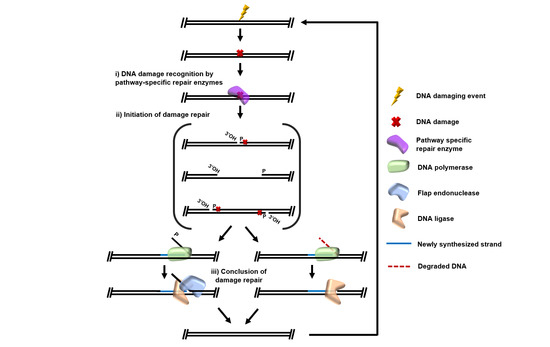
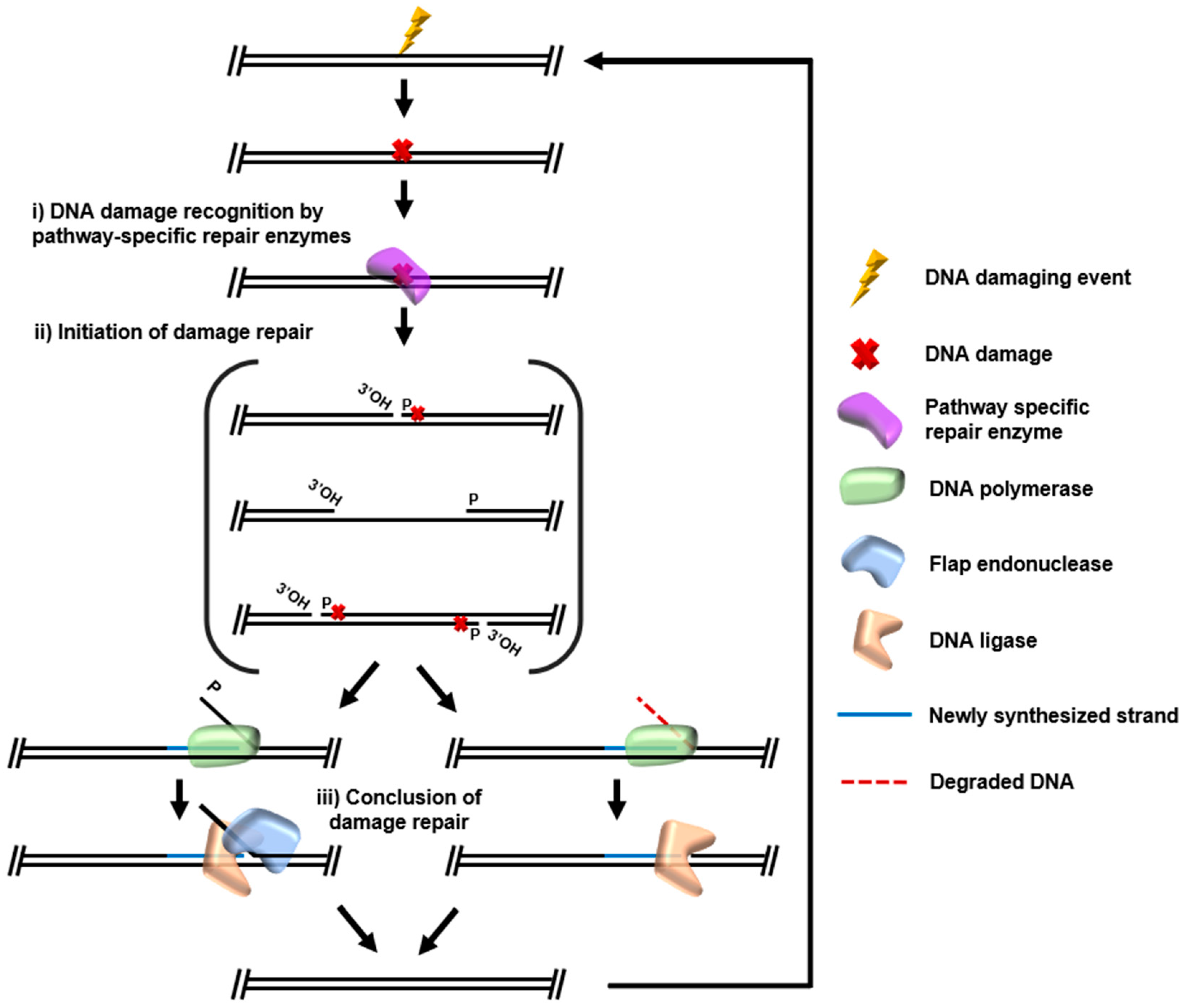
Archaeal DNA Repair Mechanisms
Abstract
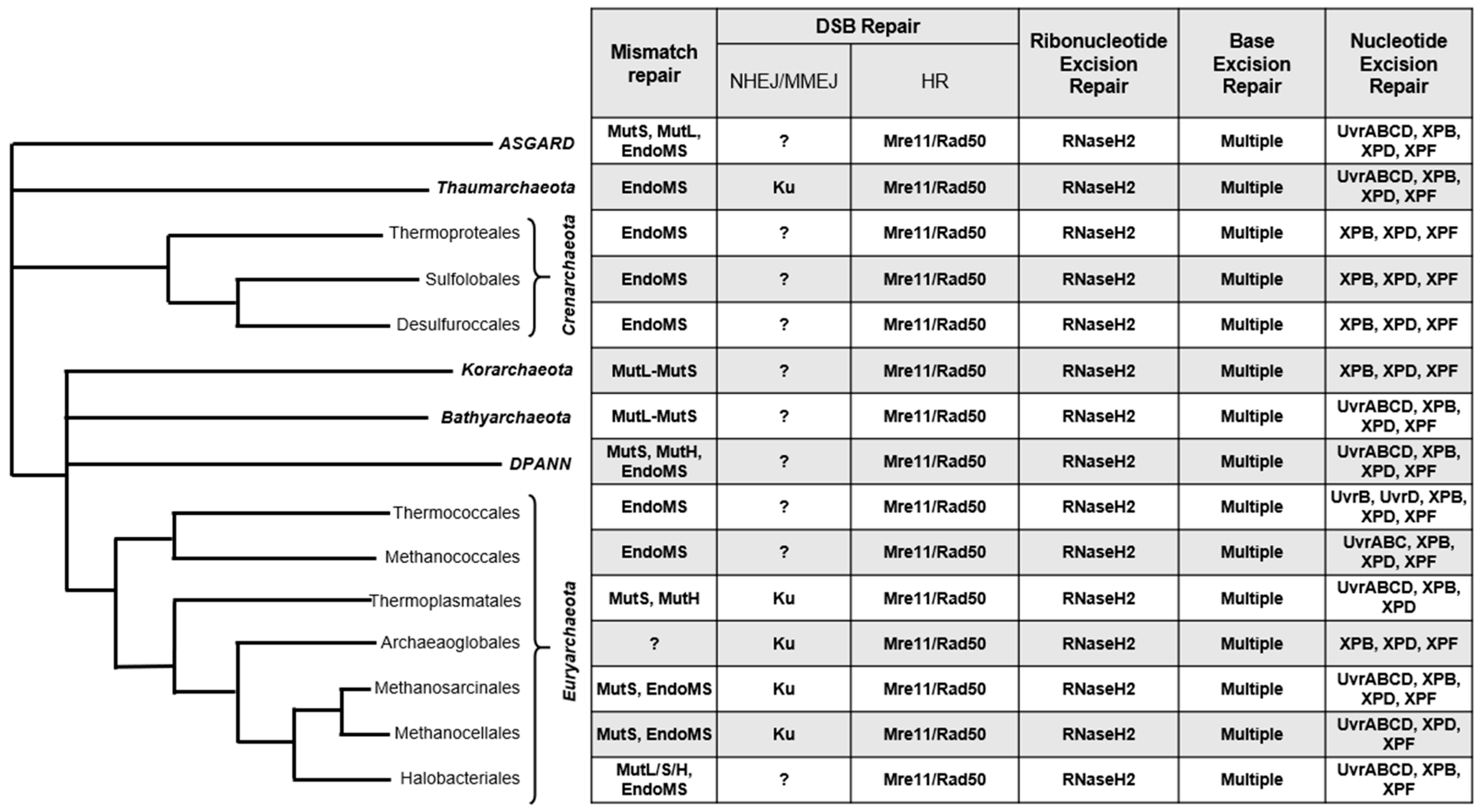
1. Introduction
2. Double-Strand Break (DSB) Repair
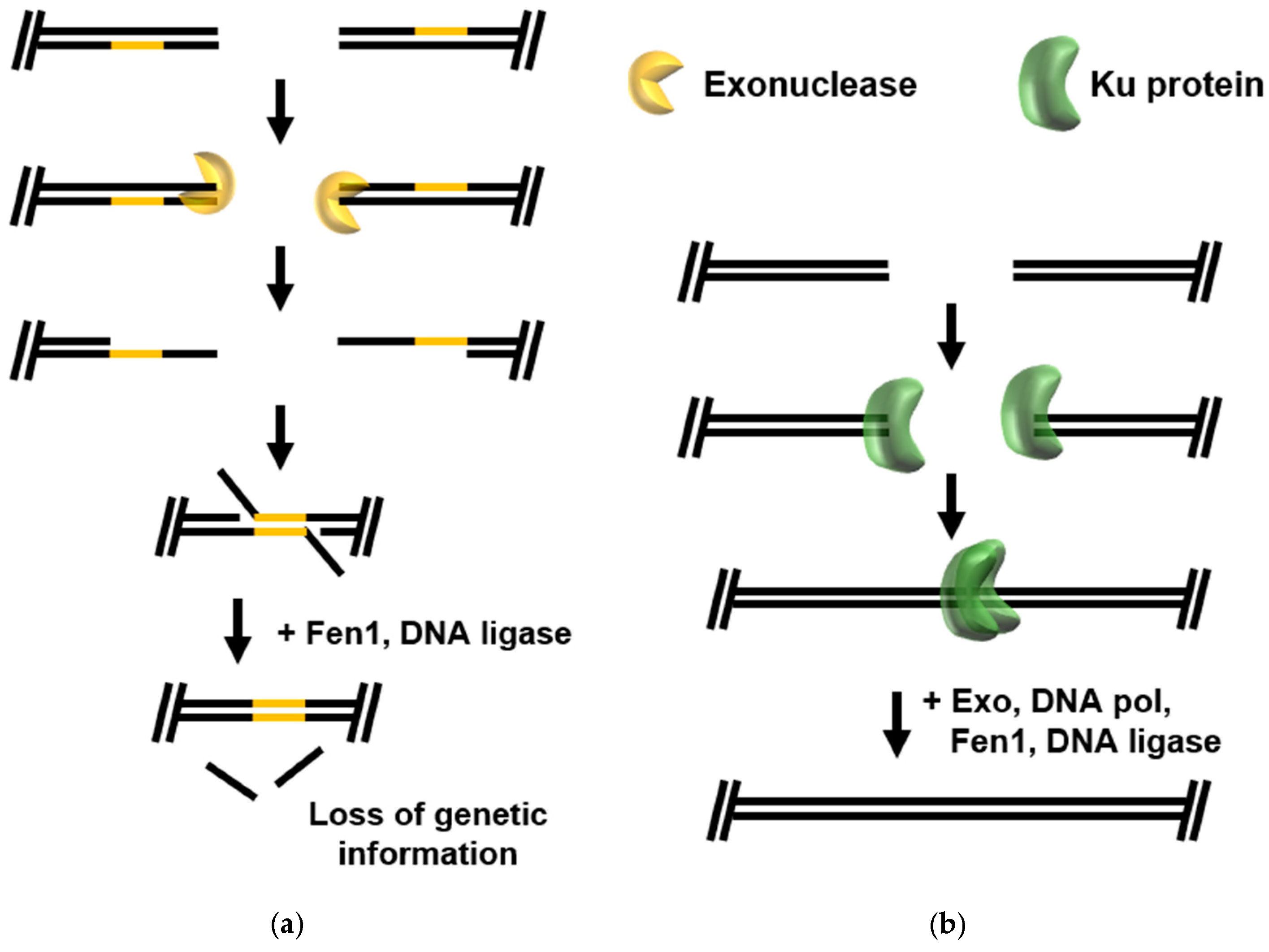
2.1. Error-Prone DSB Repair Pathways
2.2. Homologous Recombination (HR)-DSB Repair
2.3. New Resources Emerging from DSB Repair Pathways
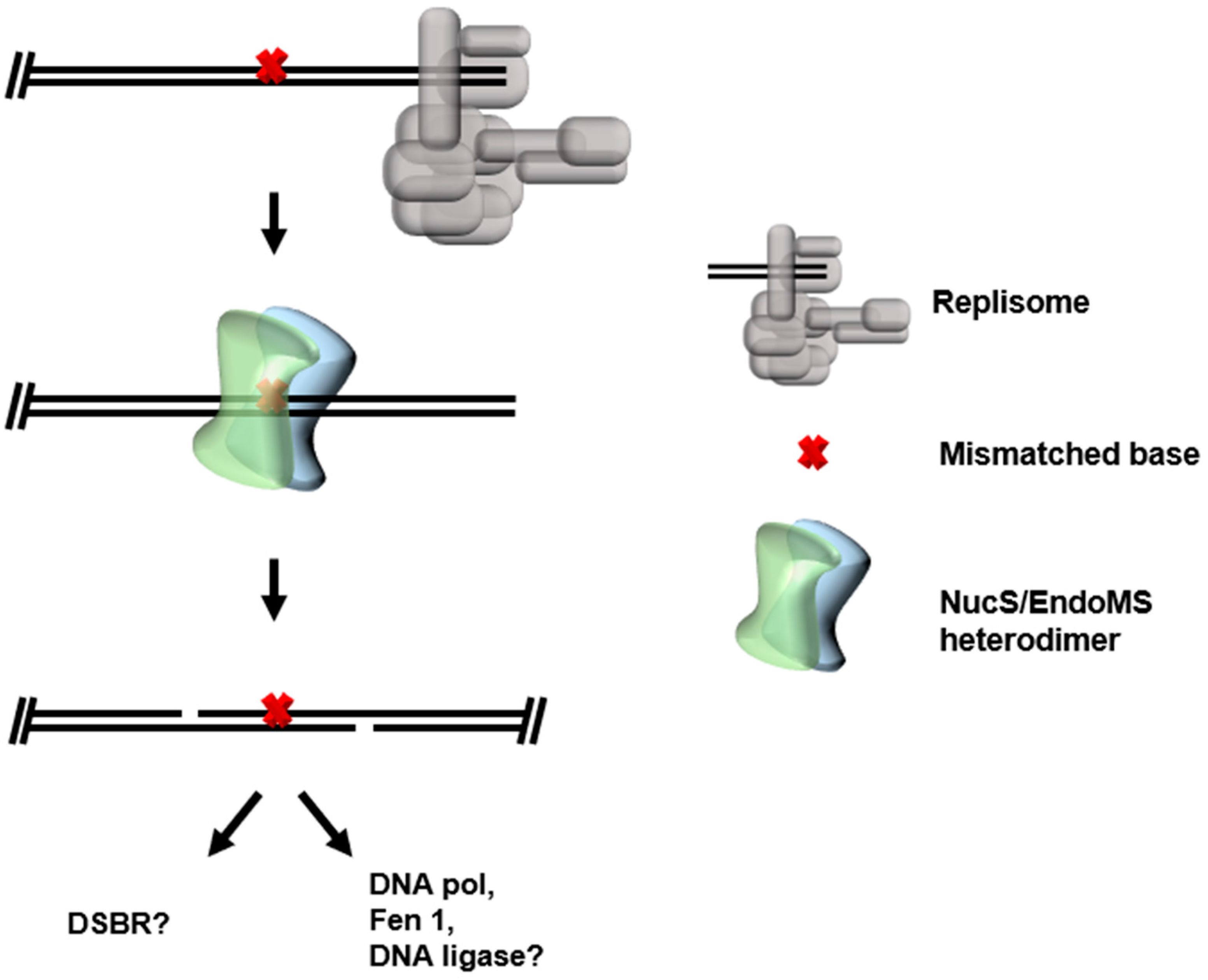
3. Mismatch Repair (MMR)
3.1. MutL/MutS
3.2. NucS/EndoMS
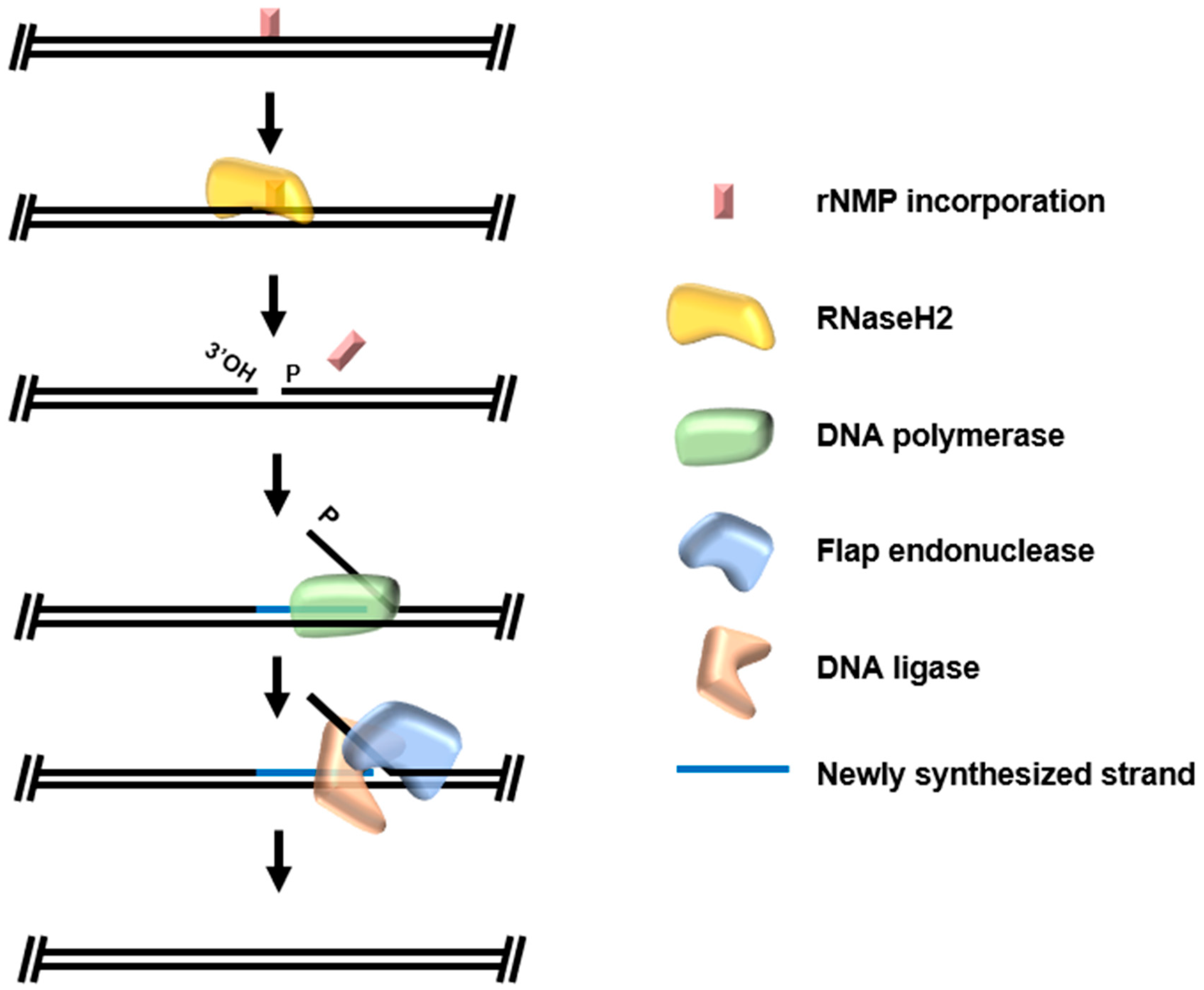
4. Ribonucleotide Excision Repair (RER)
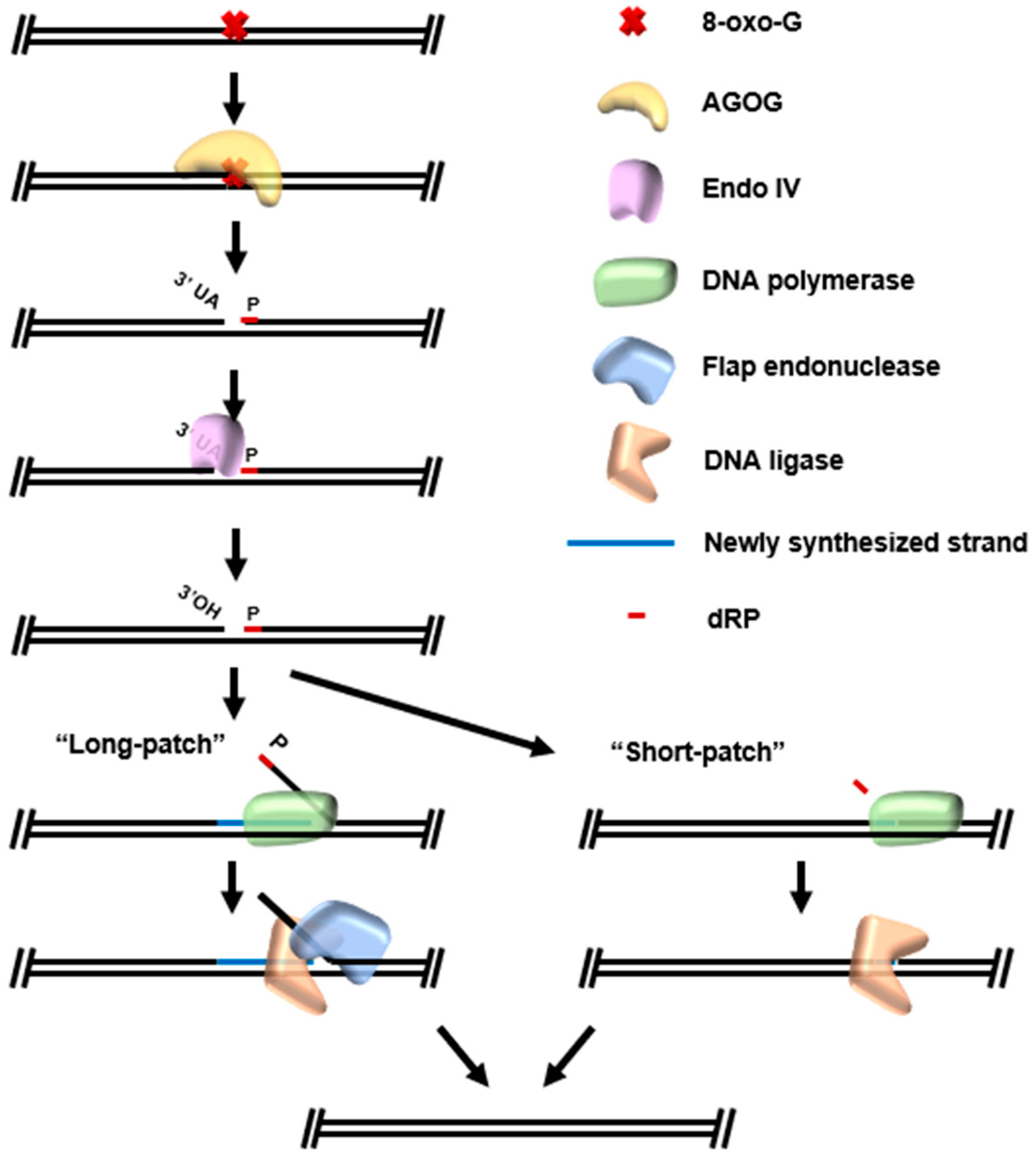
5. Base Excision Repair (BER)
New Resources Emerging from BER Pathways
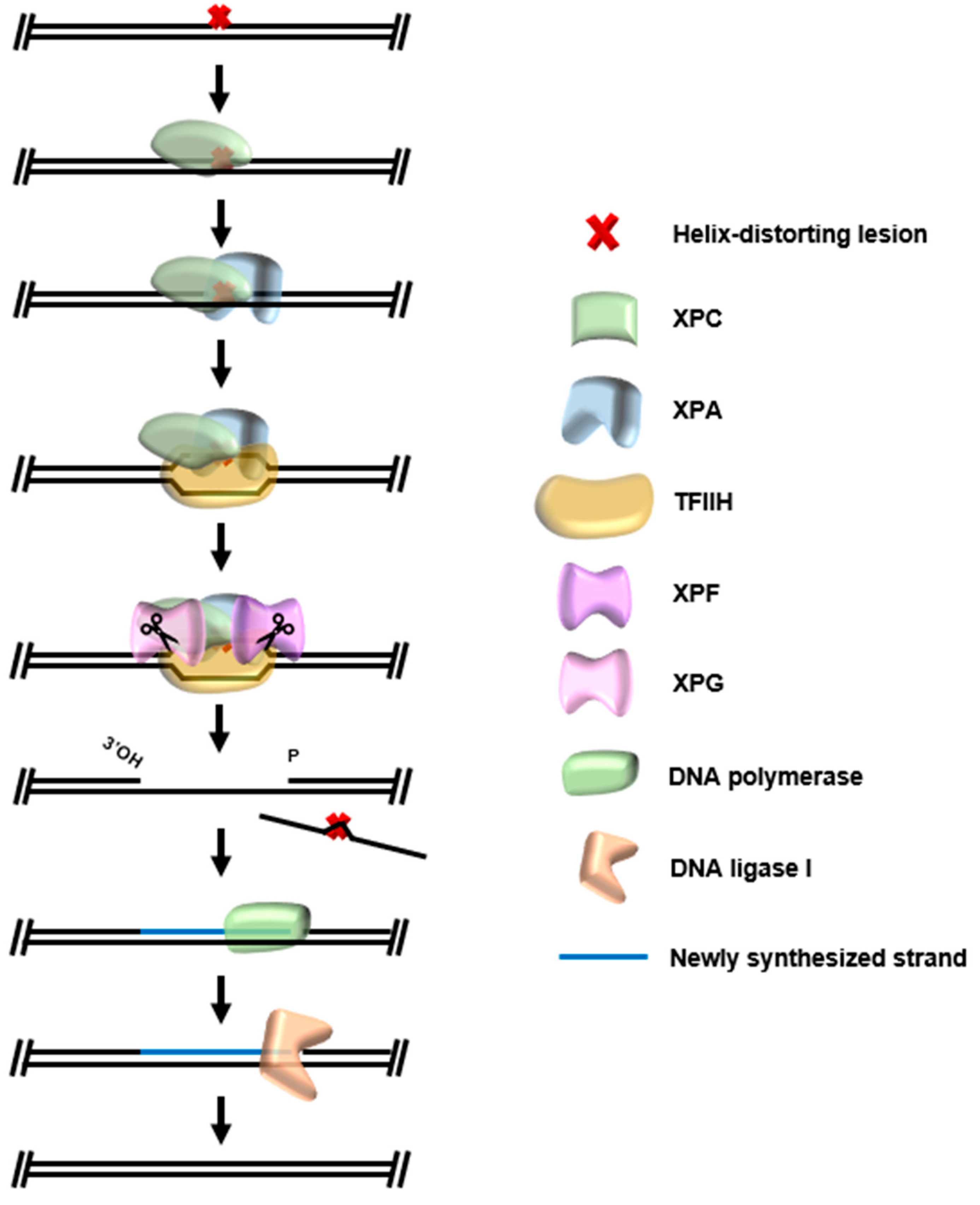
6. Global Genomic Nucleotide Excision Repair (GG-NER)
7. Transcription Coupled Nucleotide Excision Repair (TC-NER)
8. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jones, D.L.; Baxter, B.K. DNA Repair and Photoprotection: Mechanisms of Overcoming Environmental Ultraviolet Radiation Exposure in Halophilic Archaea. Front. Microbiol. 2017, 8, 1882. [Google Scholar] [CrossRef] [PubMed]
- Atomi, H.; Reeve, J.N. Microbe Profile: Thermococcus kodakarensis: The model hyperthermophilic archaeon. Microbiology 2019, 165, 1166–1168. [Google Scholar] [CrossRef] [PubMed]
- Vieille, C.; Zeikus, J.G. Hyperthermophilic Enzymes: Sources, Uses, and Molecular Mechanisms for Thermostability. Microbiol. Mol. Biol. Rev. 2001, 65, 1–43. [Google Scholar] [CrossRef]
- Lindahl, T. Instability and decay of the primary structure of DNA. Nat. Cell Biol. 1993, 362, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.C.A.; Crayle, J.; Zhou, S.; Swanstrom, R.; Wolfenden, R. Cytosine deamination and the precipitous decline of spontaneous mutation during Earth’s history. Proc. Natl. Acad. Sci. USA 2016, 113, 8194–8199. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, M.; Norris, K.F.; Wang, R.Y.; Kuo, K.C.; Gehrke, C.W. DNA cytosine methylation and heat-induced deamination. Biosci. Rep. 1986, 6, 387–393. [Google Scholar] [CrossRef]
- Ishino, Y.; Narumi, I. DNA repair in hyperthermophilic and hyperradioresistant microorganisms. Curr. Opin. Microbiol. 2015, 25, 103–112. [Google Scholar] [CrossRef]
- Grogan, D.W.; Carver, G.T.; Drake, J.W. Genetic fidelity under harsh conditions: Analysis of spontaneous mutation in the thermoacidophilic archaeon Sulfolobus acidocaldarius. Proc. Natl. Acad. Sci. USA 2001, 98, 7928–7933. [Google Scholar] [CrossRef]
- Palud, A.; Villani, G.; L’Haridon, S.; Querellou, J.; Henneke, G.; Raffin, J.-P. Intrinsic properties of the two replicative DNA polymerases of Pyrococcus abyssi in replicating abasic sites: Possible role in DNA damage tolerance? Mol. Microbiol. 2008, 70, 746–761. [Google Scholar] [CrossRef]
- Baker, B.J.; De Anda, V.; Seitz, K.W.; Dombrowski, N.; Santoro, A.E.; Lloyd, K.G. Diversity, ecology and evolution of Archaea. Nat. Microbiol. 2020, 5, 887–900. [Google Scholar] [CrossRef]
- Yi, C.; He, C. DNA Repair by Reversal of DNA Damage. Cold Spring Harb. Perspect. Biol. 2013, 5, a012575. [Google Scholar] [CrossRef] [PubMed]
- Sancar, A. Mechanisms of DNA Repair by Photolyase and Excision Nuclease (Nobel Lecture). Angew. Chem. Int. Ed. 2016, 55, 8502–8527. [Google Scholar] [CrossRef] [PubMed]
- Sinha, A.K.; Possoz, C.; Durand, A.; Desfontaines, J.-M.; Barre, F.-X.; Leach, D.; Michel, B. Broken replication forks trigger heritable DNA breaks in the terminus of a circular chromosome. PLoS Genet. 2018, 14, e1007256. [Google Scholar] [CrossRef] [PubMed]
- Michel, B.; Sinha, A.K.; Leach, D. Replication Fork Breakage and Restart in Escherichia coli. Microbiol. Mol. Biol. Rev. 2018, 82, e00013. [Google Scholar] [CrossRef]
- Keijzers, G.; Bakula, D.; Petr, M.A.; Madsen, N.G.K.; Teklu, A.; Mkrtchyan, G.; Osborne, B.; Scheibye-Knudsen, M. Human Exonuclease 1 (EXO1) Regulatory Functions in DNA Replication with Putative Roles in Cancer. Int. J. Mol. Sci. 2019, 20, 74. [Google Scholar] [CrossRef]
- Bouwman, B.A.M.; Crosetto, N. Endogenous DNA Double-Strand Breaks during DNA Transactions: Emerging Insights and Methods for Genome-Wide Profiling. Genes 2018, 9, 632. [Google Scholar] [CrossRef]
- Shiimori, M.; Garrett, S.C.; Chambers, D.P.; Glover, C.V.C.; Graveley, B.R.; Terns, M.P. Role of free DNA ends and protospacer adjacent motifs for CRISPR DNA uptake in Pyrococcus furiosus. Nucleic Acids Res. 2017, 45, 11281–11294. [Google Scholar] [CrossRef]
- Khan, F.A.; Ali, S.O. Physiological Roles of DNA Double-Strand Breaks. J. Nucleic Acids 2017, 2017, 6439169. [Google Scholar] [CrossRef]
- Cannan, W.J.; Pederson, D.S. Mechanisms and Consequences of Double-Strand DNA Break Formation in Chromatin. J. Cell. Physiol. 2015, 231, 3–14. [Google Scholar] [CrossRef]
- Gray, S.; Cohen, P.E. Control of Meiotic Crossovers: From Double-Strand Break Formation to Designation. Annu. Rev. Genet. 2016, 50, 175–210. [Google Scholar] [CrossRef]
- Dulmage, K.A.; Darnell, C.L.; Vreugdenhil, A.; Schmid, A.K. Copy number variation is associated with gene expression change in archaea. Microb. Genom. 2018, 4. [Google Scholar] [CrossRef] [PubMed]
- Hildenbrand, C.; Stock, T.; Lange, C.; Rother, M.; Soppa, J. Genome Copy Numbers and Gene Conversion in Methanogenic Archaea. J. Bacteriol. 2010, 193, 734–743. [Google Scholar] [CrossRef] [PubMed]
- Seol, J.-H.; Shim, E.Y.; Lee, S.E. Microhomology-mediated end joining: Good, bad and ugly. Mutat. Res. Mol. Mech. Mutagen. 2018, 809, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Javadekar, S.M.; Pandey, M.; Srivastava, M.; Kumari, R.; Raghavan, S.C. Homology and enzymatic requirements of microhomology-dependent alternative end joining. Cell Death Dis. 2015, 6, e1697. [Google Scholar] [CrossRef]
- Nayak, D.D.; Metcalf, W.W. Cas9-mediated genome editing in the methanogenic archaeon Methanosarcina acetivorans. Proc. Natl. Acad. Sci. USA 2017, 114, 2976–2981. [Google Scholar] [CrossRef]
- Zhang, C.; Whitaker, R.J. Microhomology-Mediated High-Throughput Gene Inactivation Strategy for the Hyperthermophilic Crenarchaeon Sulfolobus islandicus. Appl. Environ. Microbiol. 2017, 84, e02167-17. [Google Scholar] [CrossRef]
- Stachler, A.-E.; Turgeman-Grott, I.; Shtifman-Segal, E.; Allers, T.; Marchfelder, A.; Gophna, U. High tolerance to self-targeting of the genome by the endogenous CRISPR-Cas system in an archaeon. Nucleic Acids Res. 2017, 45, 5208–5216. [Google Scholar] [CrossRef]
- Bartlett, E.J.; Brissett, N.C.; Doherty, A.J. Ribonucleolytic resection is required for repair of strand displaced nonhomologous end-joining intermediates. Proc. Natl. Acad. Sci. USA 2013, 110, E1984–E1991. [Google Scholar] [CrossRef]
- Bartlett, E.J.; Brissett, N.C.; Płociński, P.; Carlberg, T.; Doherty, A.J. Molecular basis for DNA strand displacement by NHEJ repair polymerases. Nucleic Acids Res. 2015, 44, 2173–2186. [Google Scholar] [CrossRef]
- Gehring, A.M.; Astling, D.P.; Matsumi, R.; Burkhart, B.W.; Kelman, Z.; Reeve, J.N.; Jones, K.L.; Santangelo, T.J. Genome Replication in Thermococcus kodakarensis Independent of Cdc6 and an Origin of Replication. Front. Microbiol. 2017, 8, 2084. [Google Scholar] [CrossRef] [PubMed]
- Kelman, L.M.; Kelman, Z. Do Archaea Need an Origin of Replication? Trends Microbiol. 2018, 26, 172–174. [Google Scholar] [CrossRef] [PubMed]
- White, M.F. Homologous recombination in the archaea: The means justify the ends. Biochem. Soc. Trans. 2011, 39, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Hogrel, G.; Lu, Y.; Laurent, S.; Henry, E.; Etienne, C.; Phung, D.K.; Dulermo, R.; Bossé, A.; Pluchon, P.-F.; Clouet-D’Orval, B.; et al. Physical and functional interplay between PCNA DNA clamp and Mre11–Rad50 complex from the archaeon Pyrococcus furiosus. Nucleic Acids Res. 2018, 46, 5651–5663. [Google Scholar] [CrossRef] [PubMed]
- Sung, S.; Li, F.; Park, Y.B.; Kim, J.S.; Kim, A.; Song, O.; Kim, J.; Che, J.; Lee, S.E.; Cho, Y. DNA end recognition by the Mre11 nuclease dimer: Insights into resection and repair of damaged DNA. EMBO J. 2014, 33, 2422–2435. [Google Scholar] [CrossRef]
- Käshammer, L.; Saathoff, J.-H.; Lammens, K.; Gut, F.; Bartho, J.; Alt, A.; Kessler, B.; Hopfner, K.-P. Mechanism of DNA End Sensing and Processing by the Mre11-Rad50 Complex. Mol. Cell 2019, 76, 382–394.e6. [Google Scholar] [CrossRef]
- Wiktor, J.; Van Der Does, M.; Büller, L.; Sherratt, D.J.; Dekker, C. Direct observation of end resection by RecBCD during double-stranded DNA break repair in vivo. Nucleic Acids Res. 2018, 46, 1821–1833. [Google Scholar] [CrossRef]
- Constantinesco, F.; Forterre, P.; Koonin, E.V.; Aravind, L.; Elie, C. A bipolar DNA helicase gene, herA, clusters with rad50, mre11 and nurA genes in thermophilic archaea. Nucleic Acids Res. 2004, 32, 1439–1447. [Google Scholar] [CrossRef]
- Constantinesco, F.; Forterre, P.; Elie, C. NurA, a novel 5′–3′ nuclease gene linked to rad50 and mre11 homologs of thermophilic Archaea. EMBO Rep. 2002, 3, 537–542. [Google Scholar] [CrossRef]
- Quaiser, A.; Constantinesco, F.; White, M.F.; Forterre, P.; Elie, C. The Mre11 protein interacts with both Rad50 and the HerA bipolar helicase and is recruited to DNA following gamma irradiation in the archaeon Sulfolobus acidocaldarius. BMC Mol. Biol. 2008, 9, 25. [Google Scholar] [CrossRef]
- Paull, T.T.; Gellert, M. A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc. Natl. Acad. Sci. USA 2000, 97, 6409–6414. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Liu, L.; Liu, J.; Ni, J.; She, Q.; Shen, Y. Efficient 5′-3′ DNA end resection by HerA and NurA is essential for cell viability in the crenarchaeon Sulfolobus islandicus. BMC Mol. Biol. 2015, 16, 2. [Google Scholar] [CrossRef] [PubMed]
- Ahdash, Z.; Lau, A.M.; Byrne, R.T.; Lammens, K.; Stüetzer, A.; Urlaub, H.; Booth, P.J.; Reading, E.; Hopfner, K.-P.; Politis, A. Mechanistic insight into the assembly of the HerA–NurA helicase–nuclease DNA end resection complex. Nucleic Acids Res. 2017, 45, 12025–12038. [Google Scholar] [CrossRef]
- Rzechorzek, N.J.; Blackwood, J.K.; Bray, S.; Maman, J.D.; Pellegrini, L.; Robinson, N.P. Structure of the hexameric HerA ATPase reveals a mechanism of translocation-coupled DNA-end processing in archaea. Nat. Commun. 2014, 5, 5506. [Google Scholar] [CrossRef] [PubMed]
- De Falco, M.; Catalano, F.; Rossi, M.; Ciaramella, M.; De Felice, M. NurA Is Endowed with Endo- and Exonuclease Activities that Are Modulated by HerA: New Insight into Their Role in DNA-End Processing. PLoS ONE 2015, 10, e0142345. [Google Scholar] [CrossRef]
- Chae, J.; Kim, Y.C.; Cho, Y. Crystal structure of the NurA–dAMP–Mn2+ complex. Nucleic Acids Res. 2011, 40, 2258–2270. [Google Scholar] [CrossRef]
- Blackwood, J.K.; Rzechorzek, N.J.; Abrams, A.S.; Maman, J.D.; Pellegrini, L.; Robinson, N.P. Structural and functional insights into DNA-end processing by the archaeal HerA helicase–NurA nuclease complex. Nucleic Acids Res. 2011, 40, 3183–3196. [Google Scholar] [CrossRef]
- Haldenby, S.; White, M.F.; Allers, T. RecA family proteins in archaea: RadA and its cousins. Biochem. Soc. Trans. 2009, 37, 102–107. [Google Scholar] [CrossRef][Green Version]
- Komori, K.; Miyata, T.; DiRuggiero, J.; Holley-Shanks, R.; Hayashi, I.; Cann, I.K.O.; Mayanagi, K.; Shinagawa, H.; Ishino, Y. Both RadA and RadB Are Involved in Homologous Recombination in Pyrococcus furiosus. J. Biol. Chem. 2000, 275, 33782–33790. [Google Scholar] [CrossRef]
- Shin, D.S.; Pellegrini, L.; Daniels, D.S.; Yelent, B.; Craig, L.; Bates, D.; Yu, D.S.; Shivji, M.K.; Hitomi, C.; Arvai, A.S.; et al. Full-length archaeal Rad51 structure and mutants: Mechanisms for RAD51 assembly and control by BRCA2. EMBO J. 2003, 22, 4566–4576. [Google Scholar] [CrossRef]
- Hogrel, G.; Lu, Y.; Alexandre, N.; Bossé, A.; Dulermo, R.; Ishino, S.; Ishino, Y.; Flament, D. Role of RadA and DNA Polymerases in Recombination-Associated DNA Synthesis in Hyperthermophilic Archaea. Biomolecules 2020, 10, 1045. [Google Scholar] [CrossRef] [PubMed]
- Miura, T.; Yamana, Y.; Usui, T.; Ogawa, H.I.; Yamamoto, M.-T.; Kusano, K. Homologous Recombination via Synthesis-Dependent Strand Annealing in Yeast Requires the Irc20 and Srs2 DNA Helicases. Genetics 2012, 191, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sun, H.; Huang, Y.; Wang, Y.; Liu, Y.; Chen, X. Pathways and assays for DNA double-strand break repair by homologous recombination. Acta Biochim. Biophys. Sin. 2019, 51, 879–889. [Google Scholar] [CrossRef] [PubMed]
- Guy, C.P. Archaeal Hel308 helicase targets replication forks in vivo and in vitro and unwinds lagging strands. Nucleic Acids Res. 2005, 33, 3678–3690. [Google Scholar] [CrossRef]
- Northall, S.J.; Buckley, R.; Jones, N.; Penedo-Esteiro, J.C.; Soultanas, P.; Bolt, E. DNA binding and unwinding by Hel308 helicase requires dual functions of a winged helix domain. DNA Repair 2017, 57, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Wu, X.; Tompkins, J.D.; Her, C. Assessment of Anti-recombination and Double-strand Break-induced Gene Conversion in Human Cells by a Chromosomal Reporter. J. Biol. Chem. 2012, 287, 29543–29553. [Google Scholar] [CrossRef]
- Komori, K.; Sakae, S.; Fujikane, R.; Morikawa, K.; Shinagawa, H.; Ishino, Y. Biochemical characterization of the Hjc Holliday junction resolvase of Pyrococcus furiosus. Nucleic Acids Res. 2000, 28, 4544–4551. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Komori, K.; Sakae, S.; Shinagawa, H.; Morikawa, K.; Ishino, Y. A Holliday junction resolvase from Pyrococcus furiosus: Functional similarity to Escherichia coli RuvC provides evidence for conserved mechanism of homologous recombination in Bacteria, Eukarya, and Archaea. Proc. Natl. Acad. Sci. USA 1999, 96, 8873–8878. [Google Scholar] [CrossRef] [PubMed]
- Blanco, M.G.; Matos, J.; West, S.C. Dual Control of Yen1 Nuclease Activity and Cellular Localization by Cdk and Cdc14 Prevents Genome Instability. Mol. Cell 2014, 54, 94–106. [Google Scholar] [CrossRef]
- Dehé, P.-M.; Coulon, S.; Scaglione, S.; Shanahan, P.; Takedachi, A.; Wohlschlegel, J.A.; Yates, J.R.; Llorente, B.; Russell, P.; Gaillard, P.-H.L. Regulation of Mus81–Eme1 Holliday junction resolvase in response to DNA damage. Nat. Struct. Mol. Biol. 2013, 20, 598–603. [Google Scholar] [CrossRef]
- Huang, Q.; Mayaka, J.B.; Zhong, Q.; Zhang, C.; Hou, G.; Ni, J.; Shen, Y. Phosphorylation of the Archaeal Holliday Junction Resolvase Hjc Inhibits Its Catalytic Activity and Facilitates DNA Repair in Sulfolobus islandicus REY15A. Front. Microbiol. 2019, 10, 1214. [Google Scholar] [CrossRef] [PubMed]
- Delmas, S.; Shunburne, L.; Ngo, H.-P.; Allers, T. Mre11-Rad50 Promotes Rapid Repair of DNA Damage in the Polyploid Archaeon Haloferax volcanii by Restraining Homologous Recombination. PLoS Genet. 2009, 5, e1000552. [Google Scholar] [CrossRef] [PubMed]
- Kish, A.; Gaillard, J.; Armengaud, J.; Elie, C. Post-translational methylations of the archaeal Mre11:Rad50 complex throughout the DNA damage response. Mol. Microbiol. 2016, 100, 362–378. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, B.B.; Paull, T.T. The P. furiosus Mre11/Rad50 Complex Promotes 5′ Strand Resection at a DNA Double-Strand Break. Cell 2008, 135, 250–260. [Google Scholar] [CrossRef] [PubMed]
- Woodman, I.L.; Brammer, K.; Bolt, E. Physical interaction between archaeal DNA repair helicase Hel308 and Replication Protein A (RPA). DNA Repair 2011, 10, 306–313. [Google Scholar] [CrossRef]
- Dorazi, R.; Parker, J.L.; White, M.F. PCNA Activates the Holliday Junction Endonuclease Hjc. J. Mol. Biol. 2006, 364, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Craig, J.M.; Laszlo, A.H.; Nova, I.C.; Brinkerhoff, H.; Noakes, M.T.; Baker, K.S.; Bowman, J.L.; Higinbotham, H.R.; Mount, J.W.; Gundlach, J.H. Determining the effects of DNA sequence on Hel308 helicase translocation along single-stranded DNA using nanopore tweezers. Nucleic Acids Res. 2019, 47, 2506–2513. [Google Scholar] [CrossRef]
- Gophna, U.; Allers, T.; Marchfelder, A. Finally, Archaea Get Their CRISPR-Cas Toolbox. Trends Microbiol. 2017, 25, 430–432. [Google Scholar] [CrossRef]
- Castañeda-García, A.; Prieto, A.I.; Rodríguez-Beltrán, J.; Alonso, N.; Cantillon, D.; Costas, C.; Pérez-Lago, L.; Zegeye, E.D.; Herranz, M.; Płociński, P.; et al. A non-canonical mismatch repair pathway in prokaryotes. Nat. Commun. 2017, 8, 14246. [Google Scholar] [CrossRef]
- Takemoto, N.; Numata, I.; Su’Etsugu, M.; Miyoshi-Akiyama, T. Bacterial EndoMS/NucS acts as a clamp-mediated mismatch endonuclease to prevent asymmetric accumulation of replication errors. Nucleic Acids Res. 2018, 46, 6152–6165. [Google Scholar] [CrossRef]
- Meier, B.; Volkova, N.V.; Hong, Y.; Schofield, P.; Campbell, P.J.; Gerstung, M.; Gartner, A. Mutational signatures of DNA mismatch repair deficiency in C. elegans and human cancers. Genome Res. 2018, 28, 666–675. [Google Scholar] [CrossRef] [PubMed]
- Modrich, P. Mechanisms in E. coli and Human Mismatch Repair (Nobel Lecture). Angew. Chem. Int. Ed. 2016, 55, 8490–8501. [Google Scholar] [CrossRef] [PubMed]
- Lenhart, J.S.; Pillon, M.C.; Guarné, A.; Biteen, J.S.; Simmons, L.A. Mismatch repair in Gram-positive bacteria. Res. Microbiol. 2016, 167, 4–12. [Google Scholar] [CrossRef]
- Li, Z.; Pearlman, A.H.; Hsieh, P. DNA mismatch repair and the DNA damage response. DNA Repair 2016, 38, 94–101. [Google Scholar] [CrossRef]
- Kunkel, T.A.; Erie, D.A. Eukaryotic Mismatch Repair in Relation to DNA Replication. Annu. Rev. Genet. 2015, 49, 291–313. [Google Scholar] [CrossRef] [PubMed]
- Minobe, A.; Fukui, K.; Yonezu, H.; Ohshita, K.; Mizobuchi, S.; Morisawa, T.; Hakumai, Y.; Yano, T.; Ashiuchi, M.; Wakamatsu, T. Biochemical characterization of mismatch-binding protein MutS1 and nicking endonuclease MutL from a euryarchaeon Methanosaeta thermophila. DNA Repair 2019, 75, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Busch, C.R.; DiRuggiero, J. MutS and MutL Are Dispensable for Maintenance of the Genomic Mutation Rate in the Halophilic Archaeon Halobacterium salinarum NRC-1. PLoS ONE 2010, 5, e9045. [Google Scholar] [CrossRef]
- Ishino, S.; Nishi, Y.; Oda, S.; Uemori, T.; Sagara, T.; Takatsu, N.; Yamagami, T.; Shirai, T.; Ishino, Y. Identification of a mismatch-specific endonuclease in hyperthermophilic Archaea. Nucleic Acids Res. 2016, 44, 2977–2986. [Google Scholar] [CrossRef]
- Rezgui, R.; Lestini, R.; Kuhn, J.; Fave, X.; McLeod, L.; Myllykallio, H.; Alexandrou, A.; Bouzigues, C. Differential Interaction Kinetics of a Bipolar Structure-Specific Endonuclease with DNA Flaps Revealed by Single-Molecule Imaging. PLoS ONE 2014, 9, e113493. [Google Scholar] [CrossRef][Green Version]
- Ren, B.; Kühn, J.; Meslet-Cladiere, L.; Briffotaux, J.; Norais, C.; Lavigne, R.; Flament, D.; Ladenstein, R.; Myllykallio, H. Structure and function of a novel endonuclease acting on branched DNA substrates. EMBO J. 2009, 28, 2479–2489. [Google Scholar] [CrossRef]
- Ishino, S.; Skouloubris, S.; Kudo, H.; L’Hermitte-Stead, C.; Es-Sadik, A.; Lambry, J.C.; Ishino, Y.; Myllykallio, H. Activation of the mismatch-specific endonuclease EndoMS/NucS by the replication clamp is required for high fidelity DNA replication. Nucleic Acids Res. 2018, 46, 6206–6217. [Google Scholar] [CrossRef] [PubMed]
- Ariyoshi, M.; Morikawa, K. A Dual Base Flipping Mechanism for Archaeal Mismatch Repair. Structure 2016, 24, 1859–1861. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nakae, S.; Hijikata, A.; Tsuji, T.; Yonezawa, K.; Kouyama, K.-I.; Mayanagi, K.; Ishino, S.; Ishino, Y.; Shirai, T. Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease. Structure 2016, 24, 1960–1971. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jiang, D.; Wu, M.; Yang, Z.; Oger, P.M. New Insights into DNA Repair Revealed by NucS Endonucleases from Hyperthermophilic Archaea. Front. Microbiol. 2020, 11, 1263. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Huang, S.-Y. Exploring the Binding Mechanism and Dynamics of EndoMS/NucS to Mismatched dsDNA. Int. J. Mol. Sci. 2019, 20, 5142. [Google Scholar] [CrossRef]
- Creze, C.; Ligabue, A.; Laurent, S.; Lestini, R.; Laptenok, S.P.; Khun, J.; Vos, M.H.; Czjzek, M.; Myllykallio, H.; Flament, D. Modulation of the Pyrococcus abyssi NucS Endonuclease Activity by Replication Clamp at Functional and Structural Levels. J. Biol. Chem. 2012, 287, 15648–15660. [Google Scholar] [CrossRef]
- Barillà, D. Driving Apart and Segregating Genomes in Archaea. Trends Microbiol. 2016, 24, 957–967. [Google Scholar] [CrossRef]
- Lemor, M.; Kong, Z.; Henry, E.; Brizard, R.; Laurent, S.; Bossé, A.; Henneke, G. Differential Activities of DNA Polymerases in Processing Ribonucleotides during DNA Synthesis in Archaea. J. Mol. Biol. 2018, 430, 4908–4924. [Google Scholar] [CrossRef]
- Ban, C.; Ramakrishnan, B.; Sundaralingam, M. A Single 2′-Hydroxyl Group Converts B-DNA to A-DNA. J. Mol. Biol. 1994, 236, 275–285. [Google Scholar] [CrossRef]
- Heider, M.R.; Burkhart, B.W.; Santangelo, T.J.; Gardner, A.F. Defining the RNaseH2 enzyme-initiated ribonucleotide excision repair pathway in Archaea. J. Biol. Chem. 2017, 292, 8835–8845. [Google Scholar] [CrossRef]
- Gardner, A.F. Determinants of nucleotide sugar recognition in an archaeon DNA polymerase. Nucleic Acids Res. 1999, 27, 2545–2553. [Google Scholar] [CrossRef] [PubMed]
- Hiller, B.; Hoppe, A.; Haase, C.; Hiller, C.; Schubert, N.; Müller, W.; Reijns, M.A.M.; Jackson, A.P.; Kunkel, T.A.; Wenzel, J.; et al. Ribonucleotide excision repair is essential to prevent squamous cell carcinoma of the skin. Cancer Res. 2018, 78, 5917–5926. [Google Scholar] [CrossRef] [PubMed]
- Vaisman, A.; McDonald, J.P.; Noll, S.; Huston, D.; Loeb, G.; Goodman, M.F.; Woodgate, R. Investigating the mechanisms of ribonucleotide excision repair in Escherichia coli. Mutat. Res. Mol. Mech. Mutagen. 2014, 761, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.S.; Kunkel, T.A. Ribonucleotides in DNA: Origins, repair and consequences. DNA Repair 2014, 19, 27–37. [Google Scholar] [CrossRef]
- Burkhart, B.W.; Cubonova, L.; Heider, M.R.; Kelman, Z.; Reeve, J.N.; Santangelo, T.J. The GAN Exonuclease or the Flap Endonuclease Fen1 and RNase HII Are Necessary for Viability of Thermococcus kodakarensis. J. Bacteriol. 2017, 199, e00141. [Google Scholar] [CrossRef]
- Reijns, M.A.; Rabe, B.; Rigby, R.E.; Mill, P.; Astell, K.R.; Lettice, L.A.; Boyle, S.; Leitch, A.; Keighren, M.; Kilanowski, F.; et al. Enzymatic Removal of Ribonucleotides from DNA Is Essential for Mammalian Genome Integrity and Development. Cell 2012, 149, 1008–1022. [Google Scholar] [CrossRef]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef]
- Dizdaroglu, M.; Jaruga, P. Mechanisms of free radical-induced damage to DNA. Free Radic. Res. 2012, 46, 382–419. [Google Scholar] [CrossRef]
- Schuch, A.P.; Moreno, N.C.; Schuch, N.J.; Menck, C.F.M.; Garcia, C.C.M. Sunlight damage to cellular DNA: Focus on oxidatively generated lesions. Free Radic. Biol. Med. 2017, 107, 110–124. [Google Scholar] [CrossRef]
- Wallace, S.S. Base excision repair: A critical player in many games. DNA Repair 2014, 19, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Gehring, A.M.; Zatopek, K.M.; Burkhart, B.W.; Potapov, V.; Santangelo, T.J.; Gardner, A.F. Biochemical reconstitution and genetic characterization of the major oxidative damage base excision DNA repair pathway in Thermococcus kodakarensis. DNA Repair 2020, 86, 102767. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, Y.; Shi, H.; Zhang, D.; Yang, Z.-H.; Oger, P.M.; Zheng, J. Biochemical characterization and mutational studies of the 8-oxoguanine DNA glycosylase from the hyperthermophilic and radioresistant archaeon Thermococcus gammatolerans. Appl. Microbiol. Biotechnol. 2019, 103, 8021–8033. [Google Scholar] [CrossRef] [PubMed]
- Kanugula, S.; Pauly, G.T.; Moschel, R.C.; Pegg, A.E. A bifunctional DNA repair protein from Ferroplasma acidarmanus exhibits O6-alkylguanine-DNA alkyltransferase and endonuclease V activities. Proc. Natl. Acad. Sci. USA 2005, 102, 3617–3622. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, L.; Zhu, X.; Li, Y.; Shi, H.; Oger, P.M.; Yang, Z. Biochemical characterization of a thermostable endonuclease V from the hyperthermophilic euryarchaeon Thermococcus barophilus Ch5. Int. J. Biol. Macromol. 2018, 117, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Kiyonari, S.; Egashira, Y.; Ishino, S.; Ishino, Y. Biochemical characterization of endonuclease V from the hyperthermophilic archaeon, Pyrococcus furiosus. J. Biochem. 2014, 155, 325–333. [Google Scholar] [CrossRef]
- Shiraishi, M.; Ishino, S.; Yamagami, T.; Egashira, Y.; Kiyonari, S.; Ishino, Y. A novel endonuclease that may be responsible for damaged DNA base repair in Pyrococcus furiosus. Nucleic Acids Res. 2015, 43, 2853–2863. [Google Scholar] [CrossRef]
- Miyazono, K.-I.; Ishino, S.; Makita, N.; Ito, T.; Ishino, Y.; Tanokura, M. Crystal structure of the novel lesion-specific endonuclease PfuEndoQ from Pyrococcus furiosus. Nucleic Acids Res. 2018, 46, 4807–4818. [Google Scholar] [CrossRef]
- Shiraishi, M.; Iwai, S. Molecular Basis of Substrate Recognition of Endonuclease Q from the Euryarchaeon Pyrococcus furiosus. J. Bacteriol. 2020, 202, e00542. [Google Scholar] [CrossRef]
- Zatopek, K.M.; Potapov, V.; Maduzia, L.L.; Alpaslan, E.; Chen, L.; Evans, T.C.; Ong, J.L.; Ettwiller, L.M.; Gardner, A.F. RADAR-seq: A RAre DAmage and Repair sequencing method for detecting DNA damage on a genome-wide scale. DNA Repair 2019, 80, 36–44. [Google Scholar] [CrossRef]
- Eid, J.; Fehr, A.; Gray, J.; Luong, K.; Lyle, J.; Otto, G.; Peluso, P.; Rank, D.; Baybayan, P.; Bettman, B.; et al. Real-Time DNA Sequencing from Single Polymerase Molecules. Science 2009, 323, 133–138. [Google Scholar] [CrossRef]
- Truglio, J.J.; Croteau, D.L.; Van Houten, B.; Kisker, C. Prokaryotic Nucleotide Excision Repair: The UvrABC System. Chem. Rev. 2006, 106, 233–252. [Google Scholar] [CrossRef] [PubMed]
- Kemp, M.G.; Reardon, J.T.; Lindsey-Boltz, L.A.; Sancar, A. Mechanism of Release and Fate of Excised Oligonucleotides during Nucleotide Excision Repair. J. Biol. Chem. 2012, 287, 22889–22899. [Google Scholar] [CrossRef] [PubMed]
- Jaciuk, M.; Nowak, E.; Skowronek, K.; Tańska, A.; Nowotny, M. Structure of UvrA nucleotide excision repair protein in complex with modified DNA. Nat. Struct. Mol. Biol. 2011, 18, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Jaciuk, M.; Swuec, P.; Gaur, V.; Kasprzak, J.M.; Renault, L.; Dobrychłop, M.; Nirwal, S.; Bujnicki, J.M.; Costa, A.; Nowotny, M. A combined structural and biochemical approach reveals translocation and stalling of UvrB on the DNA lesion as a mechanism of damage verification in bacterial nucleotide excision repair. DNA Repair 2019, 85, 102746. [Google Scholar] [CrossRef] [PubMed]
- Sancar, A.; Reardon, J.T. Nucleotide Excision Repair in E. Coli and Man. In Advances in Protein Chemistry; Elsevier: Amsterdam, The Netherlands, 2004; Volume 69, pp. 43–71. [Google Scholar]
- Spivak, G. Nucleotide excision repair in humans. DNA Repair 2015, 36, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Fuss, J.O.; Cheng, Q.J.; Arvai, A.S.; Hammel, M.; Roberts, V.A.; Cooper, P.K.; Tainer, J.A. XPD Helicase Structures and Activities: Insights into the Cancer and Aging Phenotypes from XPD Mutations. Cell 2008, 133, 789–800. [Google Scholar] [CrossRef]
- Shuck, S.C.; Short, E.A.; Turchi, J.J. Eukaryotic nucleotide excision repair: From understanding mechanisms to influencing biology. Cell Res. 2008, 18, 64–72. [Google Scholar] [CrossRef]
- Crowley, D.J.; Boubriak, I.; Berquist, B.R.; Clark, M.; Richard, E.; Sullivan, L.; DasSarma, S.; McCready, S. The uvrA, uvrB and uvrC genes are required for repair of ultraviolet light induced DNA photoproducts in Halobacterium sp. NRC-1. Saline Syst. 2006, 2, 11. [Google Scholar] [CrossRef][Green Version]
- Lestini, R.; Duan, Z.; Allers, T. The archaeal Xpf/Mus81/FANCM homolog Hef and the Holliday junction resolvase Hjc define alternative pathways that are essential for cell viability in Haloferax volcanii. DNA Repair 2010, 9, 994–1002. [Google Scholar] [CrossRef]
- White, M.F.; Allers, T. DNA repair in the archaea—An emerging picture. FEMS Microbiol. Rev. 2018, 514–526. [Google Scholar] [CrossRef]
- Kuper, J.; Wolski, S.C.; Michels, G.; Kisker, C. Functional and structural studies of the nucleotide excision repair helicase XPD suggest a polarity for DNA translocation. EMBO J. 2011, 31, 494–502. [Google Scholar] [CrossRef]
- Wolski, S.C.; Kuper, J.; Hänzelmann, P.; Truglio, J.J.; Croteau, D.L.; Van Houten, B.; Kisker, C. Crystal Structure of the FeS Cluster–Containing Nucleotide Excision Repair Helicase XPD. PLoS Biol. 2008, 6, e149. [Google Scholar] [CrossRef] [PubMed]
- Fujikane, R.; Ishino, S.; Ishino, Y.; Forterre, P. Genetic analysis of DNA repair in the hyperthermophilic archaeon, Thermococcus kodakaraensis. Genes Genet. Syst. 2010, 85, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Rouillon, C.; White, M.F. The XBP-Bax1 Helicase-Nuclease Complex Unwinds and Cleaves DNA. J. Biol. Chem. 2010, 285, 11013–11022. [Google Scholar] [CrossRef]
- Feng, L.; Chang, C.-C.; Song, N.; Jiang, C.; Song, Y.; Wang, C.-F.; Deng, W.; Zou, Y.-J.; Chen, H.-F.; Xiao, X.; et al. The trimeric Hef-associated nuclease HAN is a 3′→5′ exonuclease and is probably involved in DNA repair. Nucleic Acids Res. 2018, 46, 9027–9043. [Google Scholar] [CrossRef] [PubMed]
- Lestini, R.; Delpech, F.; Myllykallio, H. DNA replication restart and cellular dynamics of Hef helicase/nuclease protein in Haloferax volcanii. Biochimie 2015, 118, 254–263. [Google Scholar] [CrossRef]
- Lestini, R.; Laptenok, S.P.; Kühn, J.; Hink, M.A.; Schanne-Klein, M.-C.; Liebl, U.; Myllykallio, H. Intracellular dynamics of archaeal FANCM homologue Hef in response to halted DNA replication. Nucleic Acids Res. 2013, 41, 10358–10370. [Google Scholar] [CrossRef]
- Dorazi, R.; Götz, D.; Munro, S.; Bernander, R.; White, M.F. Equal rates of repair of DNA photoproducts in transcribed and non-transcribed strands in Sulfolobus solfataricus. Mol. Microbiol. 2006, 63, 521–529. [Google Scholar] [CrossRef]
- Stantial, N.; Dumpe, J.; Pietrosimone, K.; Baltazar, F.; Crowley, D.J. Transcription-coupled repair of UV damage in the halophilic archaea. DNA Repair 2016, 41, 63–68. [Google Scholar] [CrossRef]
- Gehring, A.M.; Santangelo, T.J. Archaeal RNA polymerase arrests transcription at DNA lesions. Transcription 2017, 8, 288–296. [Google Scholar] [CrossRef]
- Xu, J.; Lahiri, I.; Wang, W.; Wier, A.; Cianfrocco, M.A.; Chong, J.; Hare, A.A.; Dervan, P.B.; DiMaio, F.; Leschziner, A.E.; et al. Structural basis for the initiation of eukaryotic transcription-coupled DNA repair. Nature 2017, 551, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Deaconescu, A.M.; Suhanovsky, M.M. From Mfd to TRCF and Back Again-A Perspective on Bacterial Transcription-coupled Nucleotide Excision Repair. Photochem. Photobiol. 2017, 93, 268–279. [Google Scholar] [CrossRef]
- Deaconescu, A.M.; Sevostyanova, A.; Artsimovitch, I.; Grigorieff, N. Nucleotide excision repair (NER) machinery recruitment by the transcription-repair coupling factor involves unmasking of a conserved intramolecular interface. Proc. Natl. Acad. Sci. USA 2012, 109, 3353–3358. [Google Scholar] [CrossRef] [PubMed]
- Adebali, O.; Chiou, Y.-Y.; Hu, J.; Sancar, A.; Selby, C.P. Genome-wide transcription-coupled repair in Escherichia coli is mediated by the Mfd translocase. Proc. Natl. Acad. Sci. USA 2017, 114, E2116–E2125. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.E.; Luyties, O.; Santangelo, T.J. Factor-dependent archaeal transcription termination. Proc. Natl. Acad. Sci. USA 2017, 114, E6767–E6773. [Google Scholar] [CrossRef] [PubMed]
- Phung, D.K.; Etienne, C.; Batista, M.; Langendijk-Genevaux, P.; Moalic, Y.; Laurent, S.; Liuu, S.; Morales, V.; Jebbar, M.; Fichant, G.; et al. RNA processing machineries in Archaea: The 5′-3′ exoribonuclease aRNase J of the β-CASP family is engaged specifically with the helicase ASH-Ski2 and the 3’-5’ exoribonucleolytic RNA exosome machinery. Nucleic Acids Res. 2020, 48, 3832–3847. [Google Scholar] [CrossRef] [PubMed]
- Le, T.T.; Yang, Y.; Tan, C.; Suhanovsky, M.M.; Fulbright, R.M.; Inman, J.T.; Li, M.; Lee, J.; Perelman, S.; Roberts, J.W.; et al. Mfd Dynamically Regulates Transcription via a Release and Catch-Up Mechanism. Cell 2018, 172, 344–357.e15. [Google Scholar] [CrossRef]
- Schalow, B.J.; Courcelle, C.T.; Courcelle, J. Mfd Is Required for Rapid Recovery of Transcription following UV-Induced DNA Damage but Not Oxidative DNA Damage in Escherichia coli. J. Bacteriol. 2012, 194, 2637–2645. [Google Scholar] [CrossRef]
- Delpech, F.; Collien, Y.; Mahou, P.; Beaurepaire, E.; Myllykallio, H.; Lestini, R. Snapshots of archaeal DNA replication and repair in living cells using super-resolution imaging. Nucleic Acids Res. 2018, 46, 10757–10770. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marshall, C.J.; Santangelo, T.J. Archaeal DNA Repair Mechanisms. Biomolecules 2020, 10, 1472. https://doi.org/10.3390/biom10111472
Marshall CJ, Santangelo TJ. Archaeal DNA Repair Mechanisms. Biomolecules. 2020; 10(11):1472. https://doi.org/10.3390/biom10111472
Chicago/Turabian StyleMarshall, Craig J., and Thomas J. Santangelo. 2020. "Archaeal DNA Repair Mechanisms" Biomolecules 10, no. 11: 1472. https://doi.org/10.3390/biom10111472
APA StyleMarshall, C. J., & Santangelo, T. J. (2020). Archaeal DNA Repair Mechanisms. Biomolecules, 10(11), 1472. https://doi.org/10.3390/biom10111472