The aim of this study was to identify previously unknown apoptotic caspase substrates with a high functional significance in the apoptotic program using different parameters of conservation of the substrate as an indicator of importance. Each selected criterion will be explained in more detail below.
We collected all experimentally derived human apoptotic caspase targets—3363 cleavage sites from 2040 proteins (
Table S1)—and used them as a query to find orthologs in vertebrates (the details of selection are described in the Methods and
Figure 1a). Similar approaches were previously used by Pearlman et al. [
21] to trace the evolution of phosphorylation sites and by Seaman et al. [
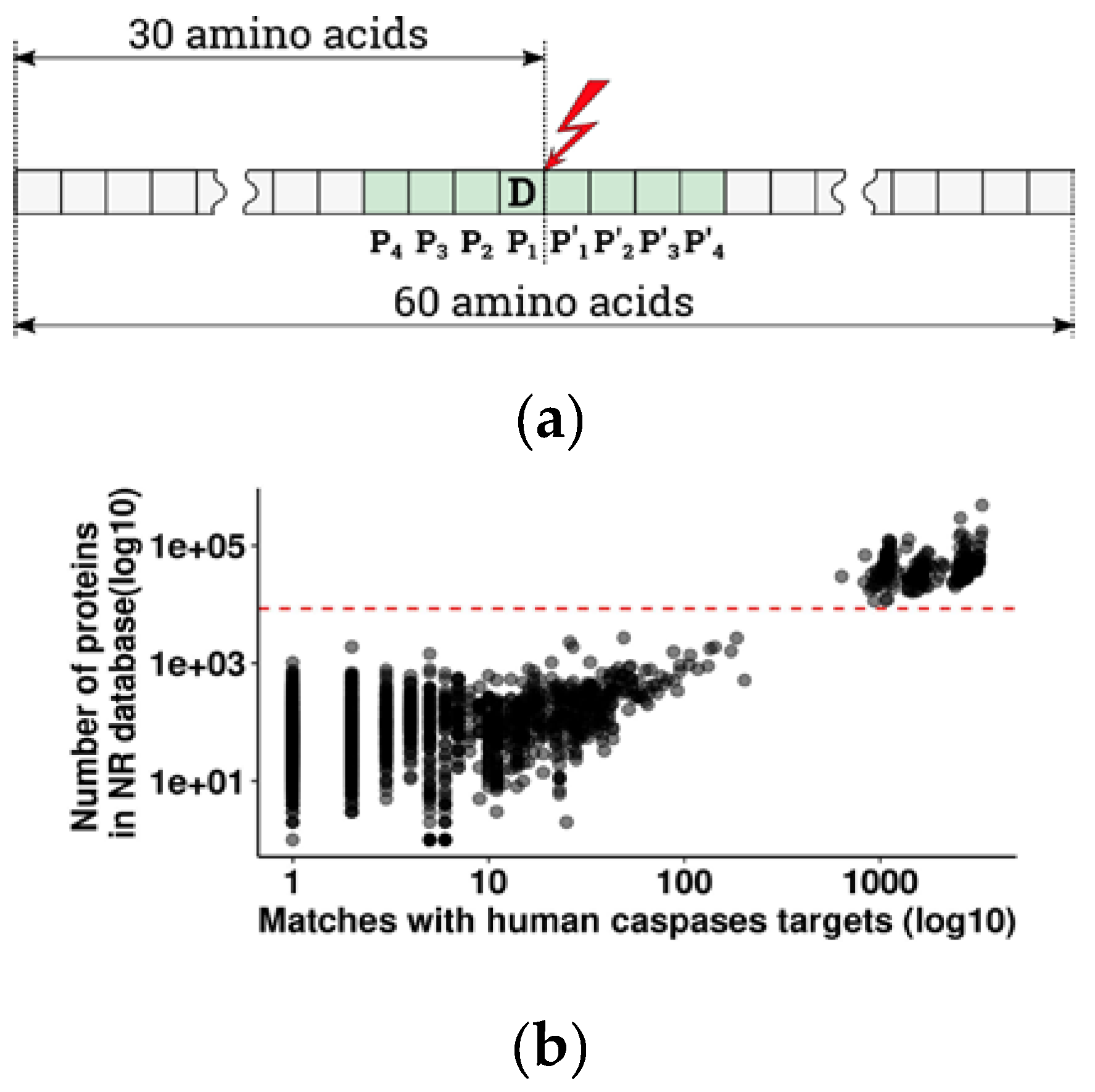
13] to estimate the significance and evolutionary conservation of cleavage sites with glutamate in the P1 position, in Metazoans. From the overall 44 885 vertebrate species present in the non-redundant protein database with at least one known protein, either sequenced or predicted from DNA and RNAseq data, 2875 species had at least one caspase cleavage target orthologous to human (pBLAST e-value < 1 × 10
−16). Since the number of orthologous targets depends on the number of known proteins for a given species (
Figure 1b), we chose to exclude species with poorly and unevenly represented proteomes in order to avoid false negative results. For further analysis, we selected 328 species with a well-represented proteome (more than 8000 annotated proteins per species) (
Figure 1b). Due to misrepresentation, four large taxa are totally absent from that group: Myxinidae (hagfishes), Petromyzontidae (lampreys), Ceratodontimorpha (lungfishes), and Cladistia (Polypterus and reedfish). The refined results are listed in
Table S2 (with a legend in
Table A2) and represent the basis for the following search of conserved caspase substrates.
3.1. Vertebrate Caspase Targets are Highly Conserved at the Level of P1 Aspartate.
Caspases cleave proteins at the scissile bond between P1-P1′ of an octamer amino acid sequence P4-P3-P2-P1-P1′-P2′-P3′-P4′, with a strict requirement for aspartate in position P1 (
Figure 1a) [
34]. Indeed, analysis of amino acid distribution in the P1 position in vertebrates has shown that in 92% of orthologous targets, the P1 position is occupied by an aspartate residue (
Table 1,
Table S2). In most cases, substitution of the D in position P1 prevents or slows down the cleavage [
13,
35,
36], and such proteins can be considered as non-conserved within the apoptotic pathway. Therefore, we decided to take the presence of aspartate in the P1 position as a prerequisite for cleavage conservation and as a threshold for further selection of the caspase targets most important for apoptosis.
Interestingly, around 5% of vertebrate cleavage site orthologs have glutamate in the P1 position instead of aspartate (
Table 1). Previous in vitro reports have shown that caspases-3, -6, and -7 are able to accommodate P1 glutamate in the active site and cleave substrates with P1 E cut sites with the same affinity but with a twofold slower rate than substrates with aspartate in the same position [
13,
37]. The same study by the Wells group demonstrated that substitution of P1 glutamate by aspartate actually results in a higher rate of protein cleavage by caspases. The shared ability of caspases-3, -6, and -7 to cleave substrates after glutamate could be explained by their very similar structure, which originates from a single ancestral effector caspase gene [
38]. Both P1 aspartate and glutamate appear to be conserved throughout Metazoans in general [
13]; however, a taxon-oriented study showed that mammals have a lower incidence of P1 E-cut sites (3%) compared to birds (7%) and ray-finned fish (9%) (
Table S3, Figure S1). Further, although P1 E cut sites exist in all taxa, the physiological presence and role of these substrates in vivo are unclear. We suggest that a transition from glutamate to aspartate in the P1 position during the evolution of Vertebrata is most likely an adaptive feature intended to make apoptosis more aggressive, which would ensure a faster elimination of damaged cells.
3.2. Low Level of Conservation of Apoptotic Caspase Cleavage Sites in Vertebrates
The numerical estimates of cleavage site similarity were calculated using the Hamming distance metric (HD) [
24]. This metric is position-dependent and determines the difference between elements of two sequences for each position as True and False, or 0 and 1. For eight-amino acid sequences, the HD ranges from 0 to 8, where 0 corresponds to an absolute identity of sequences and 8 to no amino acid matches.
Unlike for the key aspartate, the whole cleavage site sequences are not well preserved in vertebrates: although 92% of all found cleavage sites in vertebrates have aspartate in the P1 position, only 57% of orthologs have an ideal caspase cleavage site where all eight amino acids match their human counterparts, and 18% differ in one amino acid (
Table 2). This finding correlates with earlier observations on smaller datasets showing that conservation of the entire cleavage site is weaker than conservation measured by the retention of aspartate in position P1 or by the primary structure [
12,
13,
39].
The non-conserved nature of cleavage site sequences in caspase targets allowed us to recapitulate the phylogenetic relationship among vertebrates at the level of classes using the orthologous cleavage sites with almost the same accuracy as using the 60-amino acid orthologous sequences found by pBLAST (Baker’s Gamma correlation coefficient [
33] was 0.913,
Figure S2). However, this method of evolutionary analysis did not have enough resolution to track the phylogenetic tree back to orders, families, and genera (
Figures S3 and S4). This may be explained by the observation that precise determination of phylogenetic relationship usually requires using much longer sequences, such as whole genes or whole genomes [
40]. Nevertheless, the variability observed in the eight-amino acid sequences was enough to separate the species into classes.
In summary, we calculated the median HD for each caspase target present in
Table S1 among all orthologous D-cuts (
Table S4) and used the resultant numerical range as a conservation parameter for further selection of caspase targets that are most important for apoptosis.
3.3. Human Caspase Cleavage Sites are Located Preferentially in Coiled Regions
Structure analysis of cleavage sites in both human [
39] and
Escherichia coli proteins [
41] revealed that caspases cut proteins predominantly in disordered regions or coils, to a lower extent in α-helices and rarely in β-sheets. This happens because substrates can be cleaved only when in an extended conformation [
42], and the loop regions are easier to unfold locally, compared to α-helices and β-sheets which often require global unfolding of the protein [
43]. We calculated secondary structures for all human 60-amino acid sequences from
Table S2 and characterized them using Q3 accuracy symbols: α-helix (H), β-sheet (E), and coil (C) [
28]. The results are detailed in
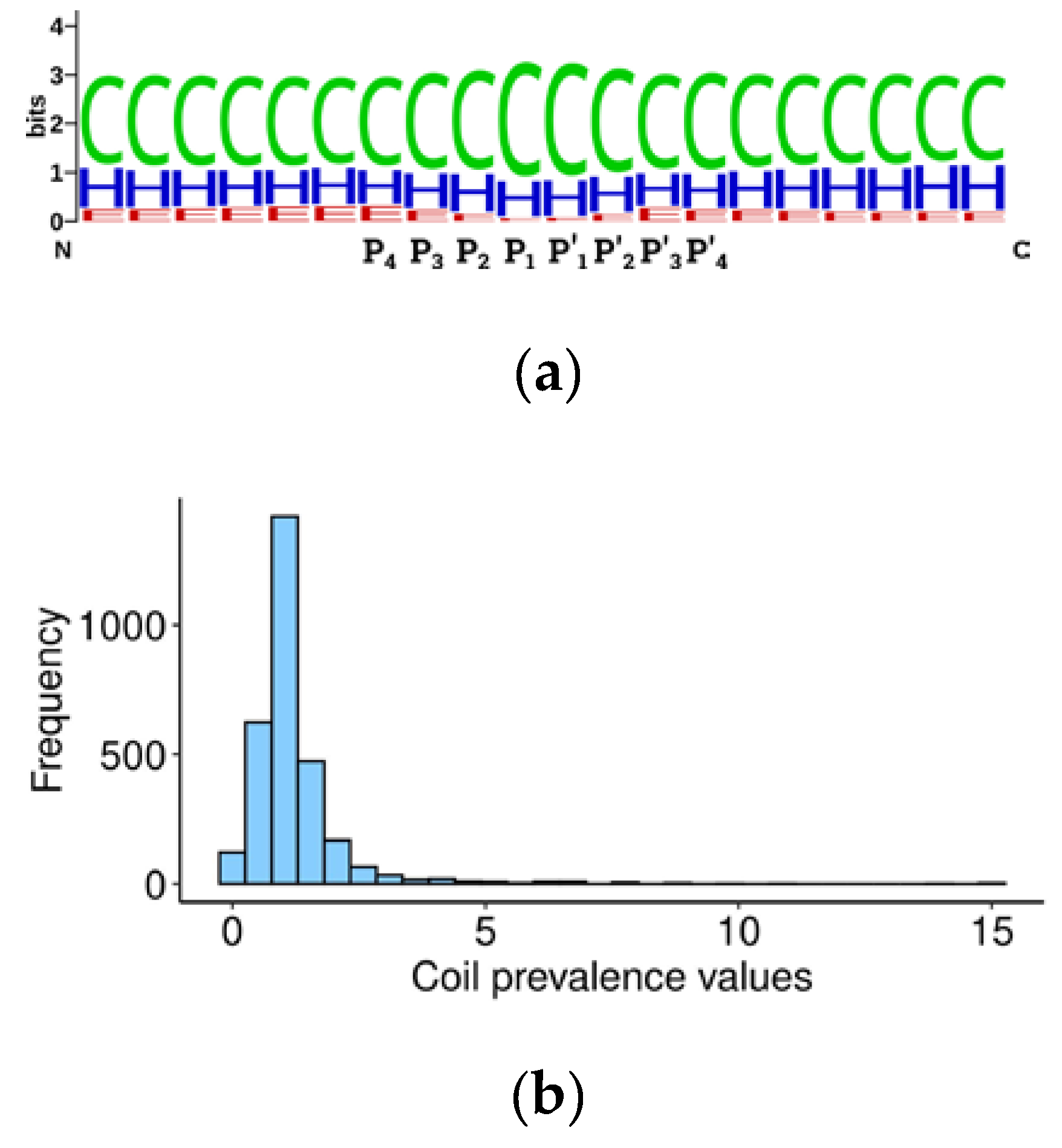
Table S5. The Weblogo alignment [
29] of secondary structure elements around cleavage sites (20 amino acids) showed that 60% of elements are represented by coils, 30% by α-helices, and around 10% by β-sheets (
Figure 2), in accordance with earlier observations [
39,
41,
44] (
Table S6).
We further developed the idea of caspase structural preferences and hypothesized that the substrate cleavage site should be more accessible for proteolysis if it is located not only in an unstructured region, but within the loop between two structured regions. Accordingly, we calculated the prevalence of coils in the central 20 amino acid sequences surrounding the P1 aspartate over marginal 20 amino acid sequences (
Table S4) using the following formula:
where ‘c’ is the percentage of coils in the central 20 amino acids, ‘l’ in the left, and ‘r’ in the right 20 amino acids. The obtained value should be proportional to the probability of the cleavage site location being in an unstructured loop between domains and, hypothetically, to the efficiency of cleavage. Consequently, this value should contribute to the overall significance of the substrate in programmed cell death. Curiously, the prevalence value for the two thirds of the human caspase targets is around 1, suggesting that there is mostly no difference between the percentage of coils in regions immediately surrounding the cleavage site and in more distant sequences (
Figure 2b). A coil prevalence value higher than 1 will be tested later as a criterion to select caspase targets which are most relevant for apoptosis.
3.4. Most Vertebrate Caspase Cleavage Sites are Located within Hydrophilic Surroundings
Proteins in aqueous conditions, such as the cytosol, tend to have hydrophobic residues hidden within their structure and hydrophilic amino acids exposed to the surface [
45]. This feature suggests that proteolysis will most likely happen within the hydrophilic portions of proteins, because these exposed parts would be more accessible for cleavage. Thereafter, hydrophilicity of cleavage sites facilitating caspase digestion of the protein would make the respective substrates more important for apoptosis.
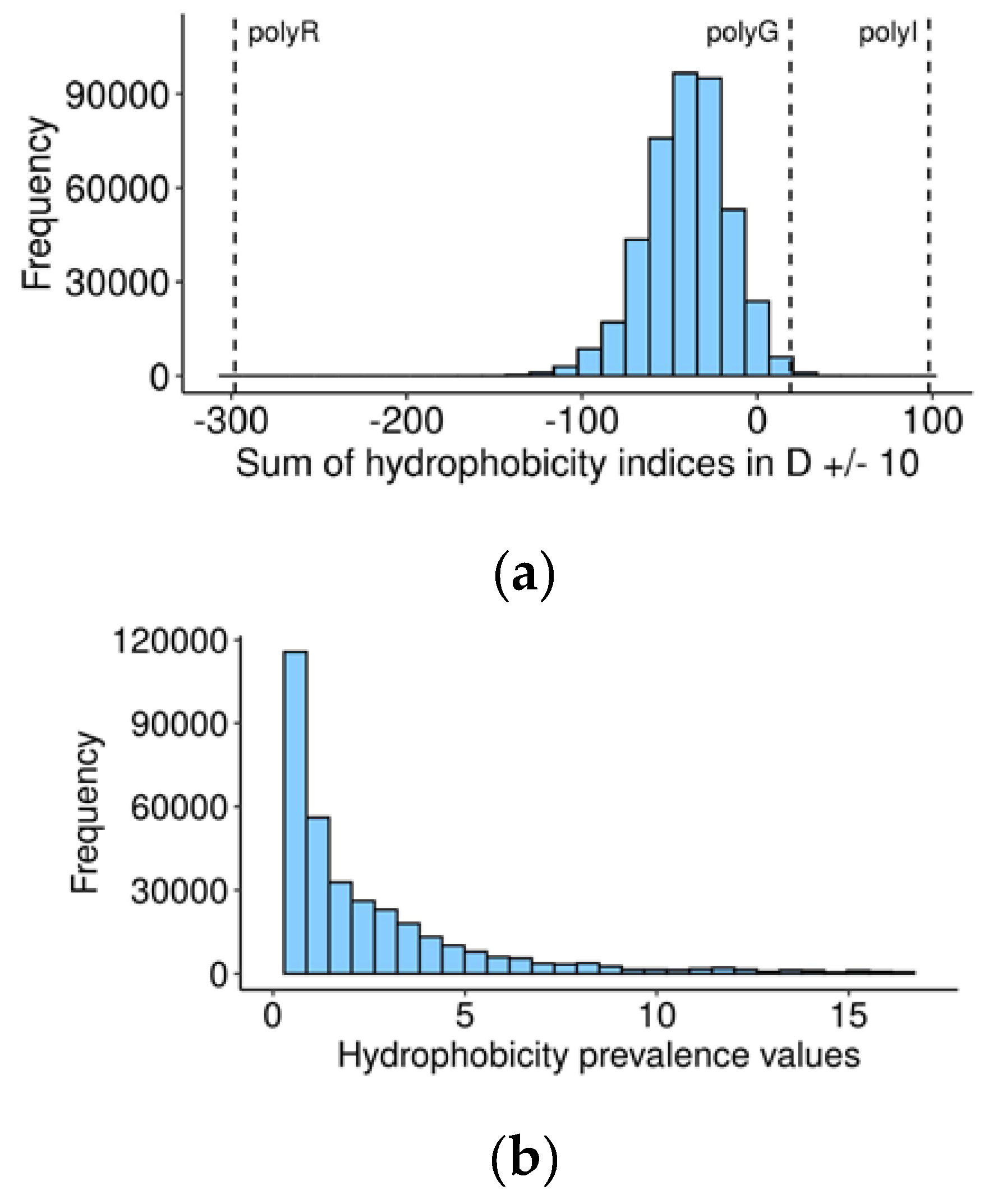
Exploring this possibility, we calculated the sum of hydrophobicity indices (kilo-calories per mole for each of the 20 amino acids at a pH of 7) [
30] for the central 20-amino acid sequences with P1 aspartate in the middle, for every human and orthologous 60-amino acid sequences (
Table S2). On the suggested scale, a negative value represents hydrophilicity, while positive suggests hydrophobicity. Most of the vertebrate caspase cleavage sites are located in a hydrophilic environment, as expected (
Figure 3a), and are likely exposed at the surface of the protein. Nevertheless, non-hydrophilic cleavage sites may relocate to the surface and become available to caspases under certain conditions, leading to a change in conformation, for example, oxidative stress and the unfolded protein response, which often precedes cell death [
46,
47].
Additionally, we found a negative correlation between the hydrophobicity of human cleavage sites and cleavage rates of the corresponding caspase targets in human cells [
48], so that hydrophilic sites tend to be cut faster than hydrophobic sites (
Figure S5, Kendall rank correlation coefficient = −0.233,
p-value = 1.082173 × 10
−5). As it was previously demonstrated that a shorter half-life is associated with a more potent regulatory outcome [
49], the higher hydrophilicity of the cleavage site could contribute to the overall importance of the substrate for apoptosis by facilitating cleavage.
As in the case of secondary structures, we hypothesized that localization of the cleavage site in a hydrophilic loop between less hydrophilic segments would increase the chances of the substrate to be cut, and calculated for each vertebrate caspase target the hydrophobicity prevalence in the central 20 amino acids over averaged marginal fragments with the same formula used to calculate the prevalence of coils (1), where ‘c’ is the sum of hydrophobicity indices in the central 20 amino acids, ‘l’ in the left, and ‘r’ in the right 20 amino acids. Prior to calculating this value, we shifted the distribution of (l + r)/2 and c values to make them all positive. The magnitude of the shift was calculated using the following formula:
where we selected the minimum of (l + r)/2 and c values calculated for all vertebrate targets and added 1 to this minimum, to avoid possibly dividing the final formula by zero. The resultant shift value was added to each (l + r)/2 and c. Hydrophobicity prevalence values for all vertebrate orthologs of human caspase cleavage sites are presented in
Table S2. In the resultant range, a value less than 1 indicates that the cleavage site area is more hydrophilic than flanking regions. Remarkably, the distribution of these values (
Figure 3b) has a peak around 1, indicating that in most human caspase targets, the cleavage site does not differ in hydrophilicity from flanking regions.
Further, we calculated the median hydrophobicity prevalence for each caspase target (
Table S4). A median hydrophobicity prevalence < 1 will be used in further analysis as a criterion to separate the most important apoptotic caspase targets from the by-standers.
3.5. Twenty Percent of Vertebrate Apoptotic Caspase Targets Have an Exposed N-degron after Cleavage
Apoptosis is a tightly regulated process and is highly influenced by the creation and degradation of apoptotic mediators generated by activated caspases. One of the degradation mechanisms for the caspase substrates is proteolysis mediated by the N-degron pathway [
14]. The N-degron pathway selectively degrades protein fragments based on the recognition of specific destabilizing amino acid residues at their N-termini [
10]. It was shown that several established proapoptotic factors activated by caspase cleavage have destabilizing amino acids in their nascent N-ends, and their ability to propagate cell death is counteracted by the N-degron pathway [
14]. Therefore, evolutionary conservation of destabilizing amino acid residues at the N-end of caspase cleavage products, corresponding to the P1′ position in the caspase cleavage site, may indicate a potential regulatory function for the protein fragment and can be used to separate caspase targets most important for cell death.
Destabilizing residues formed after protein cleavage are recognized by the Arg/N-degron pathway directly (R, K, H, L, F, Y, W, I) or after being arginylated by the ATE1 arginyl-transferase (D, E, C directly, N, Q after tertiary modifications) [
10]. Small residues such as P and G, or residues M, V, S, A, and T, that become destabilizing only after posttranslational Nt-acetylation [
50], are not recognized by the Arg/N-degron pathway and are considered stabilizing residues [
10]. We closely examined the identity of the P1′ position in all studied cleavage sites and found that both in humans and in 328 non-human vertebrates, the distribution of amino acids in the P1′ position of substrates is the same: ~30% Gly, 24% Ser, 14% Ala, with 32% distributed among the other amino acids (
Table 3). Similar results were obtained in a peptide library study [
51] and in apoptotic Jurkat cells [
39], where G, S, and A were the most common residues observed after cleavage by caspases-1, -3, -6, -7, and -8. These amino acids are stabilizing, and only 20 percent of caspase targets in 328 vertebrate species are destabilized after cleavage (
Table 3). However, nearly 100% of these 20 percent have conservation of the destabilizing nature of the P1′ residue among 328 non-human vertebrates (
Table S4, Figure S6a).
Curiously, among three classes of vertebrates, mammals have a lower proportion of destabilizing P1′ residues compared to birds and lower vertebrates (
Table S7, Figure S6b). Similar trends were observed for the percentage of P1 glutamate cut sites (
Table S3, Figure S1). Both of these evolutionary trends tend to make the apoptotic process more aggressive in higher vertebrates, by switching from glutamate to aspartate in the P1 site, which increases the rate of caspase substrate cleavage, and by shifting from a P1′ destabilizing to stabilizing amino acid, preventing the fragment from being degraded by the N-degron pathway, making it persist more and function longer. These trends can be explained by the increased complexity and redundancy of mechanisms of apoptosis regulation in higher vertebrates, reducing the need to rely on a less specific regulators of apoptosis such as the N-degron pathway [
52,
53,
54]
In further analysis, maintenance of the destabilizing nature of P1′ in more than 50% of vertebrate orthologs will be used as a criterion of target importance for apoptosis.
3.6. New Potential Apoptotic Regulators and Candidates for Cancer Therapy
The ultimate goal of this work was to identify the subset of caspase targets that are the most important in the apoptotic program, based on conservation throughout evolution. In order to reach this goal, we first selected highly similar orthologous sequences, which have aspartate in the P1 position, and then elaborated four separating parameters: cleavage site similarity, structural disordinance, hydrophobicity of the cleavage site area, and conservation of the destabilizing nature of the P1′ position.
We examined the predictive power of these parameters when applied to a reference set of caspase targets of which the newly generated C-terminal fragments possess known proapoptotic activity: RIPK1, TRAF1 [
14], CASP-2, -3, -6, -7, -8, -9, PARP1, PARP2, and ICAD [
55] (
Table 4). Only four of the 24 reference targets had a conserved destabilizing effect of the P’ amino acid (
Table 3) and because of the limited compliancy of the reference set substrates to this particular criteria, we opted not to impose it on future analysis. On the other hand, most of the reference caspase targets have well conserved cleavage sites in hydrophilic loops and in unstructured regions, which makes these three criteria predictive enough to separate the most important caspase substrates.
To refine the results, we added two more criteria. First, as the Vertebrata subphylum encompasses seven classes, the presence of a caspase target in every class was considered an additional measure of conservation. Second, since there are 328 species in the filtered pBLAST output, we included having more than 96% orthologs for a caspase target as another indicator of conservation. Applying all five thresholds to vertebrate orthologs of human caspase targets (
Table S2), we generated a final list of 107 caspase targets in 99 proteins that are most conserved among vertebrates and therefore should be the most important in the apoptotic program (
Table S8).
The main limitation of this approach resides precisely in the last two criteria: strict adherence to conservation and presence of the substrate in all seven classes implies that we are filtering out substrates that have evolved later over time. For instance, any substrate appearing after Actinopterygii would be eliminated. However, further pair-wise analysis of caspase substrates could shed light on class-specific apoptotic pathways.
Approximately 51% of the caspase targets from the list (55/107) are known to be involved in apoptosis (
Table S8). Some proteins, such as Caspase-7 (CASP7), Rho associated coiled-coil containing protein kinase 2 (ROCK2), and Protein Kinase N1 (PKN1), become activated upon caspase cleavage and proceed to propagate apoptosis [
56,
57,
58]. Others, such as aryl hydrocarbon receptor nuclear translocator (ARNT) and ubiquitin specific peptidase 19 (USP19) are anti-apoptotic regulators, which are deactivated by caspase cleavage [
59,
60]. These results validated our method and confirmed our hypothesis that important proteins involved in apoptosis would be conserved throughout evolution and that conversely, evolutionary conserved caspase targets would be important in apoptosis. Further pathway analysis of the 107 conserved caspase targets using the DAVID free online software [
31] with default parameters and an EASE threshold of 0.05%, followed by annotation using two databases, Gene Ontology [
32] and Uniprot [
22], allowed us to highlight the more prevalent pathways where these conserved caspase targets are involved. Significantly enriched terms include nucleus-related processes such as alternative splicing and RNA processing, phosphorylation, methylation, and acetylation, ATP binding, and protein interaction (
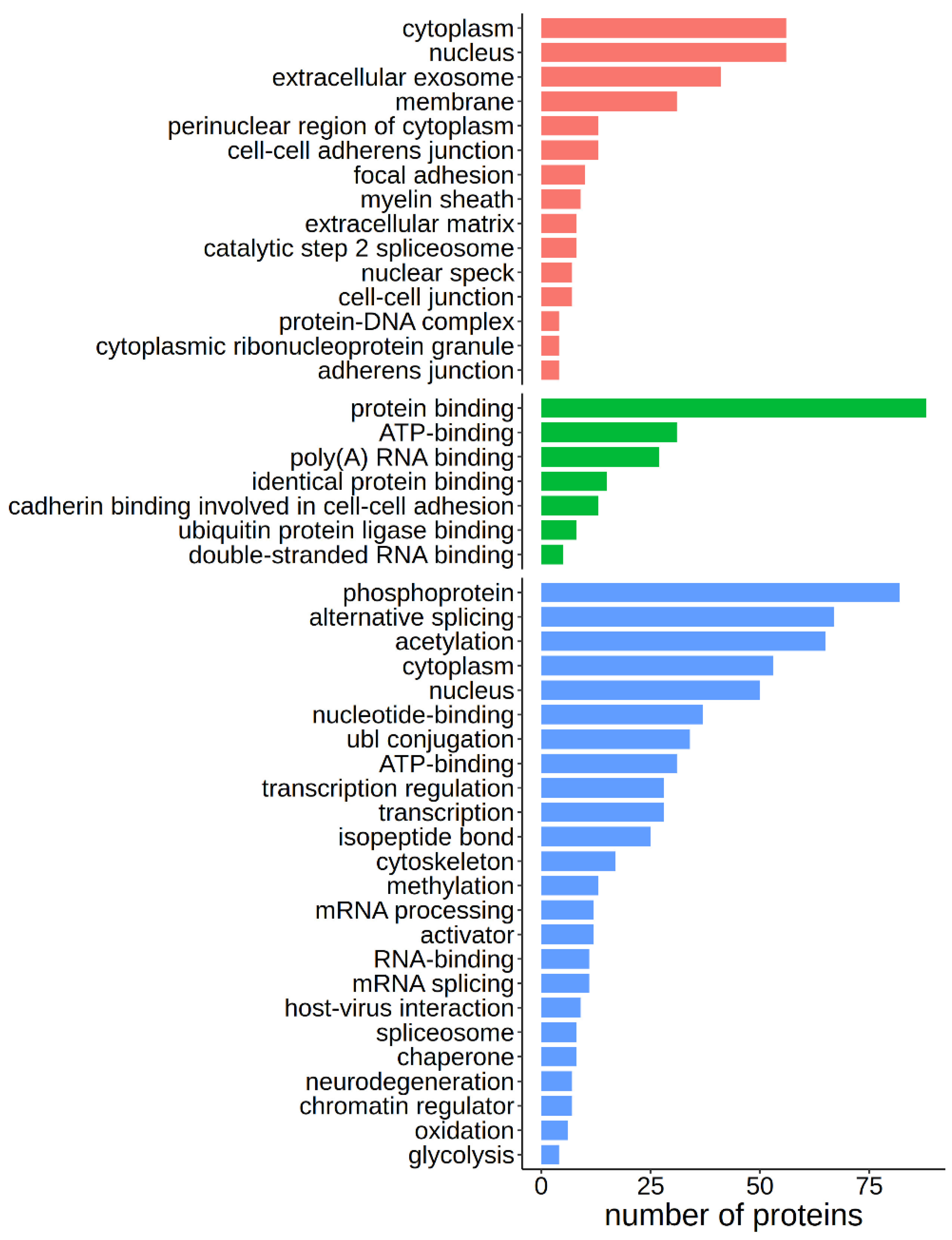
Figure 4). The 107 caspase targets can be equally found in the cytoplasm or the nucleus, and enriched cellular locations include membranes and extracellular exosomes (
Figure 4).
From the remaining targets (52/107), 22 were found to be involved in cancer, and an extensive literature search exposed 30 conserved caspase targets that have not been previously associated with apoptosis or cancer (
Table 5). From those, many targets—ATXN2L, ESS2, QRICH1, RFC1, SF3A1, and more—are related to regulation of RNA transcription, splicing, and processing, RRFM2 and SRP54 are related to protein biogenesis, ENO1 and 3 are implicated in gluconeogenesis, NAA15 participates in angiogenesis, and MAP15 is involved in microtubule organisation (
Table S8). Another interesting caspase target is CCT5. While the exact role of this protein in apoptosis has not been elucidated, recent studies indicated that this protein is significantly upregulated in several cancers [
61,
62], can serve as a tumor-associated antigen [
63], and has been implicated in an anti-viral apoptotic response [
64], suggesting an opportunity for the development of a new proapoptotic therapy.