The Role of Stress Proteins in Haloarchaea and Their Adaptive Response to Environmental Shifts
Abstract
1. Introduction
2. Stress in Halophilic Archaea
3. Response to Stress in Halophilic Archaea
3.1. Heat Shock and Stress Proteins
3.1.1. Molecular Chaperones
3.1.2. Peptidyl-Propyl Cis–Trans Isomerases
3.2. Thermoprotectants
3.3. Multicellular Structures
3.4. Other Stress Proteins
3.4.1. DNA Repair Proteins
3.4.2. Proteasome
3.4.3. Survival Proteins
3.4.4. Transcriptional Factors
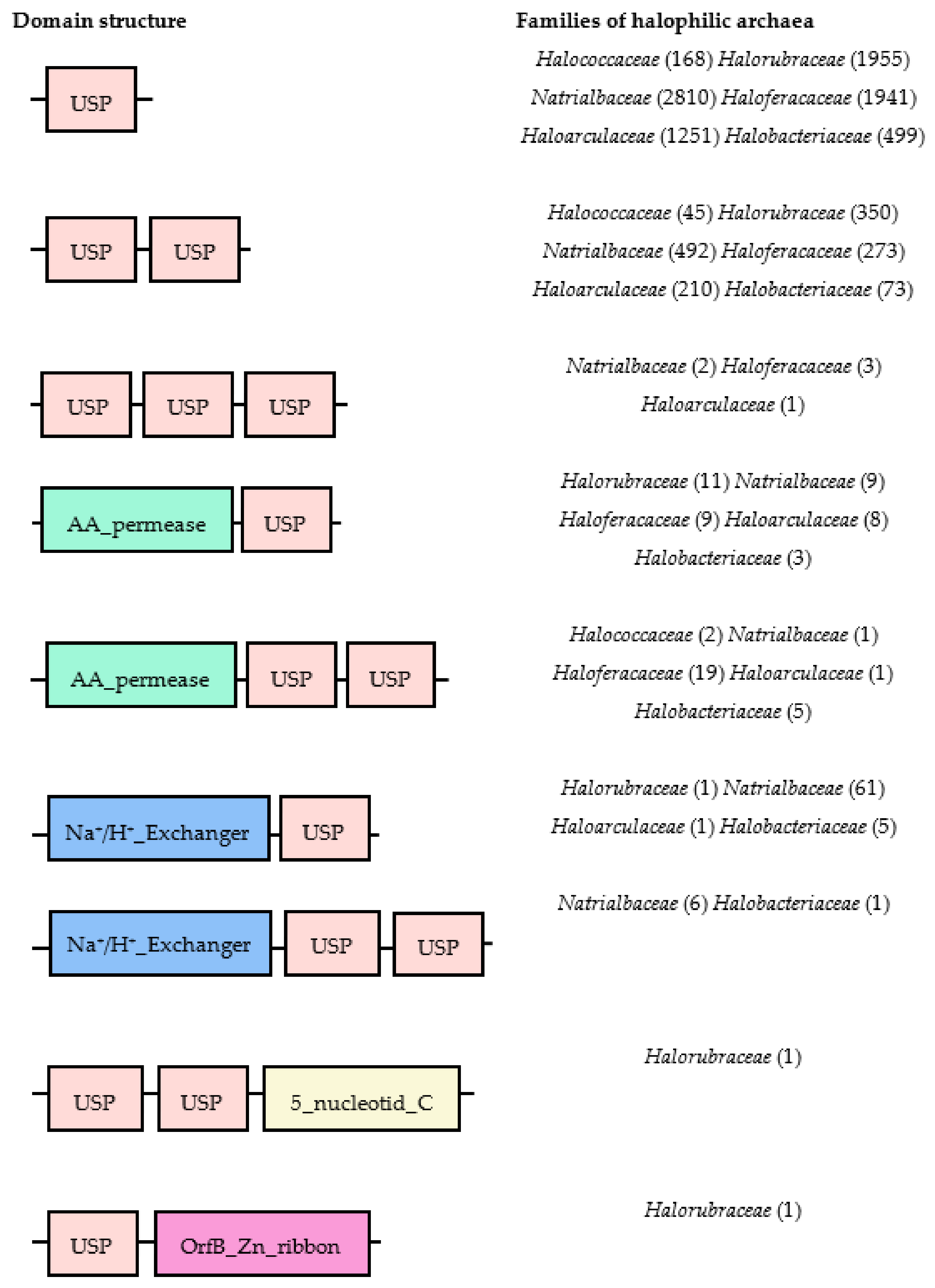
4. Universal Stress Proteins among Three Domains of Life
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Madigan, M.; Bender, K.S.; Buckley, D.H.; Sattley, W.M.; Stahl, D.A. Brock Biology of Microorganisms, 15th ed.; Pearson: London, UK, 2017; ISBN 9780134261928. [Google Scholar]
- Conway de Macario, E.; Macario, A.J. Stressors, stress and survival: Overview. Front. Biosci. 2000, 5, D780–D786. [Google Scholar] [CrossRef] [PubMed]
- Macario, A.J.L.; Conway de Macario, E. Stress and molecular chaperones in disease. Int. J. Clin. Lab. Res. 2000, 30, 49–66. [Google Scholar] [CrossRef] [PubMed]
- Macario, A.J.L.; Conway de Macario, E. Heat Shock Response, Overview. In The Encyclopedia of Stress; Academic Press: San Diego, CA, USA, 2000; p. 350. ISBN 9780123739476. [Google Scholar]
- Macario, A.J.; Conway De Macario, E. The molecular chaperone system and other anti-stress mechanisms in archaea. Front. Biosci. 2001, 6. [Google Scholar] [CrossRef] [PubMed]
- Baliga, N.S.; Bjork, S.J.; Bonneau, R.; Pan, M.; Iloanusi, C.; Kottemann, M.C.H.; Hood, L.; DiRuggiero, J. Systems level insights into the stress response to UV radiation in the halophilic archaeon Halobacterium NRC-1. Genome Res. 2004, 14, 1025–1035. [Google Scholar] [CrossRef]
- Thombre, R.S.; Shinde, V.D.; Oke, R.S.; Dhar, S.K.; Shouche, Y.S. Biology and survival of extremely halophilic archaeon Haloarcula marismortui RR12 isolated from Mumbai salterns, India in response to salinity stress. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Grant, W.D.; Kamekura, M.; McGenity, T.J.; Ventosa, A.; Holt, J.G. The History of Bergey’s ManualClass III. Halobacteria class. nov. In Bergey’s Manual of Systematic Bacteriology; Springer: New York, NY, USA, 2001; p. 294. [Google Scholar]
- Torregrosa-Crespo, J.; Bergaust, L.; Pire, C.; Martínez-Espinosa, R.M. Denitrifying haloarchaea: Sources and sinks of nitrogenous gases. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef]
- Sánchez, E.; Domínguez, M.; Romera, R.; de la Franca, N.L.; Gaertner, M.A.; Gallardo, C.; Castro, M. Regional modeling of dry spells over the Iberian Peninsula for present climate and climate change conditions. Clim. Chang. 2011, 107, 625–634. [Google Scholar] [CrossRef]
- Field, C.B.; Barros, V.; Stocker, T.F.; Dahe, Q.; Dokken, D.J.; Plattner, G.-K.; Ebi, K.L.; Allen, S.K.; Mastrandrea, M.D.; Tignor, M.; et al. Managing the Risks of Extreme Events and Disasters to Advance Climate Change Adaptation. A Special Report of Working Groups I and II of the Intergovernmental Panel on Climate Change; Cambridge University: Cambridge, UK, 2012. [Google Scholar]
- Gornall, J.; Betts, R.; Burke, E.; Clark, R.; Camp, J.; Willett, K.; Wiltshire, A. Implications of climate change for agricultural productivity in the early twenty-first century. Phil. Trans. R. Soc. B 2010, 365, 2973–2989. [Google Scholar] [CrossRef]
- Griffiths, B.S.; Philippot, L. Insights into the resistance and resilience of the soil microbial community. FEMS Microbiol. Rev. 2013, 37, 112–129. [Google Scholar] [CrossRef]
- Zafrilla, B.; Martínez-Espinosa, R.M.; Alonso, M.A.; Bonete, M.J. Biodiversity of Archaea and floral of two inland saltern ecosystems in the Alto Vinalopó Valley, Spain. Saline Syst. 2010, 6, 10. [Google Scholar] [CrossRef]
- Oren, A. Molecular ecology of extremely halophilic Archaea and Bacteria. FEMS Microbiol. Ecol. 2002, 39, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Andrei, A.Ş.; Banciu, H.L.; Oren, A. Living with salt: Metabolic and phylogenetic diversity of archaea inhabiting saline ecosystems. FEMS Microbiol. Lett. 2012, 330, 1–9. [Google Scholar] [CrossRef] [PubMed]
- DasSarma, P.; Capes, M.D.; DasSarma, S. Comparative Genomics of Halobacterium Strains From Diverse Locations. In Microbial Diversity in the Genomic Era; Elsevier: Cambridge, MA, USA, 2019; pp. 285–322. [Google Scholar]
- Delgado-Baquerizo, M.; Maestre, F.T.; Gallardo, A.; Bowker, M.A.; Wallenstein, M.D.; Quero, J.L.; Ochoa, V.; Gozalo, B.; García-Gómez, M.; Soliveres, S.; et al. Decoupling of soil nutrient cycles as a function of aridity in global drylands. Nature 2013, 502, 672–676. [Google Scholar] [CrossRef]
- Oren, A. Microbial life at high salt concentrations: Phylogenetic and metabolic diversity. Saline Syst. 2008, 4, 2. [Google Scholar] [CrossRef]
- Kennedy, S.P.; Ng, W.V.; Salzberg, S.L.; Hood, L.; DasSarma, S. Understanding the adaptation of Halobacterium species NRC-1 to its extreme environment through computational analysis of its genome sequence. Genome Res. 2001, 11, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Britton, K.L.; Baker, P.J.; Fisher, M.; Ruzheinikov, S.; Gilmour, D.J.; Bonete, M.J.; Ferrer, J.; Pire, C.; Esclapez, J.; Rice, D.W. Analysis of protein solvent interactions in glucose dehydrogenase from the extreme halophile Haloferax mediterranei. Proc. Natl. Acad. Sci. USA 2006, 103, 4846–4851. [Google Scholar] [CrossRef]
- Esclapez, J.; Pire, C.; Bautista, V.; Martínez-Espinosa, R.M.; Ferrer, J.; Bonete, M.J. Analysis of acidic surface of Haloferax mediterranei glucose dehydrogenase by site-directed mutagenesis. FEBS Lett. 2007, 581, 837–842. [Google Scholar] [CrossRef]
- Williams, W.D. Anthropogenic salinisation of inland waters. In Saline Lakes; Springer: Dordrecht, The Netherlands, 2001; pp. 329–337. [Google Scholar]
- Oren, A. Solar salterns as model systems for the study of halophilic microorganisms in their natural environments. In Model Ecosystems in Extreme Environments; Elsevier: Cambridge, MA, USA, 2019; pp. 41–56. [Google Scholar]
- Coker, J.; DasSarma, P.; Kumar, J.; Müller, J.A.; DasSarma, S. Transcriptional profiling of the model Archaeon Halobacterium sp. NRC-1: responses to changes in salinity and temperature. Saline Syst. 2007, 3, 6. [Google Scholar] [CrossRef]
- Leuko, S.; Raftery, M.J.; Burns, B.P.; Walter, M.R.; Neilan, B.A. Global Protein-Level Responses of Halobacterium salinarum NRC-1 to Prolonged Changes in External Sodium Chloride Concentrations. J. Proteome Res. 2009, 8, 2218–2225. [Google Scholar] [CrossRef]
- Mojica, F.J.; Juez, G.; Rodriguez-Valera, F. Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites. Mol. Microbiol. 1993, 9, 613–621. [Google Scholar] [CrossRef]
- Ferrer, C.; Mojica, F.J.; Juez, G.; Rodriguez-Valera, F. Differentially transcribed regions of Haloferax volcanii genome depending on the medium salinity. J. Bacteriol. 1996, 178, 309–313. [Google Scholar] [CrossRef] [PubMed]
- Kurt-Kızıldoğan, A.; Abanoz, B.; Okay, S. Global transcriptome analysis of Halolamina sp. to decipher the salt tolerance in extremely halophilic archaea. Gene 2017, 601, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Jevtic, Z.; Stoll, B.; Pfeiffer, F.; Sharma, K.; Urlaub, H.; Marchfelder, A.; Lenz, C. The Response of Haloferax volcanii to Salt and Temperature Stress: A Proteome Study by Label-Free Mass Spectrometry. Proteom. 2019, 19, e1800491. [Google Scholar] [CrossRef] [PubMed]
- Bidle, K.A.; Kirkland, P.A.; Nannen, J.L.; Maupin-Furlow, J.A. Proteomic analysis of Haloferax volcanii reveals salinity-mediated regulation of the stress response protein PspA. Microbiology 2008, 154, 1436–1443. [Google Scholar] [CrossRef] [PubMed]
- Leuko, S.; Neilan, B.A.; Burns, B.P.; Walter, M.R.; Rothschild, L.J. Molecular assessment of UVC radiation-induced DNA damage repair in the stromatolitic halophilic archaeon, Halococcus hamelinensis. J. Photochem. Photobiol. B: Biol. 2011, 102, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Stantial, N.; Dumpe, J.; Pietrosimone, K.; Baltazar, F.; Crowley, D.J. Transcription-coupled repair of UV damage in the halophilic archaea. DNA Repair 2016, 41, 63–68. [Google Scholar] [CrossRef]
- Moran-Reyna, A.; A Coker, J. The effects of extremes of pH on the growth and transcriptomic profiles of three haloarchaea. F1000Research 2014, 3, 168. [Google Scholar] [CrossRef]
- Sharma, K.; Gillum, N.; Boyd, J.L.; Schmid, A.K. The RosR transcription factor is required for gene expression dynamics in response to extreme oxidative stress in a hypersaline-adapted archaeon. BMC Genom. 2012, 13, 351. [Google Scholar] [CrossRef]
- Gelsinger, D.R.; DiRuggiero, J. Transcriptional Landscape and Regulatory Roles of Small Noncoding RNAs in the Oxidative Stress Response of the Haloarchaeon Haloferax volcanii. J. Bacteriol. 2018, 200, e00779-17. [Google Scholar] [CrossRef]
- McMillan, L.J.; Hwang, S.; Farah, R.E.; Koh, J.; Chen, S.; Maupin-Furlow, J.A. Multiplex quantitative SILAC for analysis of archaeal proteomes: a case study of oxidative stress responses. Environ. Microbiol. 2017, 20, 385–401. [Google Scholar] [CrossRef]
- Gomez, M.; Leung, W.; Dantuluri, S.; Pillai, A.; Gani, Z.; Hwang, S.; McMillan, L.J.; Kiljunen, S.; Savilahti, H.; Maupin-Furlow, J.A. Molecular Factors of Hypochlorite Tolerance in the Hypersaline Archaeon Haloferax volcanii. Genes 2018, 9, 562. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Kennedy, S.; Fasiludeen, S.; Rensing, C.; DasSarma, S. Arsenic Resistance in Halobacterium sp. Strain NRC-1 Examined by Using an Improved Gene Knockout System. J. Bacteriol. 2004, 186, 3187–3194. [Google Scholar] [CrossRef] [PubMed]
- Buda, D.-M.; Bulzu, P.-A.; Barbu-Tudoran, L.; Porfire, A.; Pătraș, L.; Sesărman, A.; Tripon, S.; Șenilă, M.; Ionescu, M.I.; Banciu, H.L.; et al. Physiological response to silver toxicity in the extremely halophilic archaeon Halomicrobium mukohataei. FEMS Microbiol. Lett. 2019, 366. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, A.; Imran Pasha, M.; Salgaonkar, B.B.; Bragança, J.M. Cadmium Tolerance by Haloarchaeal Strains Isolated from Solar Salterns of Goa, India. Int. J. Biosci. Biochem. Bioinform. 2014, 1–6. [Google Scholar] [CrossRef]
- Sehgal, S.N.; Gibbons, N.E. Effect of some metal ions on the growth of Halobacterium cutirubrum. Can. J. Microbiol. 1960, 6, 165–169. [Google Scholar] [CrossRef]
- Nieto, J.J.; Ventosa, A.; Ruiz-Berraquero, F. Susceptibility of Halobacteria to Heavy Metals. Appl. Environ. Microbiol. 1987, 53, 1199–1202. [Google Scholar] [CrossRef]
- Weng, R.R.; Shu, H.-W.; Chin, S.-W.; Kao, Y.; Chen, T.-W.; Liao, C.-C.; Tsay, Y.-G.; Ng, W.V. OMICS in Ecology: Systems Level Analyses of Halobacterium salinarum Reveal Large-scale Temperature-Mediated Changes and a Requirement of CctA for Thermotolerance. OMICS J. Integr. Biol. 2014, 18, 65–80. [Google Scholar] [CrossRef]
- Gruber, C.; Legat, A.; Pfaffenhuemer, M.; Radax, C.; Weidler, G.; Stan-Lotter, H. Halobacterium noricense sp. nov., an archaeal isolate from a bore core of an alpine Permian salt deposit, classification of Halobacterium sp. NRC-1 as a strain of H. salinarum and emended description of H. salinarum. Extremophiles 2004, 8, 431–439. [Google Scholar] [CrossRef]
- Lledó, B.; Martı́nez-Espinosa, R.; Marhuenda-Egea, F.; Bonete, M.-J.; Marhuenda-Egea, F.C. Respiratory nitrate reductase from haloarchaeon Haloferax mediterranei: biochemical and genetic analysis. Biochim. Biophys. Acta (BBA) - Gen. Subj. 2004, 1674, 50–59. [Google Scholar] [CrossRef]
- Martínez-Espinosa, R.M.; Lledó, B.; Marhuenda-Egea, F.C.; Bonete, M.-J. The effect of ammonium on assimilatory nitrate reduction in the haloarchaeon Haloferax mediterranei. Extremophiles 2007, 11, 759–767. [Google Scholar] [CrossRef]
- Oren, A.; Bardavid, R.E.; Mana, L. Perchlorate and halophilic prokaryotes: implications for possible halophilic life on Mars. Extremophiles 2013, 18, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Becker, E.; Seitzer, P.M.; Tritt, A.; Larsen, D.; Krusor, M.; Yao, A.I.; Wu, N.; Madern, M.; Eisen, J.A.; E Darling, A.; et al. Phylogenetically Driven Sequencing of Extremely Halophilic Archaea Reveals Strategies for Static and Dynamic Osmo-response. PLoS Genet. 2014, 10, e1004784. [Google Scholar] [CrossRef] [PubMed]
- Oren, A. Life at High Salt Concentrations. In The Prokaryotes; Springer Science and Business Media LLC: New York, NY, USA, 2013; Volume 9783642301230, pp. 421–440. [Google Scholar]
- Schäfer, G.; Purschke, W.; Schmidt, C.L. On the origin of respiration: electron transport proteins from archaea to man. FEMS Microbiol. Rev. 1996, 18, 173–188. [Google Scholar] [CrossRef][Green Version]
- Grant, W.D. Life at low water activity. Philos. Trans. R. Soc. B Biol. Sci. 2004, 359, 1249–1267. [Google Scholar] [CrossRef] [PubMed]
- Ali, I.; Prasongsuk, S.; Akbar, A.; Aslam, M.; Lotrakul, P.; Punnapayak, H.; Rakshit, S.K. Hypersaline habitats and halophilic microorganisms. Maejo Int. J. Sci. Technol 2015, 10, 330–345. [Google Scholar]
- Ng, W.V.; Kennedy, S.; Mahairas, G.G.; Berquist, B.; Pan, M.; Shukla, H.D.; Lasky, S.R.; Baliga, N.S.; Thorsson, V.; Sbrogna, J.; et al. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA 2000, 97, 12176–12181. [Google Scholar] [CrossRef]
- DasSarma, S. Genome sequence of an extremely halophilic archaeon. In Microbial Genomes; Fraser, C.M., Ed.; Humana Press Inc.: Totowa, NJ, USA, 2004; pp. 383–399. [Google Scholar]
- Bowers, K.J.; Wiegel, J. Temperature and pH optima of extremely halophilic archaea: a mini-review. Extremophiles 2011, 15, 119–128. [Google Scholar] [CrossRef]
- Ellis, R.J. Chaperone Function: The Orthodox View. In Molecular Chaperones and Cell Signalling; Cambridge University Press (CUP): Cambridge, MA, USA, 2005; pp. 3–21. [Google Scholar]
- Shukla, H.D. Proteomic analysis of acidic chaperones, and stress proteins in extreme halophile Halobacterium NRC-1: a comparative proteomic approach to study heat shock response. Proteome Sci. 2006, 4, 6. [Google Scholar] [CrossRef]
- Mormile, M.R.; Hong, B.-Y.; Benison, K.C. Molecular Analysis of the Microbial Communities of Mars Analog Lakes in Western Australia. Astrobiology 2009, 9, 919–930. [Google Scholar] [CrossRef]
- Kulp, T.R.; Han, S.; Saltikov, C.W.; Lanoil, B.D.; Zargar, K.; Oremland, R.S. Effects of Imposed Salinity Gradients on Dissimilatory Arsenate Reduction, Sulfate Reduction, and Other Microbial Processes in Sediments from Two California Soda Lakes. Appl. Environ. Microbiol. 2007, 73, 5130–5137. [Google Scholar] [CrossRef]
- Soliman, G.S.; Trüper, H.G. Halobacterium pharaonis sp. nov., a New, Extremely Haloalkaliphilic Archaebacterium with Low Magnesium Requirement. Zentralblatt für Bakteriologie Mikrobiologie und Hygiene: I. Abt. Originale C: Allgemeine, angewandte und ökologische Mikrobiologie 1982, 3, 318–329. [Google Scholar] [CrossRef]
- Tabak, H.H.; Lens, P.N.L.; Van Hullebusch, E.D.; Dejonghe, W. Developments in Bioremediation of Soils and Sediments Polluted with Metals and Radionuclides. Microbial Processes and Mechanisms Affecting Bioremediation of Metal Contamination and Influencing Metal Toxicity and Transport. Rev. Environ. Sci. Bio/Technology 2005, 4, 115–156. [Google Scholar] [CrossRef]
- Popescu, G.; Dumitru, L. Biosorption of Some Heavy Metals from Media with High Salt Concentrations by Halophilic Archaea. Biotechnol. Biotechnol. Equip. 2009, 23, 791–795. [Google Scholar] [CrossRef]
- Kaur, A.; Pan, M.; Meislin, M.; Facciotti, M.T.; El-Gewely, R.; Baliga, N.S. A systems view of haloarchaeal strategies to withstand stress from transition metals. Genome Res. 2006, 16, 841–854. [Google Scholar] [CrossRef] [PubMed]
- DasSarma, S. Extreme Microbes. Am. Sci. 2007, 95, 224–231. [Google Scholar] [CrossRef]
- Srivastava, P.; Kowshik, M. Mechanisms of Metal Resistance and Homeostasis in Haloarchaea. Archaea 2013, 2013, 1–16. [Google Scholar] [CrossRef]
- Oren, A. The Function of Gas Vesicles in Halophilic Archaea and Bacteria: Theories and Experimental Evidence. Life 2012, 3, 1–20. [Google Scholar] [CrossRef]
- Pfeifer, F. Haloarchaea and the Formation of Gas Vesicles. Life 2015, 5, 385–402. [Google Scholar] [CrossRef]
- Oren, A.; Trüper, H.G. Anaerobic growth of halophilic archaeobacteria by reduction of dimethysulfoxide and trimethylamineN-oxide. FEMS Microbiol. Lett. 1990, 70, 33–36. [Google Scholar] [CrossRef]
- Oren, A. Anaerobic growth of halophilic archaeobacteria by reduction of fumarate. J. Gen. Microbiol. 1991, 137, 1387–1390. [Google Scholar] [CrossRef]
- Müller, J.A.; DasSarma, S. Genomic Analysis of Anaerobic Respiration in the Archaeon Halobacterium sp. Strain NRC-1: Dimethyl Sulfoxide and Trimethylamine N-Oxide as Terminal Electron Acceptors. J. Bacteriol. 2005, 187, 1659–1667. [Google Scholar] [CrossRef] [PubMed]
- Burns, D.G.; Janssen, P.H.; Itoh, T.; Kamekura, M.; Li, Z.; Jensen, G.; Rodriguez-Valera, F.; Bolhuis, H.; Dyall-Smith, M.L. Haloquadratum walsbyi gen. nov., sp. nov., the square haloarchaeon of Walsby, isolated from saltern crystallizers in Australia and Spain. Int. J. Syst. Evol. Microbiol. 2007, 57, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Deveaux, L.C.; Müller, J.A.; Smith, J.; Petrisko, J.; Wells, D.P.; DasSarma, S. Extremely Radiation-Resistant Mutants of a Halophilic Archaeon with Increased Single-Stranded DNA-Binding Protein (RPA) Gene Expression. Radiat. Res. 2007, 168, 507–514. [Google Scholar] [CrossRef] [PubMed]
- McCready, S.; Müller, J.A.; Boubriak, I.; Berquist, B.R.; Ng, W.L.; DasSarma, S. UV irradiation induces homologous recombination genes in the model archaeon, Halobacterium sp. NRC-1. Saline Syst. 2005, 1, 3. [Google Scholar] [CrossRef]
- McCready, S.; Marcello, L. Repair of UV damage in Halobacterium salinarum. Biochem. Soc. Trans. 2003, 31, 694–698. [Google Scholar] [CrossRef]
- Hescox, M.A.; Carlberg, D.M. Photoreactivation in Halobacterium cutirubrum. Can. J. Microbiol. 1972, 18, 981–985. [Google Scholar] [CrossRef]
- Whitehead, K.; Kish, A.; Pan, M.; Kaur, A.; Reiss, D.J.; King, N.; Hohmann, L.; DiRuggiero, J.; Baliga, N.S. An integrated systems approach for understanding cellular responses to gamma radiation. Mol. Syst. Biol. 2006, 2, 47. [Google Scholar] [CrossRef]
- Kottemann, M.; Kish, A.; Iloanusi, C.; Bjork, S.; DiRuggiero, J. Physiological responses of the halophilic archaeon Halobacterium sp. strain NRC1 to desiccation and gamma irradiation. Extremophiles 2005, 9, 219–227. [Google Scholar] [CrossRef]
- De Macario, E.C.; Macario, A.J.L. Molecular Biology of Stress Genes in Methanogens: Potential for Bioreactor Technology. Adv. Biochem. Eng. Biotechnol. 2003, 81, 95–150. [Google Scholar] [CrossRef]
- Macario, A.J.; Lange, M.; Ahring, B.K.; de Macario, E.C. Stress Genes and Proteins in the Archaea. Microbiol Mol. Biol Rev. 1999, 64, 923–967. [Google Scholar] [CrossRef]
- Oren, A. Halophilic archaea on Earth and in space: Growth and survival under extreme conditions. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2014, 372, 20140194. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, S.; Roche, E.; Zhou, Y.; Sauer, R.T. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998, 12, 1338–1347. [Google Scholar] [CrossRef] [PubMed]
- Herman, C.; D’Ari, R. Proteolysis and chaperones: the destruction/reconstruction dilemma. Curr. Opin. Microbiol. 1998, 1, 204–209. [Google Scholar] [CrossRef]
- Martin, J.; Hartl, F.U. Chaperone-assisted protein folding. Curr. Opin. Struct. Biol. 1997, 7, 41–52. [Google Scholar] [CrossRef]
- Netzer, W.J.; Hartl, F.U. Protein folding in the cytosol: chaperonin-dependent and -independent mechanisms. Trends Biochem. Sci. 1998, 23, 68–73. [Google Scholar] [CrossRef]
- Trent, J.D.; Nimmesgern, E.; Wall, J.S.; Hartl, F.-U.; Horwich, A.L. A molecular chaperone from a thermophilic archaebacterium is related to the eukaryotic protein t-complex polypeptide-1. Nature 1991, 354, 490–493. [Google Scholar] [CrossRef]
- Condò, I.; Ruggero, D.; Reinhardt, R.; Londei, P. A novel aminopeptidase associated with the 60 kDa chaperonin in the thermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 1998, 29, 775–785. [Google Scholar] [CrossRef]
- Louvion, J.-F.; Abbas-Terki, T.; Picard, D. Hsp90 Is Required for Pheromone Signaling in Yeast. Mol. Biol. Cell 1998, 9, 3071–3083. [Google Scholar] [CrossRef]
- Ruggero, D.; Ciammaruconi, A.; Londei, P. The chaperonin of the archaeon Sulfolobus solfataricus is an RNA-binding protein that participates in ribosomal RNA processing. EMBO J. 1998, 17, 3471–3477. [Google Scholar] [CrossRef]
- Hartl, F.-U.; Martin, J. Molecular chaperones in cellular protein folding. BioEssays 1994, 16, 689–692. [Google Scholar] [CrossRef]
- Callahan, M.K.; Chaillot, D.; Jacquin, C.; Clark, P.R.; Menoret, A. Differential Acquisition of Antigenic Peptides by Hsp70 and Hsc70 under Oxidative Conditions. J. Biol. Chem. 2002, 277, 33604–33609. [Google Scholar] [CrossRef] [PubMed]
- Zmijewski, M.A.; Macario, A.J.; Lipińska, B. Functional Similarities and Differences of an Archaeal Hsp70 (DnaK) Stress Protein Compared with its Homologue from the Bacterium Escherichia coli. J. Mol. Biol. 2004, 336, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Petitjean, C.; Moreira, D.; López-García, P.; Brochier-Armanet, C. Horizontal gene transfer of a chloroplast DnaJ-Fer protein to Thaumarchaeota and the evolutionary history of the DnaK chaperone system in Archaea. BMC Evol. Biol. 2012, 12, 226. [Google Scholar] [CrossRef] [PubMed]
- Shih, C.-J.; Lai, M.-C. Differentially expressed genes after hyper- and hypo-salt stress in the halophilic archaeon Methanohalophilus portucalensis. Can. J. Microbiol. 2010, 56, 295–307. [Google Scholar] [CrossRef]
- Bukau, B.; Deuerling, E.; Pfund, C.; A Craig, E. Getting Newly Synthesized Proteins into Shape. Cell 2000, 101, 119–122. [Google Scholar] [CrossRef]
- Ellis, R.J.; Hartl, F.U. Principles of protein folding in the cellular environment. Curr. Opin. Struct. Biol. 1999, 9, 102–110. [Google Scholar] [CrossRef]
- Frydman, J. Chaperones get in touch: the Hip-Hop connection. Trends Biochem. Sci. 1997, 22, 87–92. [Google Scholar] [CrossRef]
- Hartl, F.U. Molecular chaperones in cellular protein folding. Nature 1996, 381, 571–580. [Google Scholar] [CrossRef]
- Pfanner, N. Protein folding: Who chaperones nascent chains in bacteria? Curr. Biol. 1999, 9. [Google Scholar] [CrossRef][Green Version]
- De Macario, E.C.; Macario, A.J. Heat-shock response in Archaea. Trends Biotechnol. 1994, 12, 512–518. [Google Scholar] [CrossRef]
- Lin, W.; Bonin, M.; Boden, A.; Wieduwild, R.; Murawala, P.; Wermke, M.; Andrade, H.; Bornhäuser, M.; Zhang, Y. Peptidyl prolyl cis/trans isomerase activity on the cell surface correlates with extracellular matrix development. Commun. Biol. 2019, 2, 1–11. [Google Scholar] [CrossRef]
- Fischer, G. Peptidyl-Prolyl cis/trans Isomerases and Their Effectors. Angew. Chem. Int. Ed. Engl. 1994, 33, 1415–1436. [Google Scholar] [CrossRef]
- Rudd, K.E.; Sofia, H.J.; Koonin, E.V.; Plunkett, G.; Lazar, S.; Rouvière, P.E. A new family of peptidyl-prolyl isomerases. Trends Biochem. Sci. 1995, 20, 12–14. [Google Scholar] [CrossRef]
- Boonyaratanakornkit, B.B.; Simpson, A.J.; Whitehead, T.A.; Fraser, C.; El-Sayed, N.M.; Clark, D.S. Transcriptional profiling of the hyperthermophilic methanarchaeon Methanococcus jannaschii in response to lethal heat and non-lethal cold shock. Environ. Microbiol. 2005, 7, 789–797. [Google Scholar] [CrossRef]
- Yoshinaga, K.; Yoshioka, H.; Kurosaki, H.; Hirasawa, M.; Uritani, M.; Hasegawa, K. Protection by Trehalose of DNA from Radiation Damage. Biosci. Biotechnol. Biochem. 1997, 61, 160–161. [Google Scholar] [CrossRef] [PubMed]
- Elbein, A.D.; Pan, Y.; Pastuszak, I.; Carroll, D. New insights on trehalose: a multifunctional molecule. Glycobiol. 2003, 13, 17R–27. [Google Scholar] [CrossRef] [PubMed]
- Leuko, S.; Domingos, C.; Parpart, A.; Reitz, G.; Rettberg, P. The Survival and Resistance of Halobacterium salinarum NRC-1, Halococcus hamelinensis, and Halococcus morrhuae to Simulated Outer Space Solar Radiation. Astrobiol. 2015, 15, 987–997. [Google Scholar] [CrossRef]
- Fröls, S. Archaeal biofilms: widespread and complex. Biochem. Soc. Trans. 2013, 41, 393–398. [Google Scholar] [CrossRef]
- Mayerhofer, L.E.; Macario, A.J.; De Macario, E.C. Lamina, a novel multicellular form of Methanosarcina mazei S-6. J. Bacteriol. 1992, 174, 309–314. [Google Scholar] [CrossRef][Green Version]
- Pecl, G.T.; Araújo, M.B.; Bell, J.D.; Blanchard, J.L.; Bonebrake, T.C.; Chen, I.-C.; Clark, T.D.; Colwell, R.K.; Danielsen, F.; Evengård, B.; et al. Biodiversity redistribution under climate change: Impacts on ecosystems and human well-being. Science 2017, 355, eaai9214. [Google Scholar] [CrossRef]
- Webster, N.S.; Negri, A.P. Site-specific variation in Antarctic marine biofilms established on artificial surfaces. Environ. Microbiol. 2006, 8, 1177–1190. [Google Scholar] [CrossRef]
- Justice, N.B.; Pan, C.; Mueller, R.; Spaulding, S.E.; Shah, V.; Sun, C.L.; Yelton, A.P.; Miller, C.S.; Thomas, B.C.; Shah, M.; et al. Heterotrophic Archaea Contribute to Carbon Cycling in Low-pH, Suboxic Biofilm Communities. Appl. Environ. Microbiol. 2012, 78, 8321–8330. [Google Scholar] [CrossRef] [PubMed]
- Edwards, K.J. An Archaeal Iron-Oxidizing Extreme Acidophile Important in Acid Mine Drainage. Science 2000, 287, 1796–1799. [Google Scholar] [CrossRef] [PubMed]
- Weidler, G.W.; Gerbl, F.W.; Stan-Lotter, H. Crenarchaeota and Their Role in the Nitrogen Cycle in a Subsurface Radioactive Thermal Spring in the Austrian Central Alps. Appl. Environ. Microbiol. 2008, 74, 5934–5942. [Google Scholar] [CrossRef]
- Fröls, S.; Dyall-Smith, M.; Pfeifer, F. Biofilm formation by haloarchaea. Environ. Microbiol. 2012, 14, 3159–3174. [Google Scholar] [CrossRef]
- Crowley, D.J.; Boubriak, I.; Berquist, B.R.; Clark, M.; Richard, E.; Sullivan, L.; DasSarma, S.; McCready, S. The uvrA, uvrB and uvrC genes are required for repair of ultraviolet light induced DNA photoproducts in Halobacterium sp. NRC-1. Saline Syst. 2006, 2, 11. [Google Scholar] [CrossRef][Green Version]
- Delmas, S.; Duggin, I.G.; Allers, T. DNA damage induces nucleoid compaction via the Mre11-Rad50 complex in the archaeon Haloferax volcanii. Mol. Microbiol. 2012, 87, 168–179. [Google Scholar] [CrossRef]
- Zhao, A.; Gray, F.C.; MacNeill, S.A. ATP- and NAD+-dependent DNA ligases share an essential function in the halophilic archaeon Haloferax volcanii. Mol. Microbiol. 2006, 59, 743–752. [Google Scholar] [CrossRef]
- Reuter, C.J.; Kaczowka, S.J.; Maupin-Furlow, J.A. Differential Regulation of the PanA and PanB Proteasome-Activating Nucleotidase and 20S Proteasomal Proteins of the Haloarchaeon Haloferax volcanii. J. Bacteriol. 2004, 186, 7763–7772. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Kowalczyk, D.; Humbard, M.A.; Rohatgi, S.; Maupin-Furlow, J.A. Proteasomal Components Required for Cell Growth and Stress Responses in the Haloarchaeon Haloferax volcanii. J. Bacteriol. 2008, 190, 8096–8105. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.L.; Baxter, B.K. DNA Repair and Photoprotection: Mechanisms of Overcoming Environmental Ultraviolet Radiation Exposure in Halophilic Archaea. Front. Microbiol. 2017, 8, 1882. [Google Scholar] [CrossRef] [PubMed]
- Jun, S.-H.; Kim, T.G.; Ban, C. DNA mismatch repair system. Classical and fresh roles. FEBS J. 2006, 273, 1609–1619. [Google Scholar] [CrossRef] [PubMed]
- Majumder, P.; Baumeister, W. Proteasomes: unfoldase-assisted protein degradation machines. Biol. Chem. 2019, 401, 183–199. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 2005, 6, 79–87. [Google Scholar] [CrossRef]
- Dikic, I. Proteasomal and Autophagic Degradation Systems. Annu. Rev. Biochem. 2017, 86, 193–224. [Google Scholar] [CrossRef]
- Fu, X.; Adams, Z.; Maupin-Furlow, J.A. Assays for ubiquitin-like protein ligation and proteasome function in archaea. Methods Enzymol. 2019, 619, 161–178. [Google Scholar] [CrossRef]
- Prunetti, L.; Reuter, C.J.; Hepowit, N.L.; Wu, Y.; Barrueto, L.; Miranda, H.V.; Kelly, K.; Maupin-Furlow, J.A. Structural and biochemical properties of an extreme ’salt-loving’ proteasome activating nucleotidase from the archaeon Haloferax volcanii. Extremophiles 2013, 18, 283–293. [Google Scholar] [CrossRef]
- Kvint, K.; Nachin, L.; A Diez, A.; Nyström, T. The bacterial universal stress protein: function and regulation. Curr. Opin. Microbiol. 2003, 6, 140–145. [Google Scholar] [CrossRef]
- Perez-Rueda, E.; Hernandez-Guerrero, R.; Martínez-Núñez, M.A.; Armenta-Medina, D.; Sanchez, I.; Ibarra, J.A. Abundance, diversity and domain architecture variability in prokaryotic DNA-binding transcription factors. PLoS ONE 2018, 13, e0195332. [Google Scholar] [CrossRef]
- Plaisier, C.L.; Lo, F.-Y.; Ashworth, J.; Brooks, A.N.; Beer, K.D.; Kaur, A.; Pan, M.; Reiss, D.J.; Facciotti, M.T.; Baliga, N.S. Evolution of context dependent regulation by expansion of feast/famine regulatory proteins. BMC Syst. Biol. 2014, 8, 122. [Google Scholar] [CrossRef][Green Version]
- Busenlehner, L.S.; Pennella, M.A.; Giedroc, D.P. The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 2003, 27, 131–143. [Google Scholar] [CrossRef]
- Lemmens, L.; Maklad, H.R.; Bervoets, I.; Peeters, E. Transcription Regulators in Archaea: Homologies and Differences with Bacterial Regulators. J. Mol. Biol. 2019, 431, 4132–4146. [Google Scholar] [CrossRef] [PubMed]
- Karr, E.A. The Methanogen-Specific Transcription Factor MsvR Regulates the fpaA-rlp-rub Oxidative Stress Operon Adjacent to msvR in Methanothermobacter thermautotrophicus. J. Bacteriol. 2010, 192, 5914–5922. [Google Scholar] [CrossRef] [PubMed]
- Siegele, D.A. Universal Stress Proteins in Escherichia coli. J. Bacteriol. 2005, 187, 6253–6254. [Google Scholar] [CrossRef] [PubMed]
- Hingley-Wilson, S.; Lougheed, K.; Ferguson, K.; Leiva, S.; Williams, H.D. Individual Mycobacterium tuberculosis universal stress protein homologues are dispensable in vitro. Tuberc. 2010, 90, 236–244. [Google Scholar] [CrossRef]
- Nyström, T.; Neidhardt, F.C. Cloning, mapping and nucleotide sequencing of a gene encoding a universal stress protein in Eschericha coli. Mol. Microbiol. 1992, 6, 3187–3198. [Google Scholar] [CrossRef]
- Gustavsson, N.; A Diez, A.; Nyström, T. The universal stress protein paralogues of Escherichia coli are co-ordinately regulated and co-operate in the defence against DNA damage. Mol. Microbiol. 2002, 43, 107–117. [Google Scholar] [CrossRef]
- Nyström, T.; Neidhardt, F.C. Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol. Microbiol. 1994, 11, 537–544. [Google Scholar] [CrossRef]
- Tkaczuk, K.L.; Shumilin, I.A.; Chruszcz, M.; Evdokimova, E.; Savchenko, A.; Minor, W. Structural and functional insight into the universal stress protein family. Evol. Appl. 2013, 6, 434–449. [Google Scholar] [CrossRef]
- Hensel, M. Secreted proteins and virulence in Salmonella enterica. In Bacterial Secreted Proteins: Secretory Mechanisms and Role in Pathogenesis; Caister Academic Press: Nottingham, UK, 2009. [Google Scholar]
- Liu, W.-T.; Karavolos, M.H.; Bulmer, D.M.; Allaoui, A.; Hormaeche, R.D.C.E.; Lee, J.J.; Khan, C.A. Role of the universal stress protein UspA of Salmonella in growth arrest, stress and virulence. Microb. Pathog. 2007, 42, 2–10. [Google Scholar] [CrossRef]
- Al-Maleki, A.R.; Mariappan, V.; Vellasamy, K.M.; Shankar, E.M.; Tay, S.T.; Vadivelu, J. Enhanced intracellular survival and epithelial cell adherence abilities of Burkholderia pseudomallei morphotypes are dependent on differential expression of virulence-associated proteins during mid-logarithmic growth phase. J. Proteom. 2014, 106, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, A.C.; Bark, S. Twenty-Five Years of Investigating the Universal Stress Protein: Function, Structure, and Applications. In Advances in Applied Microbiology; Elsevier BV: Amsterdam, The Netherlands, 2018; Volume 102, pp. 1–36. [Google Scholar]
- Nachin, L.; Nannmark, U.; Nyström, T. Differential Roles of the Universal Stress Proteins of Escherichia coli in Oxidative Stress Resistance, Adhesion, and Motility. J. Bacteriol. 2005, 187, 6265–6272. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.H.; Koo, S.S.; Oh, H.T.; Lee, E.S.; Park, J.H.; Phan, K.A.T.; Wi, S.D.; Bin Bae, S.; Paeng, S.K.; Chae, H.B.; et al. The Physiological Functions of Universal Stress Proteins and Their Molecular Mechanism to Protect Plants From Environmental Stresses. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
| Stressor | Species | Reference(s) |
|---|---|---|
| Hypo-osmolarity | Halobacterium sp. NRC-1, Haloferax mediterranei, Haloferax volcanii, Halolamina sp | [25,26,27,28,29,30] |
| Hyper-osmolarity | Halobacterium sp. NRC-1, Haloferax mediterranei, Haloferax volcanii, Halolamina sp. | [25,26,27,28,29,30,31] |
| Ultraviolet radiation | Halococcus hamelinensis, Halobacterium sp. NRC-1, Haloferax volcanii | [6,32,33] |
| pH | Haloferax volcanii, Halorubrum lacusprofundi, Halobacterium sp. NRC-1 | [34] |
| H2O2 | Halobacterium sp. NRC-1, Haloferax volcanii | [35,36,37,38] |
| Heavy metals | Halobacterium sp. NRC-1, Halomicrobium mukohataei, Halococcus salifodinae, Haloferax volcanii, Haloferax mediterranei, Haloarcula japónica, Halorubrum sp., Halobacterium cutirubrum | [39,40,41,42,43] |
| Temperature | Halobacterium sp. NRC-1 Haloferax volcanii | [25,30,44] |
| Oxygen | Halobacterium sp. NRC-1, Haloferax mediterranei, Haloferax denitrificans, Haloferax gibbonsii, Haloarcula marismortui, Haloarcula vallismortis | [45,46,47,48] |
| Family Name | Subunit (kDa) | Families of Haloarchaea |
|---|---|---|
| Hsp100 Heavy Hsp, high MW | ≥100 | - |
| Hsp90 | 81–99 | Halorubraceae, Natrialbaceae, Haloferacaceae, Haloarculaceae, Halobacteriaceae |
| Hsp70 Chaperones | 65–80 | Halococcaceae, Halorubraceae, Natrialbaceae, Haloferacaceae, Haloarculaceae, Halobacteriaceae |
| Hsp60 Chaperonins | 55–64 | Halococcaceae, Halorubraceae, Natrialbaceae, Haloferacaceae, Haloarculaceae, Halobacteriaceae |
| Hsp40 DnaJ | 35–54 | Halococcaceae, Halorubraceae, Natrialbaceae, Haloferacaceae, Haloarculaceae, Halobacteriaceae |
| sHsp Small Hsp | ≤34 | Halococcaceae, Halorubraceae, Natrialbaceae, Haloferacaceae, Haloarculaceae, Halobacteriaceae |
| Others (Proteases, etc.) | various | Halococcaceae, Halorubraceae, Natrialbaceae, Haloferacaceae, Haloarculaceae, Halobacteriaceae |
| Number of Proteins | |||
|---|---|---|---|
| Haloarchaea Families | Hsp70(DnaK) | Hsp40(DnaJ) | GrpE |
| Halococcaceae | 6 | 5 | 6 |
| Halorubraceae | 66 | 63 | 69 |
| Natrialbaceae | 67 | 62 | 64 |
| Haloferacaceae | 62 | 60 | 62 |
| Haloarculaceae | 46 | 44 | 48 |
| Halobacteriaceae | 24 | 22 | 23 |
| Gene(s) | Protein | Theoretical Function | Species | Ref. |
|---|---|---|---|---|
| mutS | DNA mismatch | This protein is involved in the repair of mismatches in DNA. | Methanohalophilus portucalensis | [94] |
| uvrA uvrB uvrC | UvrABC system | The UvrABC system catalyzes the recognition and processing of DNA lesions. | Halobacterium sp. NRC-1 | [116] |
| radA | DNA repair | Involved in DNA repair and in homologous recombination. | Haloferax volcanii | [117] |
| ligA ligN | DNA ligase | Essential for DNA replication and repair of damaged DNA. | Haloferax volcanii | [118] |
| psmA psmB psmC panA panB | Proteasome subunit | Involved in protein degradation. | Haloferax volcanii | [119,120] |
| Genus | Number of USPs | Genus | Number of USPs |
|---|---|---|---|
| Halapricum | 26 | Haloplanus | 244 |
| Haloarcula | 970 | Haloprofundus | 51 |
| Halomicrobium | 118 | Haloquadratum | 106 |
| Halorhabdus | 40 | Halobaculum | 21 |
| Halorientalis | 139 | Halobium | 22 |
| Halosimplex | 32 | Halohasta | 38 |
| Natronomonas | 197 | Halolamina | 88 |
| Haladaptatus | 148 | Halonotius | 105 |
| Halalkalicoccus | 83 | Halopenitus | 59 |
| Halanaeroarchaeum | 25 | Halorubrum | 1939 |
| Halarchaeum | 47 | Salinigranum | 52 |
| Haloarchaeobius | 31 | Halobiforma | 184 |
| Halobacterium | 412 | Halopiger | 165 |
| Halodesulfurarchaeum | 21 | Halostagnicola | 107 |
| Halomarina | 21 | Haloterrigena | 428 |
| Halovenus | 28 | Halovivax | 60 |
| Natronoarchaeum | 30 | Natrarchaeobius | 174 |
| Salarchaeum | 17 | Natrialba | 352 |
| Halococcus | 278 | Natrinema | 571 |
| Halobellus | 165 | Natronobacterium | 289 |
| Haloferax | 1301 | Natronococcus | 205 |
| Halogeometricum | 194 | Natronolimnobius | 362 |
| Halogranum | 160 | Natronorubrum | 557 |
| Halopelagius | 70 | Salinarchaeum | 32 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matarredona, L.; Camacho, M.; Zafrilla, B.; Bonete, M.-J.; Esclapez, J. The Role of Stress Proteins in Haloarchaea and Their Adaptive Response to Environmental Shifts. Biomolecules 2020, 10, 1390. https://doi.org/10.3390/biom10101390
Matarredona L, Camacho M, Zafrilla B, Bonete M-J, Esclapez J. The Role of Stress Proteins in Haloarchaea and Their Adaptive Response to Environmental Shifts. Biomolecules. 2020; 10(10):1390. https://doi.org/10.3390/biom10101390
Chicago/Turabian StyleMatarredona, Laura, Mónica Camacho, Basilio Zafrilla, María-José Bonete, and Julia Esclapez. 2020. "The Role of Stress Proteins in Haloarchaea and Their Adaptive Response to Environmental Shifts" Biomolecules 10, no. 10: 1390. https://doi.org/10.3390/biom10101390
APA StyleMatarredona, L., Camacho, M., Zafrilla, B., Bonete, M.-J., & Esclapez, J. (2020). The Role of Stress Proteins in Haloarchaea and Their Adaptive Response to Environmental Shifts. Biomolecules, 10(10), 1390. https://doi.org/10.3390/biom10101390








