Tetrameric Structures of Inorganic CBS-Pyrophosphatases from Various Bacterial Species Revealed by Small-Angle X-ray Scattering in Solution
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Scattering Experiments and Data Analysis
3. Results
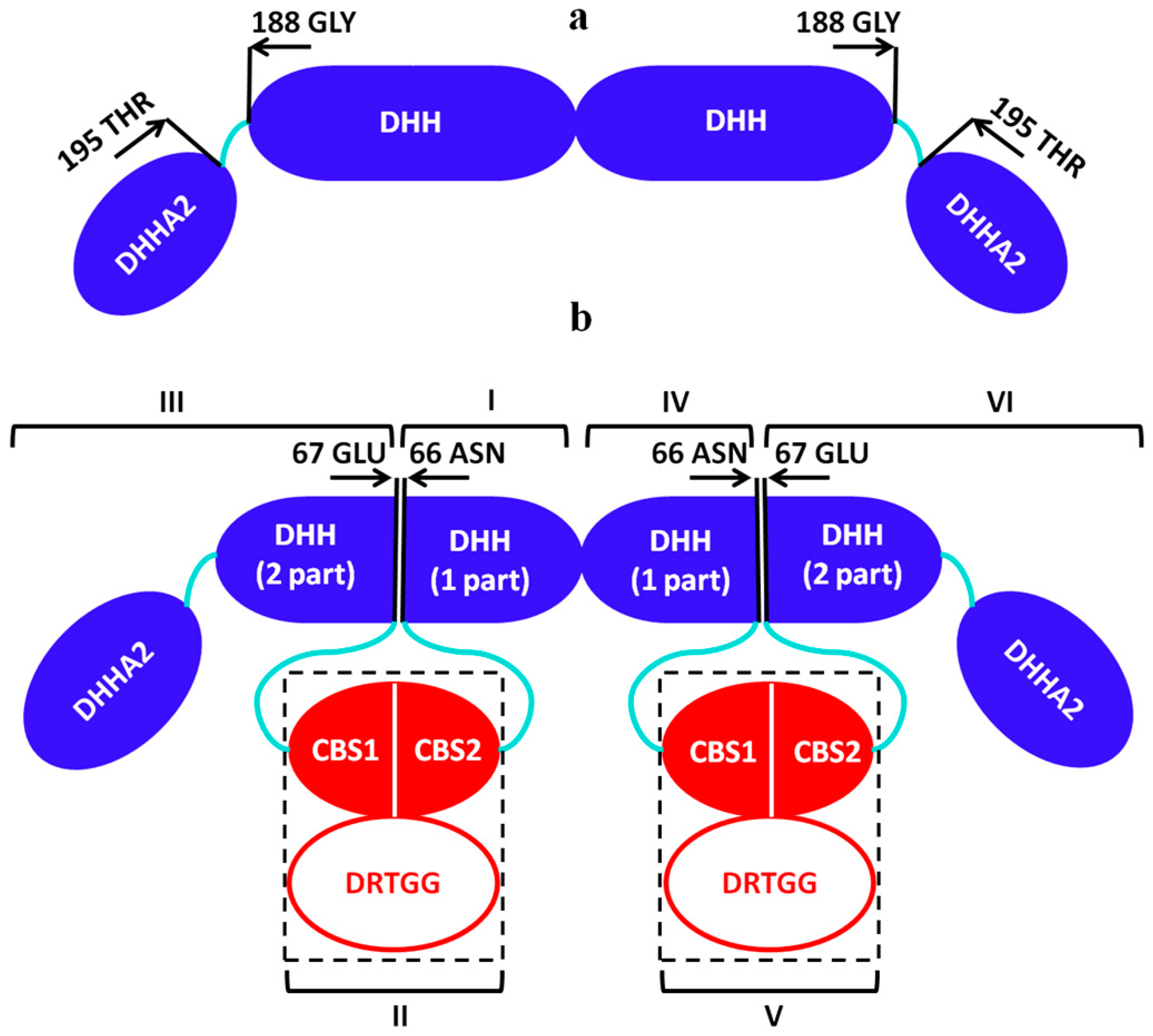
3.1. Structural Study of the Catalytical Domain of dh-PPase (dh-PPaseΔCDC) in Solution
3.2. Structural Study of Full-Length CBS-PPases in Solution
3.3. Ab Initio Modeling
3.4. Hybrid Modeling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Heinonen, J. Biological Role of Inorganic Pyrophosphate; Kluwer Academic Publishers: New York, MA, USA, 2001; pp. 1–250. [Google Scholar]
- Shintani, T.; Uchiumi, T.; Yonezawa, T.; Salminen, A.; Baykov, A.; Lahti, R.; Hachimori, A. Cloning and expression of a unique inorganic pyrophosphatase from Bacillus subtilis: Evidence for a new family of enzymes. FEBS Lett. 1998, 439, 263–266. [Google Scholar] [CrossRef]
- Young, T.; Kuhn, N.; Wadeson, A.; Ward, S.; Burges, D.; Cooke, G. Bacillus subtilis ORF yybQ encodes a manganese dependent inorganic pyrophosphatase with distinctive properties: The first of a new class of soluble pyrophosphatase? Microbiology 1998, 144, 2563–2571. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Aravind, L.; Koonin, E. A novel family of predicted phosphoesterases includes Drosophila prune protein and bacterial RecJ exonuclease. Trends Cell Biol. 1998, 23, 17–19. [Google Scholar] [CrossRef]
- Merckel, M.; Fabrichniy, I.; Salminen, A.; Kalkkinen, N.; Baykov, A.; Lahti, R.; Goldman, A. Crystal structure of Streptococcus mutans pyrophosphatase: A new fold for an old mechanism. Structure 2001, 94, 289–297. [Google Scholar] [CrossRef][Green Version]
- Ahn, S.; Milner, A.; Fütterer, K.; Konopka, M.; Ilias, M.; Young, T.; White, S.A. The “open” and “closed” structures of the type-C inorganic pyrophosphatases from Bacillus subtilis and Streptococcus gordonii. J. Mol. Biol. 2001, 313, 797–811. [Google Scholar] [CrossRef]
- Fabrichniy, I.; Lehtiö, L.; Salminen, A.; Zyryanov, A.; Baykov, A.; Lahti, R.; Goldman, A. Structural studies of metal ions in family II pyrophosphatases: The requirement for a Janus ion. Biochemistry 2004, 43, 14403–14411. [Google Scholar] [CrossRef]
- Baykov, A.A.; Anashkin, V.A.; Salminen, A.; Lahti, R. Inorganic pyrophosphatases of Family II—Two decades after their discovery. FEBS Lett. 2017, 591, 3225–3234. [Google Scholar] [CrossRef]
- Baykov, A.A.; Tuominen, H.K.; Lahti, R. The CBS domain: A protein module with an emerging prominent role in regulation. ACS Chem. Biol. 2011, 6, 1156–1163. [Google Scholar] [CrossRef]
- Parfenyev, A.; Salminen, A.; Halonen, P.; Hachimori, A.; Baykov, A.; Lahti, R. Quaternary structure and metal ion requirement of family II pyrophosphatases from Bacillus subtilis; Streptococcus gordonii; and Streptococcus mutans. J. Biol. Chem. 2001, 276, 24511–24518. [Google Scholar] [CrossRef]
- Salminen, A.; Anashkin, V.A.; Lahti, M.; Tuominen, H.K.; Lahti, R.; Baykov, A.A. Cystathionine ß-synthase (CBS) domains confer multiple forms of Mg2+-dependent co-operativity to Family II pyrophosphatases. J. Biol. Chem. 2014, 289, 22865–22876. [Google Scholar] [CrossRef]
- Meier, M.; Janosik, M.; Kery, V.; Kraus, J.; Burkhard, P. Structure of human cystathionine beta-synthase: A unique pyridoxal 5′-phosphate-dependent heme protein. EMBO J. 2001, 20, 3910–3916. [Google Scholar] [CrossRef]
- Kery, V.; Poneleit, L.; Kraus, J. Trypsin cleavage of human cystathionine betasynthase into an evolutionarily conserved active core: Structural and functional consequences. Arch. Biochem. Biophys. 1998, 355, 222–232. [Google Scholar] [CrossRef]
- Rantanen, M.; Lehtiö, L.; Rajagopal, L.; Rubens, C.; Goldman, A. Structure of the Streptococcus agalactiae family II inorganic pyrophosphatase at 2.80 Å resolution. Acta Crystallogr. D Struct. Biol. 2007, 63, 738–743. [Google Scholar] [CrossRef]
- Anashkin, V.A.; Salminen, A.; Tuominen, H.K.; Lahti, R.; Baykov, A.A. Cystathionine β-synthase (CBS) domain-containing pyrophosphatase as a target for diadenosine polyphosphates in bacteria. J. Biol. Chem. 2015, 290, 27594–27603. [Google Scholar] [CrossRef]
- Svergun, D.I.; Koch, M.H.J.; Timmins, P.A.; May, R.P. Small Angle X-ray and Neutron Scattering from Solutions of Biological Macromolecules; Oxford University Press: Oxford, UK, 2013; pp. 1–358. [Google Scholar]
- Panjkovich, A.; Svergun, D.I. CHROMIXS: Automatic and interactive analysis of chromatography-coupled small angle X-ray scattering data. Bioinformatics 2017, 34, 1944–1946. [Google Scholar] [CrossRef]
- Anashkin, V.A.; Salminen, A.; Osipova, E.; Kurilova, S.A.; Deltsov, I.D.; Lahti, R.; Baykov, A.A. Residue Network Involved in the Allosteric Regulation of Cystathionine β-Synthase Domain-Containing Pyrophosphatase by Adenine Nucleotides. ACS Omega 2019, 4, 15549–15559. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Blanchet, C.E.; Spilotros, A.; Schwemmer, F.; Graewert, M.A.; Kikhney, A.; Jeffries, C.M.; Franke, D.; Mark, D.; Zengerle, R.; Cipriani, F.; et al. Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III.; DESY). J. Appl. Crystallogr. 2015, 48, 431–443. [Google Scholar] [CrossRef]
- Jeffries, C.M.; Graewert, M.A.; Svergun, D.I.; Blanchet, C.E. Limiting radiation damage for high-brilliance biological solution scattering: Practical experience at the EMBL P12 beamline PETRA III. J. Synchrotron Radiat. 2015, 22, 273–279. [Google Scholar] [CrossRef]
- Feigin, L.A.; Svergun, D.I. Structure Analysis by Small-Angle X-ray and Neutron Scattering; Taylor, G.W., Ed.; Springer: Boston, MA, USA, 1987; pp. 1–335. [Google Scholar]
- Hajizadeh, N.R.; Franke, D.; Svergun, D.I. Integrated beamline control and data acquisition for small-angle X-ray scattering at the P12 BioSAXS beamline at PETRAIII storage ring DESY. J. Synchrotron Radiat. 2018, 25, 906–914. [Google Scholar] [CrossRef]
- Franke, D.; Kikhney, A.G.; Svergun, D.I. Automated acquisition and analysis of small angle X-ray scattering data. Nucl. Instrum. Methods Phys. Res. A 2012, 689, 52–59. [Google Scholar] [CrossRef]
- Graewert, M.A.; Franke, D.; Jeffries, C.M.; Blanchet, C.E.; Ruskule, D.; Kuhle, K.; Flieger, A.; Schaefer, B.; Tartsch, B.; Meijers, R.; et al. Automated pipeline for purification, biophysical and X-ray analysis of biomacromolecular solutions. Sci. Rep. 2015, 5, 10734. [Google Scholar] [CrossRef]
- Porod, G. Small-Angle X-Ray Scattering. In Small-Angle X-Ray Scattering; Glatter, O., Kratky, O., Eds.; Academic Press: London, UK, 1982; pp. 17–51. [Google Scholar]
- Franke, D.; Petoukhov, M.V.; Konarev, P.V.; Panjkovich, A.; Tuukkanen, A.; Mertens, H.D.T.; Kikhney, A.G.; Hajizadeh, N.R.; Franklin, J.M.; Jeffries, C.M.; et al. ATSAS 2.8: A comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 2017, 50, 1212–1225. [Google Scholar] [CrossRef]
- Hajizadeh, N.R.; Franke, D.; Jeffries, C.M.; Svergun, D.I. Consensus Bayesian assessment of protein molecular mass from solution X-ray scattering data. Sci Rep. 2018, 8, 7204. [Google Scholar] [CrossRef]
- Svergun, D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992, 25, 495–503. [Google Scholar] [CrossRef]
- Svergun, D.I.; Petoukhov, M.V.; Koch, M.H.J. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 2001, 80, 2946–2953. [Google Scholar] [CrossRef]
- Franke, D.; Jeffries, C.M.; Svergun, D.I. Correlation Map, a goodness-of-fit test for one-dimensional X-ray scattering spectra. Nat. Methods 2015, 12, 419–422. [Google Scholar] [CrossRef]
- Petoukhov, M.V.; Franke, D.; Shkumatov, A.V.; Tria, G.; Kikhney, A.G.; Gajda, M.; Gorba, C.; Mertens, H.D.T.; Konarev, P.V.; Svergun, D.I. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2012, 45, 342–350. [Google Scholar] [CrossRef]
- Svergun, D.I.; Barberato, C.; Koch, M.H.J. CRYSOL—A program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995, 28, 768–773. [Google Scholar] [CrossRef]
- Kozin, M.V.; Svergun, D.I. Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 2001, 34, 33–41. [Google Scholar] [CrossRef]
- Volkov, V.V.; Svergun, D.I. Uniqueness of ab initio shape determination in small angle scattering. J. Appl. Crystallogr. 2003, 36, 860–864. [Google Scholar] [CrossRef]
- Bernado, P.; Mylonas, E.; Petoukhov, M.V.; Blackledge, M.; Svergun, D.I. Structural Characterization of Flexible Proteins Using Small-Angle X-ray Scattering. JACS 2007, 129, 5656–5664. [Google Scholar] [CrossRef]
- Konarev, P.V.; Volkov, V.V.; Sokolova, A.V.; Koch, M.H.J.; Svergun, D.I. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003, 36, 1277–1282. [Google Scholar] [CrossRef]
- Petoukhov, M.V.; Svergun, D.I. Ambiguity assessment of small-angle scattering curves from monodisperse systems. Acta Cryst. D 2015, 71, 1051–1058. [Google Scholar] [CrossRef]
- Nakabayashi, M.; Shibata, N.; Ishido-Nakai, E.; Kanagawa, M.; Iio, Y.; Komori, H.; Ueda, Y.; Nakagawa, N.; Kuramitsu, S.; Higuchi, Y. Crystal structure of a hypothetical protein, TTHA0829113 from Thermus thermophilus HB8, composed of cystathionine-β-synthase (CBS) and aspartatekinase chorismate-mutase tyrA (ACT) domains. Extremophiles 2016, 20, 275–282. [Google Scholar] [CrossRef]
- Anashkin, V.A.; Salminen, A.; Vorobjeva, N.N.; Lahti, R.; Baykov, A.A. An asparagine residue mediates intramolecular communication in nucleotide-regulated pyrophosphatase. Biochem. J. 2016, 473, 2097–2107. [Google Scholar] [CrossRef]
- Tuominen, H.; Salminen, A.; Oksanen, E.; Jämsen, J.; Heikkilä, O.; Lehtiö, L.; Magretova, N.N.; Goldman, A.; Baykov, A.A.; Lahti, R. Crystal structures of the CBS and DRTGG domains of the regulatory region of Clostridium perfringens pyrophosphatase complexed with the inhibitor, AMP, and activator, diadenosine tetraphosphate. J. Mol. Biol. 2010, 398, 400–413. [Google Scholar] [CrossRef]
- Goodsell, D.S.; Olson, A.J. Structural symmetry and protein function. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 105–153. [Google Scholar] [CrossRef]
- Ali, M.H.; Imperiali, B. Protein oligomerization: How and why. Bioorg. Med. Chem. 2005, 13, 5013–5020. [Google Scholar] [CrossRef]
- Goodsell, D.S.; Olson, A.J. Soluble proteins: Size, shape and function. Trends Biochem. Sci. 1993, 18, 65–68. [Google Scholar] [CrossRef]
- Pereira-Leal, J.B.; Levy, E.D.; Kamp, C.; Teichmann, S.A. Evolution of protein complexes by duplication of homomeric interactions. Genome Biol. 2007, 8, R51. [Google Scholar] [CrossRef]


| dh-PPase | eh-PPase | el-PPase | |
|---|---|---|---|
| Guinier analysis | |||
| Rg (nm) | 4.95 ± 0.10 | 4.75 ± 0.38 | 4.29 ± 0.29 |
| Molecular mass from Bayesian (credibility interval), kDa | 243 (195–264) | 186 (151–195) | 208 (177–264) |
| Analysis of the p(r) function | |||
| Rg (nm) | 4.94 ± 0.05 | 4.73 ± 0.05 | 4.24 ± 0.05 |
| Dmax, (nm) | 18.3 ± 1 | 13.7 ± 1 | 14.0 ± 1 |
| Vp (nm3) | 412 ± 20 | 345 ± 17 | 368 ± 18 |
| Molecular mass from Porod volume, kDa | 250 ± 10 | 209 ± 10 | 223 ± 10 |
| reciprocal-space fit to data (χ2, CorMap P) | 1.09, 0.003 | 1.11, 0.01 | 1.18, 0.037 |
| Shape classification and ambiguity | |||
| Classification/(predicted Dmax, nm) | Compact (18.4) | Compact-hollow (14.4) | Compact (14.9) |
| Ambiguity score | 1.176 | 0 | 0.30 |
| Shape topologies | 15 | 1 | 2 |
| Uniqueness | Potentially unique | Potentially unique | Potentially unique |
| Ab initio modelling | |||
| Method | GASBOR | ||
| Symmetry imposed | P2 | P2 | P2 |
| Model fits to data (χ2, CorMap P) | 1.19, 0.003 | 1.23, 0.001 | 1.17, 0.037 |
| Atomistic modelling | |||
| Method | CORAL | ||
| Symmetry imposed | P2 | P2 | P2 |
| Model Rg (nm) | 4.93 | 4.80 | 4.23 |
| Model fit to data (χ2, CorMap P) | 1.62, 0.00 | 1.13, 0.075 | 1.03, 0.084 |
| MALLS-RI-UV MM and QELS Rh1 | |||
| Calculated MM, amino acid sequence of monomer (kDa) | 60.35 | 47.68 | 49.28 |
| Average MM from MALLS/RI, kDa | 254 ± 1 | 187 ± 1 | 190 ± 1 |
| Hydrodynamic radius, Rh (nm) | 5.62 ± 0.3 | 5.41 ± 0.2 | 4.37 ± 0.3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dadinova, L.A.; Soshinskaia, E.Y.; Jeffries, C.M.; Svergun, D.I.; Shtykova, E.V. Tetrameric Structures of Inorganic CBS-Pyrophosphatases from Various Bacterial Species Revealed by Small-Angle X-ray Scattering in Solution. Biomolecules 2020, 10, 564. https://doi.org/10.3390/biom10040564
Dadinova LA, Soshinskaia EY, Jeffries CM, Svergun DI, Shtykova EV. Tetrameric Structures of Inorganic CBS-Pyrophosphatases from Various Bacterial Species Revealed by Small-Angle X-ray Scattering in Solution. Biomolecules. 2020; 10(4):564. https://doi.org/10.3390/biom10040564
Chicago/Turabian StyleDadinova, Liubov A., Ekaterina Yu. Soshinskaia, Cy M. Jeffries, Dmitri I. Svergun, and Eleonora V. Shtykova. 2020. "Tetrameric Structures of Inorganic CBS-Pyrophosphatases from Various Bacterial Species Revealed by Small-Angle X-ray Scattering in Solution" Biomolecules 10, no. 4: 564. https://doi.org/10.3390/biom10040564
APA StyleDadinova, L. A., Soshinskaia, E. Y., Jeffries, C. M., Svergun, D. I., & Shtykova, E. V. (2020). Tetrameric Structures of Inorganic CBS-Pyrophosphatases from Various Bacterial Species Revealed by Small-Angle X-ray Scattering in Solution. Biomolecules, 10(4), 564. https://doi.org/10.3390/biom10040564






