Induction of Cryptic Antifungal Pulicatin Derivatives from Pantoea Agglomerans by Microbial Co-Culture
Abstract
1. Introduction
2. Materials and Methods
2.1. General Experimental Procedures
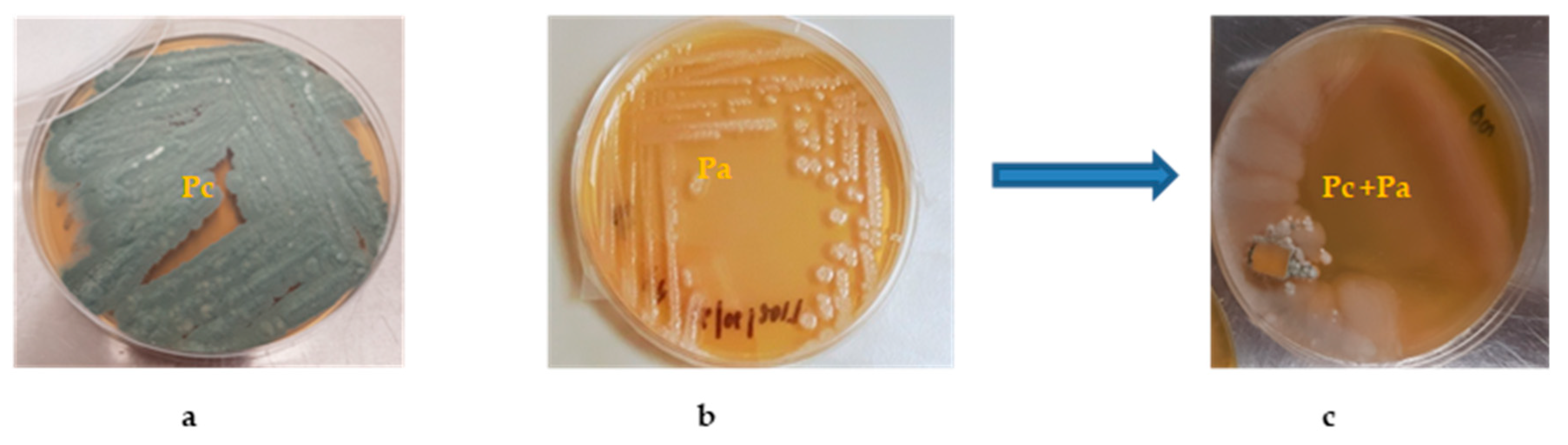
2.2. Bacterial/Fungal Isolation and Mixed Fermentation
2.3. Analysis of Raw Mass Data Using MZmine 2.3.7
2.4. Dereplication Process to Identify Known and New Hits
2.5. Isolation of Compounds
2.6. Determination of Absolute Stereochemistry of Compound 2 by Modified Mosher’s Method
2.7. Antimicrobial Screening (Antibacterial and Antifungal Activities)
2.7.1. Preparation of Samples and Microorganisms Used
2.7.2. Calculation of Minimum Inhibitory Concentration (MIC) Using Microdilution Method
3. Results
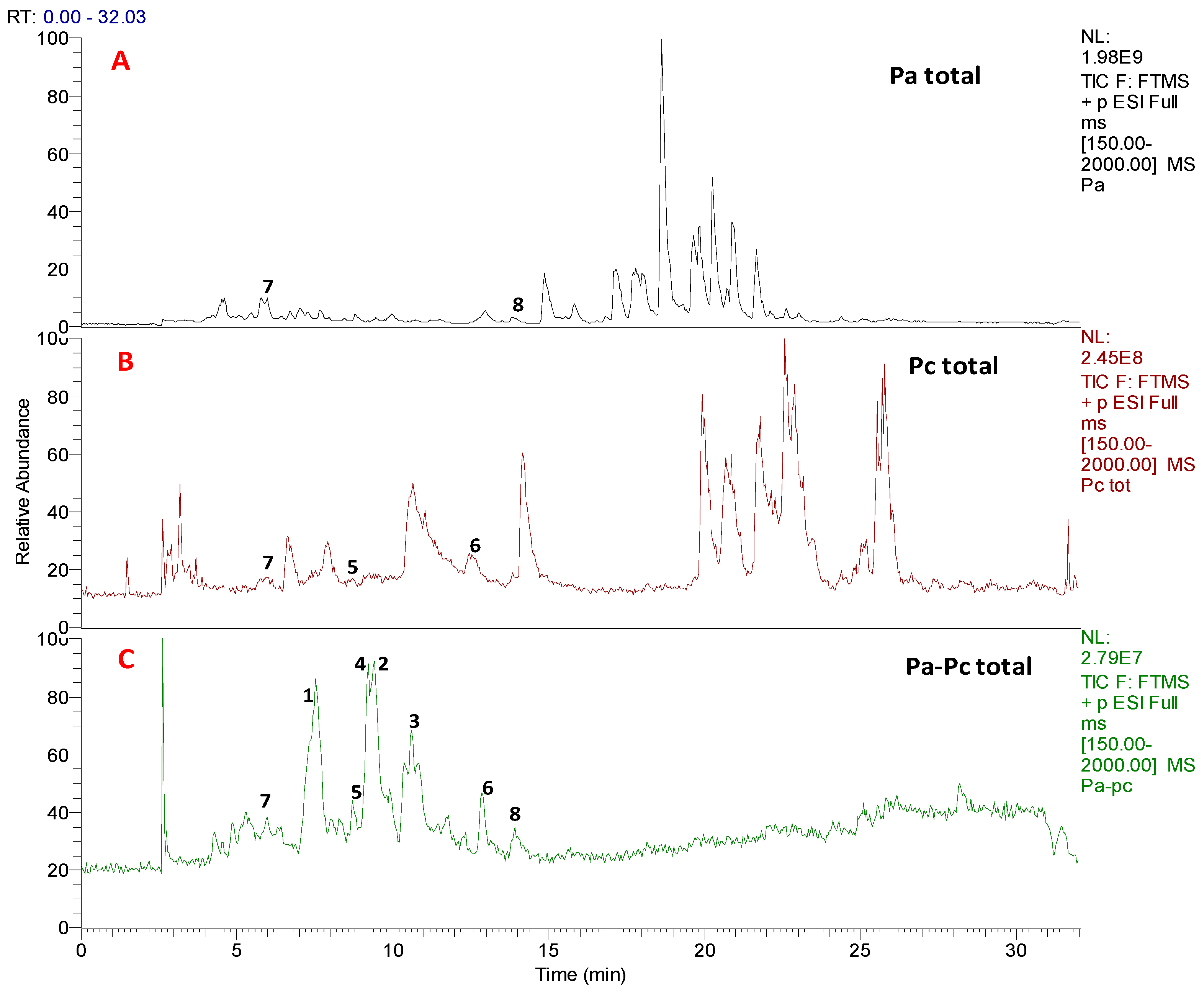
3.1. Chemical Profiling and Dereplication of Pa, Pc and Pa-Pc
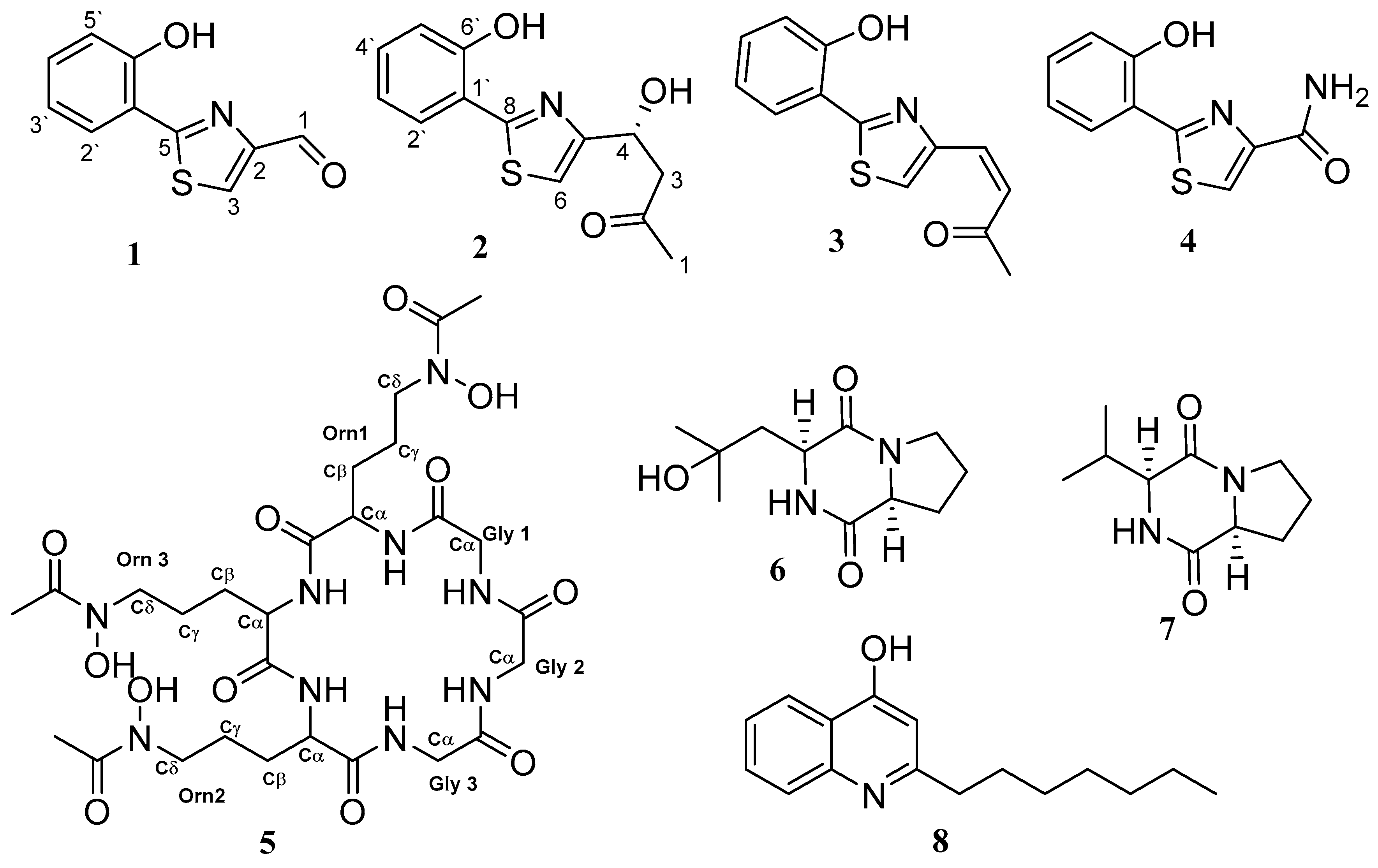
3.2. Structure Characterisation of the Isolated Compounds
3.3. Antimicrobial Screening
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Atanasov, A.G.; Yeung, A.W.K.; Banach, M. Natural products for targeted therapy in precision medicine. Biotechnol. Adv. 2018, 36, 1559–1562. [Google Scholar] [CrossRef] [PubMed]
- Carroll, A.R.; Zani, C.L. Database for Rapid Dereplication of Known Natural Products Using Data from MS and Fast NMR Experiments. J. Nat. Prod. 2017, 80, 1758–1766. [Google Scholar]
- Sayed, A.M.; Hassan, M.H.A.; Alhadrami, H.A.; Hassan, H.M.; Goodfellow, M.; Rateb, M.E. Extreme environments: Microbiology leading to specialized metabolites. J. Appl. Microbiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Meyer, V.; Nai, C. From Axenic to Mixed Cultures: Technological Advances Accelerating a Paradigm Shift in Microbiology. Trends Microbiol. 2018, 26, 538–554. [Google Scholar]
- Wakefield, J.; Hassan, H.M.; Jaspars, M.; Ebel, R.; Rateb, M.E. Dual induction of new microbial secondary metabolites by fungal bacterial co-cultivation. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Netzker, T.; Fischer, J.; Weber, J.; Mattern, D.J.; König, C.C.; Valiante, V.; Schroeckh, V.; Brakhage, A.A. Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Front. Microbiol. 2015. [Google Scholar] [CrossRef]
- Rateb, M.E.; Hallyburton, I.; Houssen, W.E.; Bull, A.T.; Goodfellow, M.; Santhanam, R.; Jaspars, M.; Ebel, R. Induction of diverse secondary metabolites in Aspergillus fumigatus by microbial co-culture. RSC Adv. 2013, 3, 14444–14450. [Google Scholar] [CrossRef]
- Abdelwahab, M.F.; Kurtán, T.; Mándi, A.; Müller, W.E.; Fouad, M.A.; Kamel, M.S.; Liu, Z.; Ebrahim, W.; Daletos, G.; Proksch, P. Induced secondary metabolites from the endophytic fungus Aspergillus versicolor through bacterial co-culture and OSMAC approaches. Tetrahedron Lett. 2018, 59, 2647–2652. [Google Scholar] [CrossRef]
- Moussa, M.; Ebrahim, W.; Bonus, M.; Gohlke, H.; Mándi, A.; Kurtán, T.; Hartmann, R.; Kalscheuer, R.; Lin, W.; Liu, Z.; et al. Co-culture of the fungus Fusarium tricinctum with Streptomyces lividans induces production of cryptic naphthoquinone dimers. RSC Adv. 2019, 9, 1491–1500. [Google Scholar] [CrossRef]
- Cherif-Silini, H.; Thissera, B.; Bouket, A.C.; Saadaoui, N.; Silini, A.; Eshelli, M.; Alenezi, F.N.; Vallat, A.; Luptakova, L.; Yahiaoui, B.; et al. Durum Wheat Stress Tolerance Induced by Endophyte Pantoea agglomerans with Genes Contributing to Plant Functions and Secondary Metabolite Arsenal. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef]
- Ben Mefteh, F.; Daoud, A.; Bouket, A.C.; Thissera, B.; Kadri, Y.; Cherif-Silini, H.; Eshelli, M.; Alenezi, F.N.; Vallat, A.; Oszako, T.; et al. Date Palm Trees Root-Derived Endophytes as Fungal Cell Factories for Diverse Bioactive Metabolites. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Macintyre, L.; Zhang, T.; Viegelmann, C.; Martinez, I.; Cheng, C.; Dowdells, C.; Abdelmohsen, U.; Gernert, C.; Hentschel, U.; Edrada-Ebel, R. Metabolomic tools for secondary metabolite discovery from marine microbial symbionts. Marine Drugs 2014, 12, 3416–3448. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, I.; Kusumi, T.; Kashman, Y.; Kakisawa, H. High-field FT NMR application of Mosher’s method. The absolute configurations of marine terpenoids. JACS 1991, 113, 4092–4096. [Google Scholar]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protocols 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Tudela, J.L.; Barchiesi, F.; Bille, J.; Chryssanthou, E.; Cuenca-Estrella, M.; Denning, D.; Donnelly, J.P.; Dupont, B.; Fegeler, W.; Moore, C.; et al. Method for the determination of minimum inhibitory concentration (MIC) by broth dilution of fermentative yeasts. Clin. Microbiol. Infect. 2003, 9, 1–8. [Google Scholar] [CrossRef]
- Anand, S.; Deighton, M.; Livanos, G.; Morrison, P.D.; Pang, E.C.; Mantri, N. Antimicrobial activity of Agastache honey and characterization of its bioactive compounds in comparison with important commercial honeys. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Ye, L.; Cornelis, P.; Guillemyn, K.; Ballet, S.; Hammerich., O. Structure revision of N-mercapto-4-formylcarbostyril produced by Pseudomonas fluorescens G308 to 2-(2-hydroxyphenyl) thiazole-4-carbaldehyde [aeruginaldehyde]. Nat. Prod. Commun. 2014, 9, 789–794. [Google Scholar] [CrossRef]
- Fakhouri, W.; Walker, F.; Vogler, B.; Armbruster, W.; Buchenauer, H. Isolation and identification of N-mercapto-4-formylcarbostyril, an antibiotic produced by Pseudomonas fluorescens. Phytochemistry 2001, 58, 1297–1303. [Google Scholar] [CrossRef]
- Lin, Z.; Antemano, R.R.; Hughen, R.W.; Tianero, M.D.B.; Peraud, O.; Haygood, M.G.; Concepcion, G.P.; Olivera, B.M.; Light, A.; Schmidt, E.W. Pulicatins A-E, Neuroactive Thiazoline Metabolites from Cone Snail-Associated Bacteria. J. Nat. Prod. 2010, 73, 1922–1926. [Google Scholar] [CrossRef]
- Schmidt, E.W.; Lin, Z.; Antemano, R.R.; Light, A.; Olivera, B.M.; Concepcion, G.P. Methods and compositions related to neuroactive thiazoline compounds. U.S. Patent US20140018400, 2017. [Google Scholar]
- Neilands, J.B. A crystalline organo-iron pigment from a rust fungus Ustilago sphaerogena. J. Amer. Chem. Soc. 1952. J. Amer. Chem. Soc. 1952, 74, 4846–4847. [Google Scholar]
- Zhen, X.; Gong, T.; Liu, F.; Zhang, P.C.; Zhou, W.Q.; Li, Y.; Zhu, P. A new analogue of echinomycin and a new cyclic dipeptide from a marine-derived Streptomyces sp. LS298. Marine Drugs 2015, 13, 6947–6961. [Google Scholar] [CrossRef]
- Chen, M.Z.; Dewis, M.L.; Kraut, K.; Merrit, D.; Reiber, L.; Trinnaman, L.; Da Costa, N.C. 2,5-Diketopiperazines (Cyclic Dipeptides) in Beef: Identification, Synthesis, and Sensory Evaluation. J. Food. Sci. 2009, 74. [Google Scholar] [CrossRef] [PubMed]
- Debitus, C.; Guella, G.; Mancini, I.; Waikedre, J.; Guemas, J.P.; Nicolas, J.L.; Pietra, F. Quinolones from a bacterium and tyrosine metabolites from its host sponge, Suberea creba from the Coral Sea. J. Mar. Biotechnol. 1998, 6, 136–141. [Google Scholar]
- Khalil, Z.G.; Cruz-Morales, P.; Licona-Cassani, C.; Marcellin, E.; Capon, R.J. Inter-Kingdom beach warfare: Microbial chemical communication activates natural chemical defences. ISME J. 2019, 13, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Rutledge, P.J.; Challis, G.L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 2015, 13, 509–523. [Google Scholar] [CrossRef]
- Pettit, R.K. Mixed fermentation for natural product drug discovery. Appl. Microbiol. Biotechnol. 2009, 83, 19–25. [Google Scholar] [CrossRef]
- Bertrand, S.; Bohni, N.; Schnee, S.; Schumpp, O.; Gindro, K.; Wolfender, J. Metabolite induction via microorganism co-culture: A potential way to enhance chemical diversity for drug discovery. Biotechnol. Adv. 2014, 32, 1180–1204. [Google Scholar] [CrossRef]
- Schroeckh, V.; Scherlach, K.; Nützmann, H.W.; Shelest, E.; Schmidt-Heck, W.; Schuemann, J.; Martin, K.; Hertweck, C.; Brakhage, A.A. Intimate bacterial–fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. PNAS 2009, 106, 14558–14563. [Google Scholar] [CrossRef]
- Abraham, E. Cephalosporins 1945–1986. Drugs 1987, 34, 1–14. [Google Scholar] [CrossRef]
- Ottenheijm, H.C.; Van Den Broek, L.A.; Ballesta, J.P.; Zylicz, Z. 6 Chemical and Biological Aspects of Sparsomycin, an Antibiotic, from Streptomyces. Progress Med. Chem. 1986, 23, 219–268. [Google Scholar]
- Lin, Z.; Smith, M.D.; Concepcion, G.P.; Haygood, M.G.; Olivera, B.M.; Light, A.; Schmidt, E.W. Modulating the Serotonin Receptor Spectrum of Pulicatin Natural Products. J. Nat. Prod. 2017, 80, 2360–2370. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, T.C.; Nolan, E.M. Beyond iron: Non-classical biological functions of bacterial siderophores. Dalton Trans. 2015, 44, 6320–6339. [Google Scholar] [CrossRef]
- Hannauer, M.; Barda, Y.; Mislin, G.L.A.; Shanzer, A.; Schalk, I.J. The Ferrichrome Uptake Pathway in Pseudomonas aeruginosa Involves an Iron Release Mechanism with Acylation of the Siderophore and Recycling of the Modified Desferrichrome. J. Bacteriol. 2010, 192, 1212–1220. [Google Scholar] [CrossRef] [PubMed]
- Adams, C.J.; Wilson, J.J.; Boros, E. Multifunctional Desferrichrome Analogues as Versatile 89Zr (IV) Chelators for ImmunoPET Probe Development. Mol. Pharm. 2017, 14, 2831–2842. [Google Scholar] [CrossRef] [PubMed]
- Borthwick, A.D. 2,5-Diketopiperazines: Synthesis, Reactions, Medicinal Chemistry, and Bioactive Natural Products. Chem. Rev. 2012, 112, 3641–3716. [Google Scholar] [CrossRef]
- Holden, M.T.G.; Chhabra, S.R.; de Nys, R.; Stead, P.; Bainton, N.J.; Hill, P.J.; Manefield, M.; Kumar, N.; Labatte, M.; England, D.; et al. Quorum-sensing cross talk: Isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other gram-negative bacteria. Mol. Microbiol. 1999, 33, 1254–1266. [Google Scholar] [CrossRef]
- Diggle, S.P.; Matthijs, S.; Wright, V.J.; Fletcher, M.P.; Chhabra, S.R.; Lamont, I.L.; Kong, X.; Hider, R.C.; Cornelis, P.; Cámara, M.; et al. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem. Biol. 2007, 14, 87–96. [Google Scholar] [CrossRef]
- Khan, S.A.; Hamayun, M.; Yoon, H.; Kim, H.; Suh, S.; Hwang, S.; Kim, J.; Lee, I.; Choo, Y.; Yoon, U.; et al. Plant growth promotion and Penicillium citrinum. BMC Microbiol. 2008, 8, 231. [Google Scholar] [CrossRef]
- Luo, H.; Qing, Z.; Deng, Y.; Deng, Z.; Tang, X.; Feng, B.; Lin, W. Two polyketides produced by endophytic Penicillium citrinum DBR-9 from medicinal plant Stephania kwangsiensis and their antifungal activity against plant pathogenic fungi. Nat. Prod. Comm. 2019. [Google Scholar] [CrossRef]
- Radhakrishnan, R.; Kang, S.; Baek, I.; Lee, I. Characterization of plant growth-promoting traits of Penicillium species against the effects of high soil salinity and root disease. J. Plant Interact. 2014, 9, 754–762. [Google Scholar] [CrossRef]
- Ting, A.S.Y.; Mah, S.W.; Tee, C.S. Evaluating the feasibility of induced host resistance by endophytic isolate Penicillium citrinum BTF08 as a control mechanism for Fusarium wilt in banana plantlets. Biol. Control 2012, 61, 155–159. [Google Scholar] [CrossRef]
- Coutinho, T.C.; Ferreira, M.C.; Rosa, L.H.; de Oliveira, A.; Júnior, E.N.O. Penicillium citrinum and Penicillium mallochii: New phytopathogens of orange fruit and their control using chitosan. Carbohydr. Polym. 2020, 234. [Google Scholar] [CrossRef]
- Dutkiewicz, J.; Mackiewicz, B.; Lemieszek, M.K.; Golec, M.; Milanowski, J. Pantoea agglomerans: A mysterious bacterium of evil and good. Part III. Deleterious effects: Infections of humans, animals and plants. Annals Agric. Environ. Med. 2016, 23, 197–205. [Google Scholar] [CrossRef] [PubMed]
 ) and HMBC (
) and HMBC ( ) correlation of compounds 1–4.
) correlation of compounds 1–4.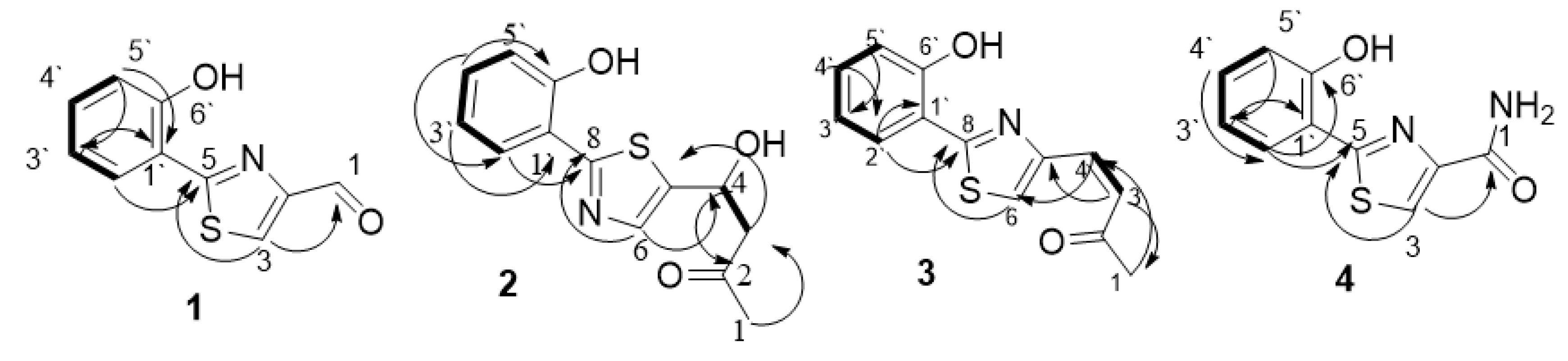
| Atom | 1 | 2 | 3 | 4 | ||||
|---|---|---|---|---|---|---|---|---|
| δC, mult. | δH, mult. (J in Hz) | δC, mult. | δH, mult. (J in Hz) | δC, mult. | δH, mult. (J in Hz) | δC, mult. | δH, mult. (J in Hz) | |
| 1 | 185.1 (CH) | 10.02, s | 30.5 (CH3) | 2.16. s | 26.7 (C) | 2.37, s | 163.3 (C) | |
| 2 | 153.5 (C) | - | 206.5 (C) | 198.7 (C) | 149.4 (C) | |||
| 3 | 131.9 (CH) | 8.72, s | 51.2 (CH2) | a. 2.84, dd (15.3, 8.2) b.2.91, dd (15.7, 4.6) | 135.3 (CH) | 7.64, d (16.9) | 124.4 (CH) | 8.19, s |
| 4 | 66.3 (CH) | 5.17, q | 127.1 (CH) | 6.93, d (17.1) | ||||
| 5 | 164.3 (C) | - | 158.6 (C) | 149.6 (C) | 163.0 (C) | |||
| 6 | 114.9 (CH) | 7.48, s | 124.2 (C) | 8.03, s | ||||
| 8 | 164.8 (C) | 164.9 (C) | ||||||
| 1′ | 118.6 (C) | - | 119.1 (C)) | - | 119.5 (C) | 116.3 (C) | - | |
| 2′ | 127.9 (CH) | 8.19, dd (6.1, 1.8) | 127.1 (CH) | 8.00, dd (7.8, 1.7) | 126.7 (CH) | 8.17, dd (8.1, 2.8) | 128.4 (CH) | 8.29, dd (8.1, 3.0) |
| 3′ | 116.9 (CH) | 7.07, dd (7.5, 1.4) | 119.8 (CH) | 6.91, t (6.9) | 118.4 (CH) | 6.80, t (7.1) | 118.5 (CH) | 6.84, t (7.9) |
| 4′ | 132.1 (CH) | 7.37, dd (7.4,1.3) | 131.2 (CH) | 7.29, t (7.6) | 131.9 (CH) | 7.28, t (7.5) | 131.8 (CH) | 7.26, t (7.1) |
| 5′ | 120.0 (CH) | 6.99, t (7.5) | 117.3 (CH) | 7.02, d (8.7) | 117.1 (CH) | 6.98, d (9.1) | 118.7 (C) | 6.89, d (8.7) |
| 6′ | 155.7 (C) | - | 155.4 (C) | 152.0 (C) | 155.1 (C) | |||
| C/H No. | Chemical Shift (1H in pyridine–d5 at 500 MHz) | |||
|---|---|---|---|---|
| 5.17 | (S)–MTPA Ester | (R)–MTPA Ester | ∆S–R | |
| 4 | 5.453 | 5.455 | 5.815 | −0.36 |
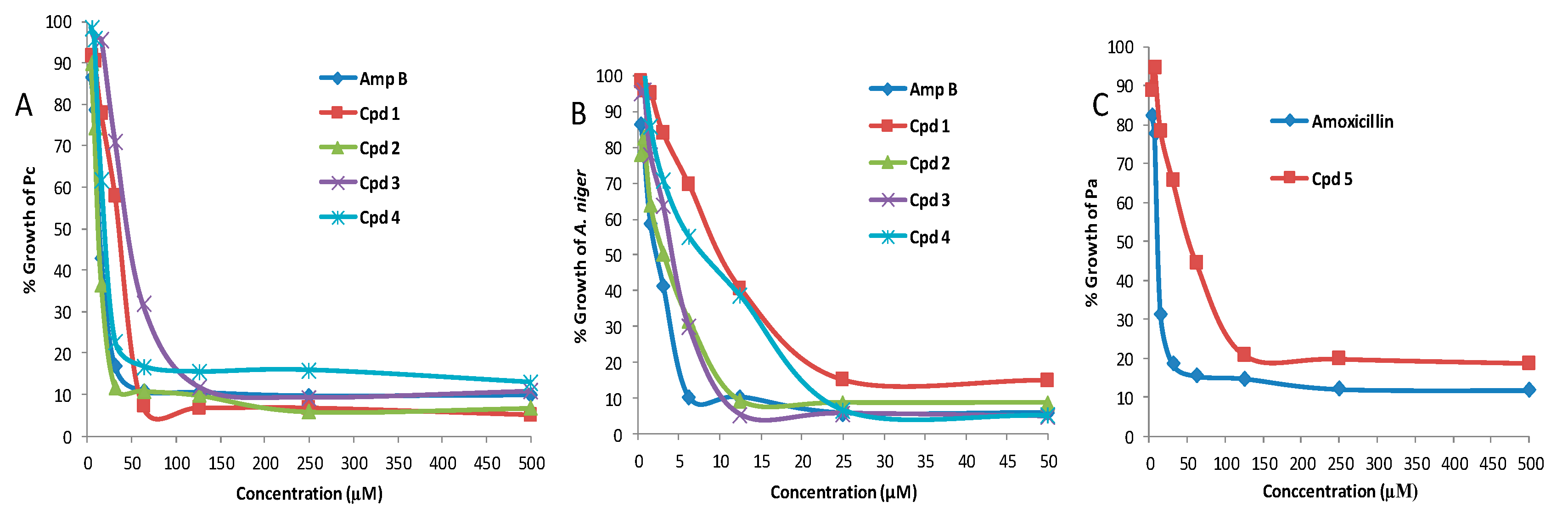
| Compound | MIC (μM) | |||
|---|---|---|---|---|
| Pc | Pa | A. niger | C. albicans | |
| 1 | 43 (±1.6) | >200 | 40.2 (±1.2) | >50 |
| 2 | 25 (±1.1) | >200 | 8.4 (±0.7) | >50 |
| 3 | 127 (±1.3) | >200 | 12.8 (±0.8) | >50 |
| 4 | 53 (±1.5) | >200 | 22.6 (±0.3) | >50 |
| 5 | >200 | 93 (±1.6) | >50 | >50 |
| 6 | >200 | >200 | >50 | >50 |
| 7 | >200 | >200 | >50 | >50 |
| 8 | >200 | >200 | >50 | >50 |
| Amphotericin B | 16.7 (±1.2) | 5.7 (±0.2) | 2.4 (±0.1) | |
| Amoxicillin | 31 (±1.1) | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thissera, B.; Alhadrami, H.A.; Hassan, M.H.A.; Hassan, H.M.; Behery, F.A.; Bawazeer, M.; Yaseen, M.; Belbahri, L.; Rateb, M.E. Induction of Cryptic Antifungal Pulicatin Derivatives from Pantoea Agglomerans by Microbial Co-Culture. Biomolecules 2020, 10, 268. https://doi.org/10.3390/biom10020268
Thissera B, Alhadrami HA, Hassan MHA, Hassan HM, Behery FA, Bawazeer M, Yaseen M, Belbahri L, Rateb ME. Induction of Cryptic Antifungal Pulicatin Derivatives from Pantoea Agglomerans by Microbial Co-Culture. Biomolecules. 2020; 10(2):268. https://doi.org/10.3390/biom10020268
Chicago/Turabian StyleThissera, Bathini, Hani A. Alhadrami, Marwa H. A. Hassan, Hossam M. Hassan, Fathy A. Behery, Majed Bawazeer, Mohammed Yaseen, Lassaad Belbahri, and Mostafa E. Rateb. 2020. "Induction of Cryptic Antifungal Pulicatin Derivatives from Pantoea Agglomerans by Microbial Co-Culture" Biomolecules 10, no. 2: 268. https://doi.org/10.3390/biom10020268
APA StyleThissera, B., Alhadrami, H. A., Hassan, M. H. A., Hassan, H. M., Behery, F. A., Bawazeer, M., Yaseen, M., Belbahri, L., & Rateb, M. E. (2020). Induction of Cryptic Antifungal Pulicatin Derivatives from Pantoea Agglomerans by Microbial Co-Culture. Biomolecules, 10(2), 268. https://doi.org/10.3390/biom10020268








