 UGU | ||
| Dihydrouridine, D | M | Asp21-22 GUC Ile21-22 GAU Ile21-22 CAU | X | |
| Epoxyqueuosine, oQ | X | |||
| Inosine, I | M | Arg34 ACG | Arg34 ACG | |
| Lysidine, k2C | M | |||
| N4-acetylcytidine, ac4C | [50] | Met34 CAU | ||
| N6-isopentenyladenosine, i6A | M | |||
| N6-methyladenosine, m6A | M | Met37 CAU | ||
| N6-threonylcarbamoyladenosine, t6A | Ile37 GAU Thr37 UGU | M3 | ||
| Pseudouridine, ψ | M | many, ψ 31/32/55 | ||
| Queuosine, Q | M | His34 GUG Asp34 GUC | ||
| rRNA modifications | ||||
| 2-methylguanosine, m2G | ||||
| N4,2’-O-dimethylcytidine, m4Cm | 16S rRNA 1410 | |||
| N2,N2-dimethylguanosine, m2,2G | putative | |||
| N6,N6-dimethyladenosine, m6,6A | Yes | 16S rRNA1527/28 | ||
| 2’-O-methyladenosine, Am | putative | |||
| Chemical or biochemical artifacts | ||||
| 1-methylinosine, m1I | ||||
| 8-oxoguanosine, oxoG |
| E.c. Mod | B.s. Mod (n° in Ref [93,94]) | E.c Protein | B.s Protein | Evidence |
|---|---|---|---|---|
| Ψ746 | U793 (791) | RluA | NA | |
| Ψ955 | U1001 (999) | RluC | NA | |
| Ψ1911 | Ψ1940 (1938) | RluD | YhcT/BSU09210 | PsiSeq |
| Ψ1915 * | Ψ1944 * (1942) | RluD | YhcT/BSU09210 | PsiSeq |
| Ψ1917 | Ψ1946 (1944) | RluD | YhcT/BSU09210 | PsiSeq |
| Ψ2457 | U2486(2484) | RluE | NA | |
| U2492 | Ψ2521 (2520) | NA | YlmL/BSU15460 | Only sequence similarity |
| Ψ2504 | U2533 (2532) | RluC | NA | |
| Ψ2580 | U2609 (2608) | RluC | NA | |
| Ψ2604 | U2633 (2632) | RluF | NA | |
| Ψ2605 | Ψ2634 (2633) | RluB | YpuL/BSU23160 | Experimental [95] |
| Organism | Modifications at Specific Positions in tRNAs | Modification Genes |
|---|---|---|
| Bacillus subtilis subsp. subtilis str. 168 | 35 | 41 + 2 missing |
| Mycoplasma capricolum | 17 | 22 |
| Escherichia coli K12 MG1655 | 45 | 59 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Crécy-Lagard, V.; Ross, R.L.; Jaroch, M.; Marchand, V.; Eisenhart, C.; Brégeon, D.; Motorin, Y.; Limbach, P.A. Survey and Validation of tRNA Modifications and Their Corresponding Genes in Bacillus subtilis sp Subtilis Strain 168. Biomolecules 2020, 10, 977. https://doi.org/10.3390/biom10070977
de Crécy-Lagard V, Ross RL, Jaroch M, Marchand V, Eisenhart C, Brégeon D, Motorin Y, Limbach PA. Survey and Validation of tRNA Modifications and Their Corresponding Genes in Bacillus subtilis sp Subtilis Strain 168. Biomolecules. 2020; 10(7):977. https://doi.org/10.3390/biom10070977
Chicago/Turabian Stylede Crécy-Lagard, Valérie, Robert L. Ross, Marshall Jaroch, Virginie Marchand, Christina Eisenhart, Damien Brégeon, Yuri Motorin, and Patrick A. Limbach. 2020. "Survey and Validation of tRNA Modifications and Their Corresponding Genes in Bacillus subtilis sp Subtilis Strain 168" Biomolecules 10, no. 7: 977. https://doi.org/10.3390/biom10070977
APA Stylede Crécy-Lagard, V., Ross, R. L., Jaroch, M., Marchand, V., Eisenhart, C., Brégeon, D., Motorin, Y., & Limbach, P. A. (2020). Survey and Validation of tRNA Modifications and Their Corresponding Genes in Bacillus subtilis sp Subtilis Strain 168. Biomolecules, 10(7), 977. https://doi.org/10.3390/biom10070977






