Voltage Sensing in Bacterial Protein Translocation
Abstract
1. Introduction
2. Materials and Methods
2.1. SecYEG Purification
2.2. SecYEG Reconstitution into Lipid Vesicles
2.3. ProOmpA-DHFR (pOD) Purification
2.4. SecA Purification
2.5. Translocation Assay
2.6. RNC Purification
2.7. Signal Peptide
2.8. Purification of Empty Ribosomes
2.9. Reconstitution of the SecYEG Complex into Planar Bilayers
2.10. Electrophysiological Measurements
2.11. Computation of Dipole Moments
3. Results
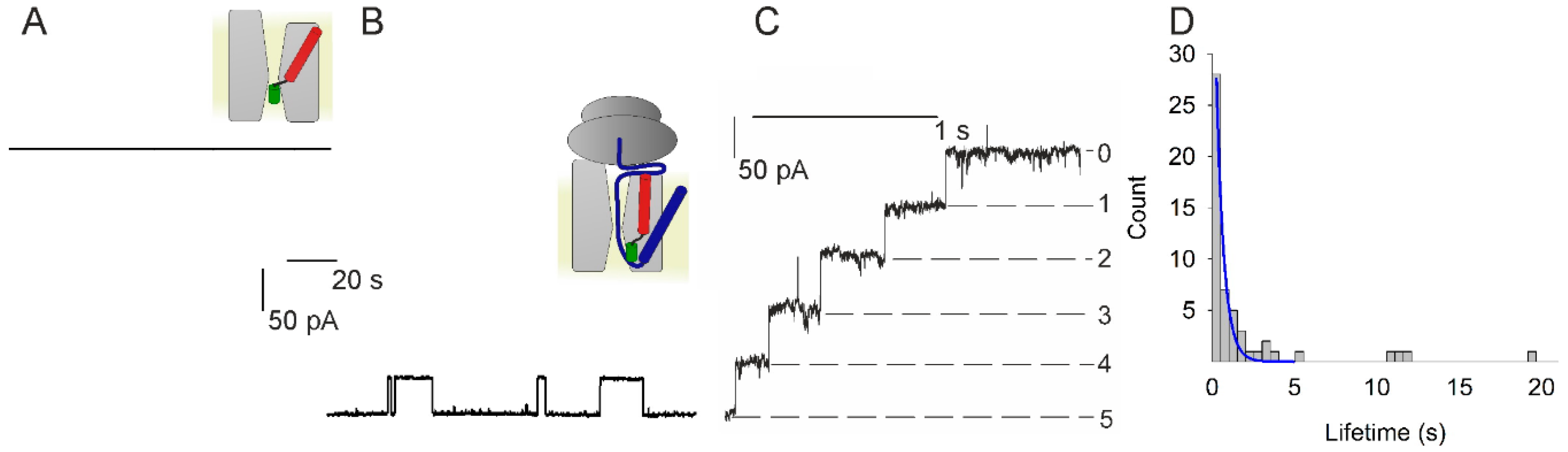
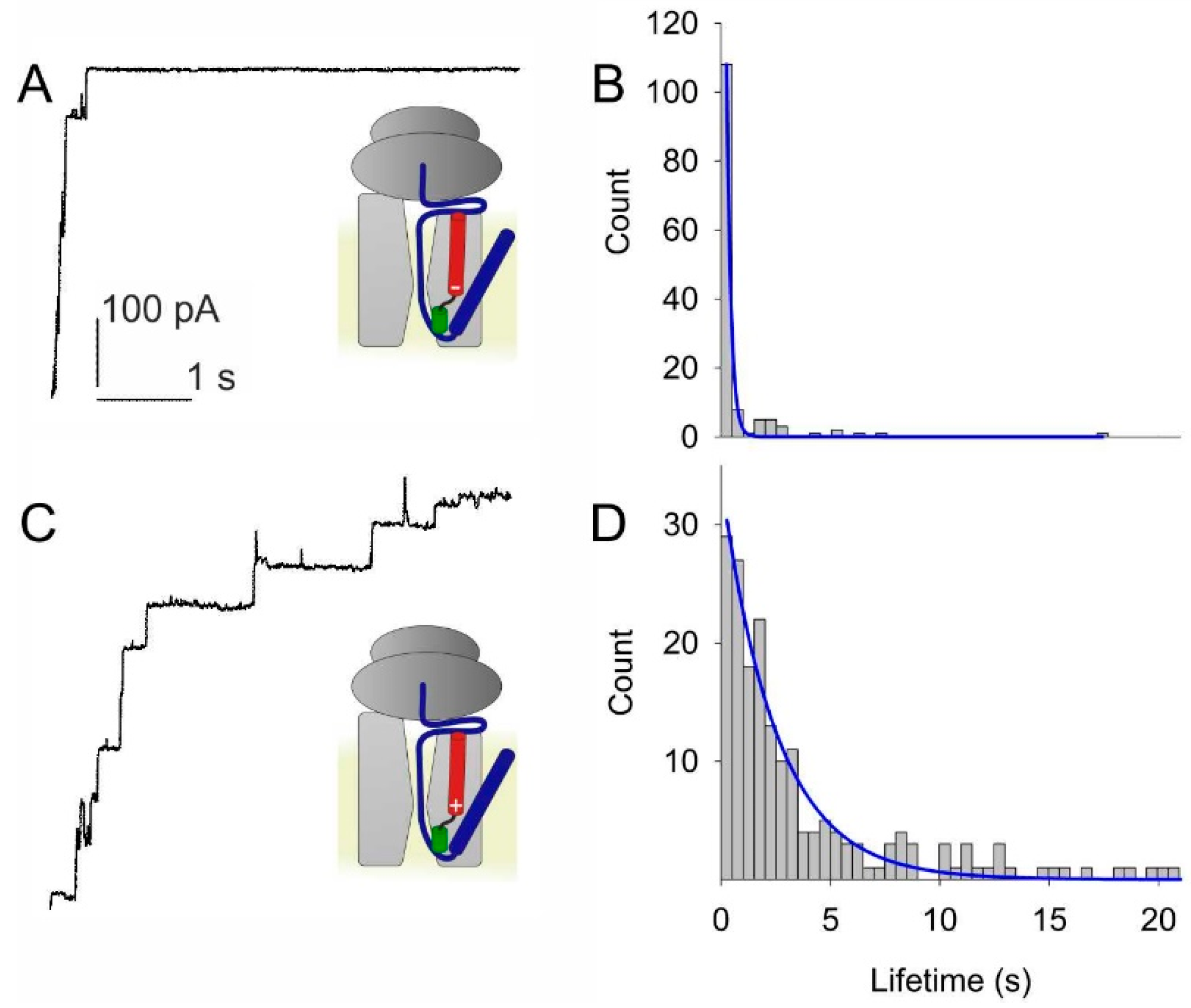
3.1. Voltage Sensitivity of SecYEG in Complex with FtsQ-RNC
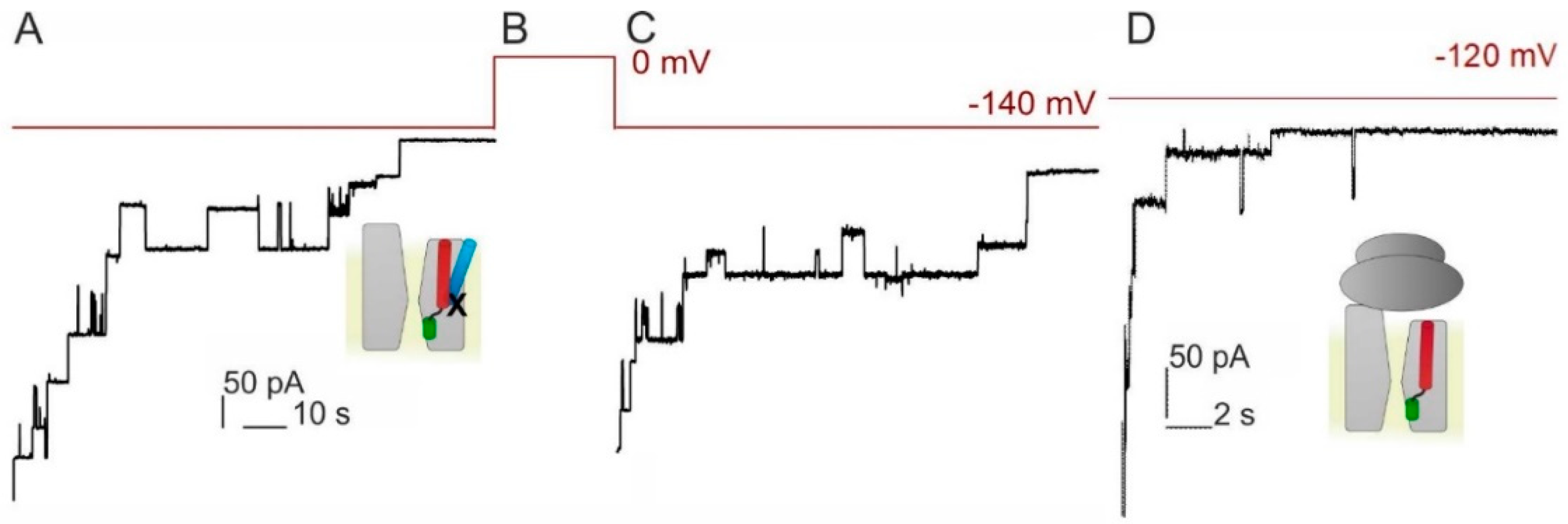
3.2. Voltage Sensitivity of SecYEG Is Not Granted by the Translocated Substrate
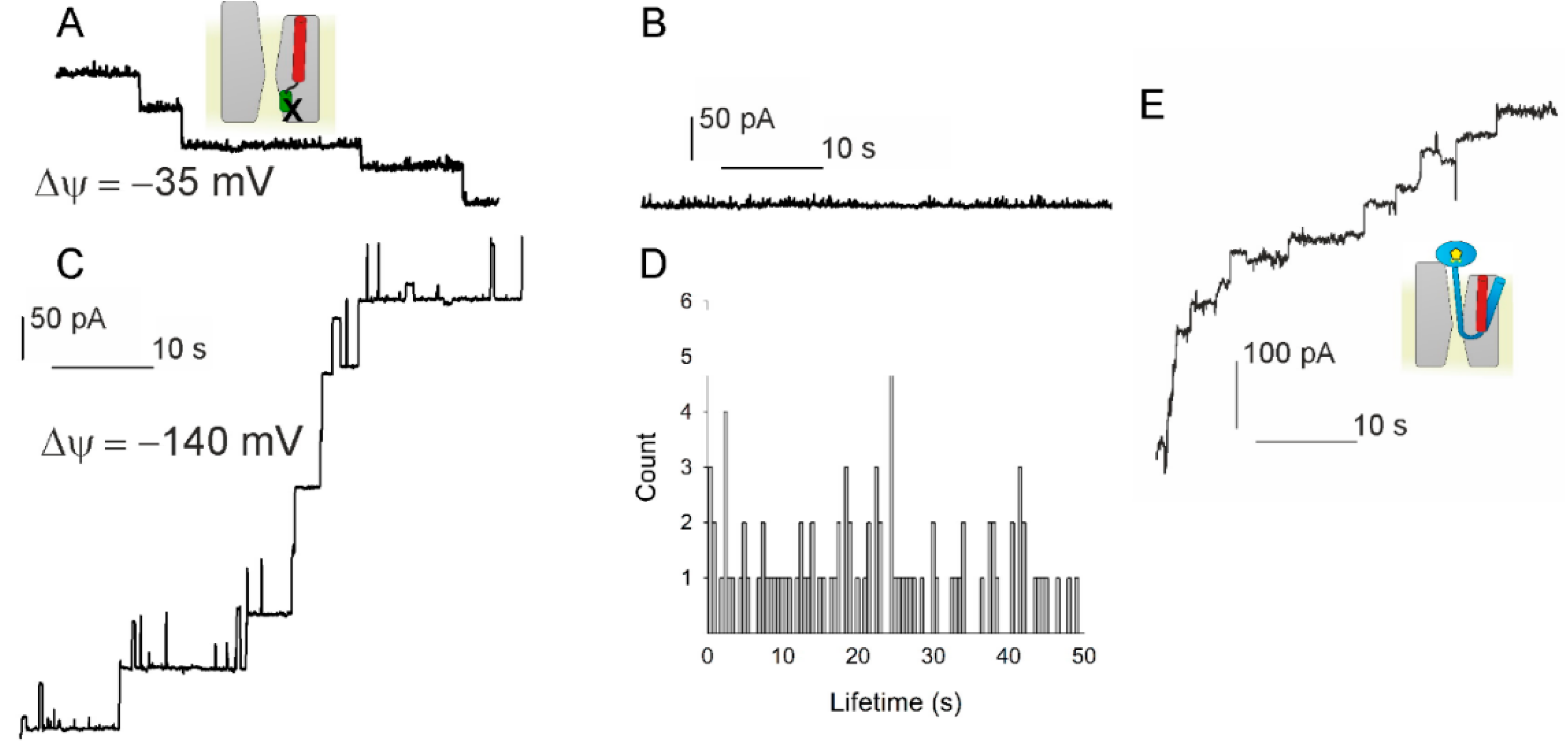
3.3. PD Affects the Voltage Sensitivity of SecYEG
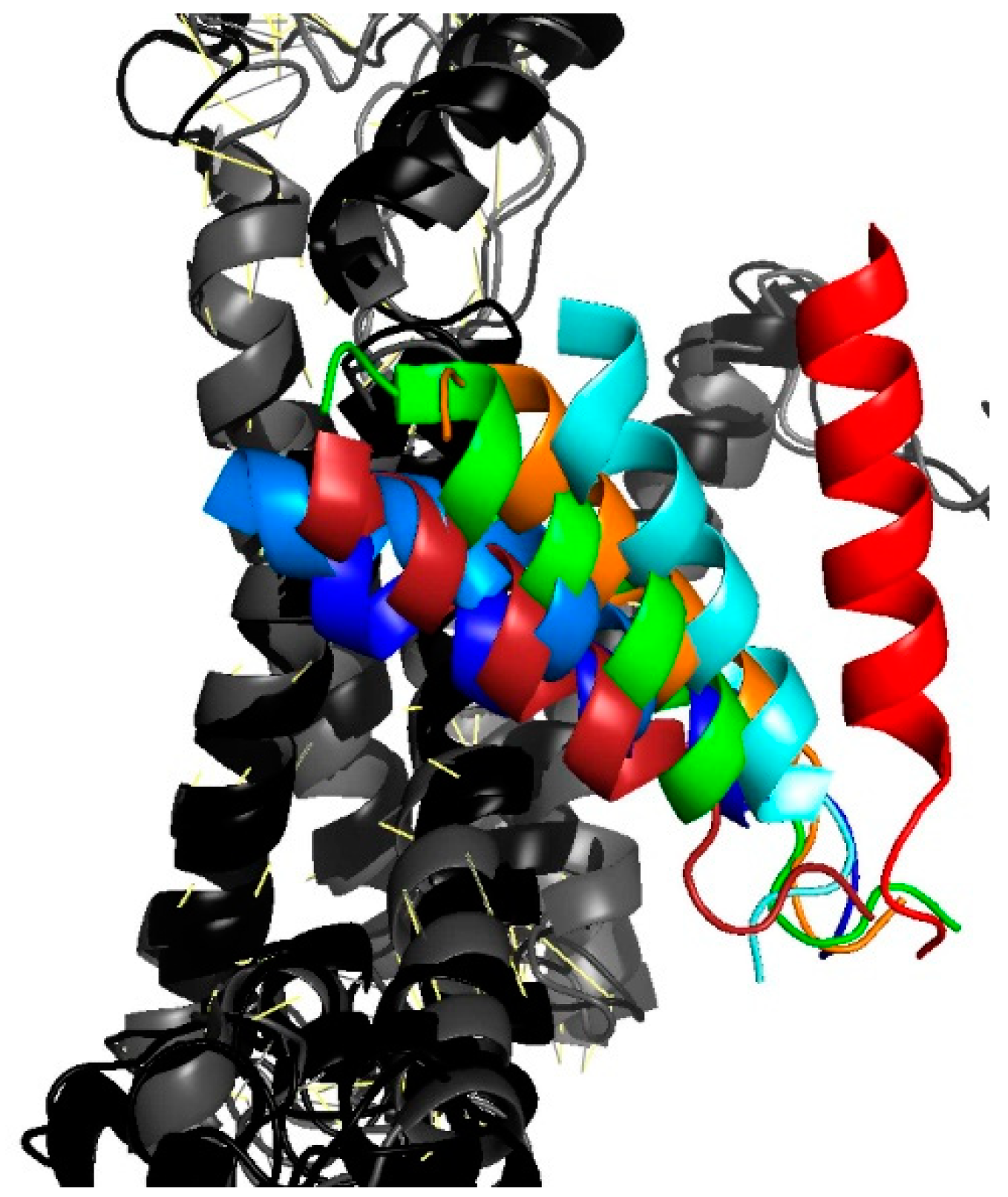
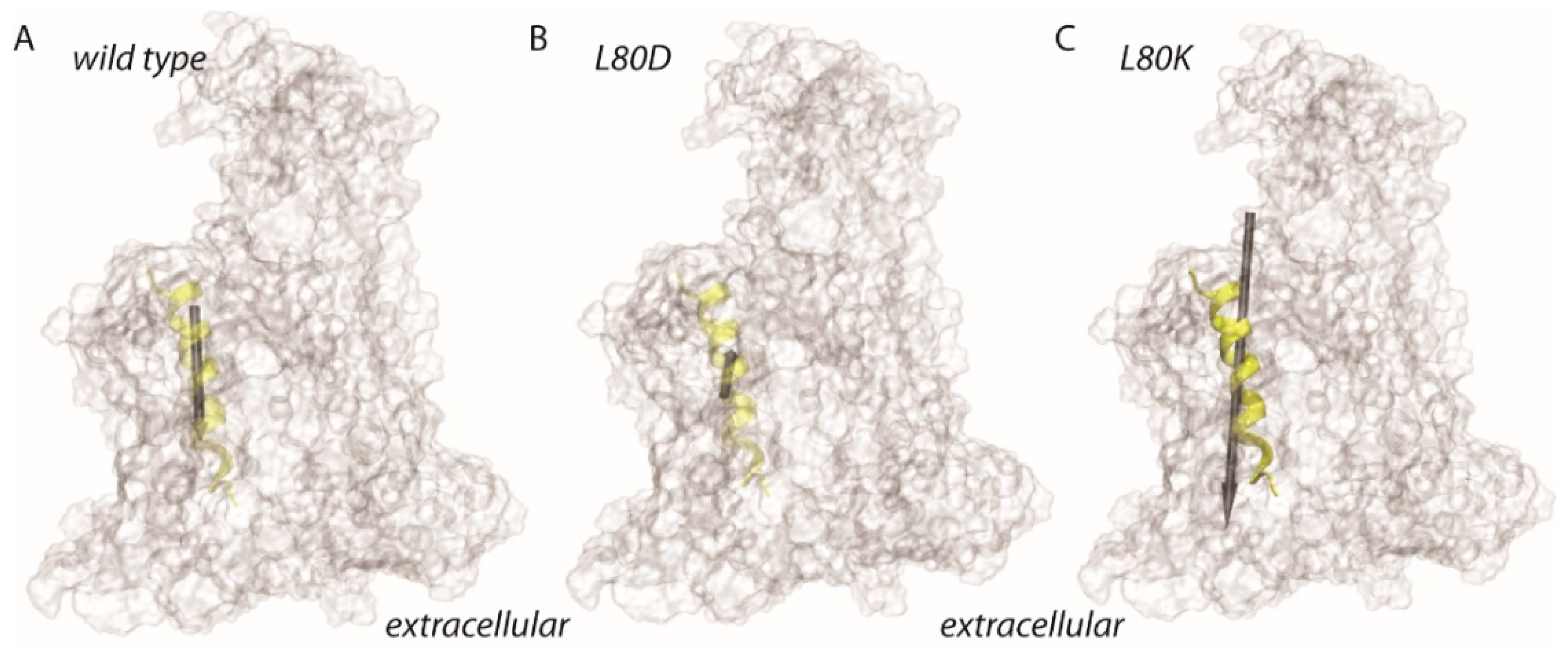
3.4. Lateral Gate Helix 2b Is Part of the Voltage Sensor
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Spiess, M.; Junne, T.; Janoschke, M. Membrane protein integration and topogenesis at the ER. Protein J. 2019, 38, 306–316. [Google Scholar] [CrossRef] [PubMed]
- Seinen, A.B.; Driessen, A.J.M. Single-molecule studies on the protein translocon. In Annual Review of Biophysics, 48; Dill, K.A., Ed.; Annual Reviews: Palo Alto, CA, USA, 2019; pp. 185–207. [Google Scholar]
- Saparov, S.M.; Erlandson, K.; Cannon, K.; Schaletzky, J.; Schulman, S.; Rapoport, T.A.; Pohl, P. Determining the conductance of the SecY protein translocation channel for small molecules. Mol. Cell 2007, 26, 501–509. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, B.; Clemons, W.M., Jr.; Collinson, I.; Modis, Y.; Hartmann, E.; Harrison, S.C.; Rapoport, T.A. X-ray structure of a protein-conducting channel. Nature 2004, 427, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Simon, S.M.; Blobel, G. Signal peptides open protein-conducting channels in e. Coli. Cell 1992, 69, 677–684. [Google Scholar] [CrossRef]
- Knyazev, D.G.; Winter, L.; Bauer, B.W.; Siligan, C.; Pohl, P. Ion conductivity of the bacterial translocation channel SecYEG engaged in translocation. J. Biol. Chem. 2014, 289, 24611–24616. [Google Scholar] [CrossRef] [PubMed]
- Cannon, K.S.; Or, E.; Clemons, W.M., Jr.; Shibata, Y.; Rapoport, T.A. Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J. Cell Biol. 2005, 169, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Rapoport, T.A.; Goder, V.; Heinrich, S.U.; Matlack, K.E. Membrane-protein integration and the role of the translocation channel. Trends Cell Biol. 2004, 14, 568–575. [Google Scholar] [CrossRef]
- Gogala, M.; Becker, T.; Beatrix, B.; Armache, J.P.; Barrio-Garcia, C.; Berninghausen, O.; Beckmann, R. Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion. Nature 2014, 506, 107–110. [Google Scholar] [CrossRef]
- Gumbart, J.; Schulten, K. The roles of pore ring and plug in the SecY protein-conducting channel. J. Gen. Physiol. 2008, 132, 709–719. [Google Scholar] [CrossRef]
- Park, E.; Rapoport, T.A. Preserving the membrane barrier for small molecules during bacterial protein translocation. Nature 2011, 473, 239–242. [Google Scholar] [CrossRef]
- Sachelaru, I.; Winter, L.; Knyazev, D.G.; Zimmermann, M.; Vogt, A.; Kuttner, R.; Ollinger, N.; Siligan, C.; Pohl, P.; Koch, H.-G. Yidc and SecYEG form a heterotetrameric protein translocation channel. Sci. Rep. 2017, 7, 101. [Google Scholar] [CrossRef] [PubMed]
- Knyazev, D.G.; Lents, A.; Krause, E.; Ollinger, N.; Siligan, C.; Papinski, D.; Winter, L.; Horner, A.; Pohl, P. The bacterial translocon SecYEG opens upon ribosome binding. J. Biol. Chem. 2013, 288, 17941–17946. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Sugano, Y.; Takemoto, M.; Mori, T.; Furukawa, A.; Kusakizako, T.; Kumazaki, K.; Kashima, A.; Ishitani, R.; Sugita, Y.; et al. Crystal structures of SecYEG in lipidic cubic phase elucidate a precise resting and a peptide-bound state. Cell Rep. 2015, 13, 1561–1568. [Google Scholar] [CrossRef]
- Osborne, A.R.; Rapoport, T.A. Protein translocation is mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell 2007, 129, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Or, E.; Navon, A.; Rapoport, T. Dissociation of the dimeric SecA atpase during protein translocation across the bacterial membrane. EMBO J. 2002, 21, 4470–4479. [Google Scholar] [CrossRef] [PubMed]
- Ederth, J.; Mandava, C.S.; Dasgupta, S.; Sanyal, S. A single-step method for purification of active his-tagged ribosomes from a genetically engineered escherichia coli. Nucleic Acids Res. 2009, 37, e15. [Google Scholar] [CrossRef] [PubMed]
- Horner, A.; Akimov, S.A.; Pohl, P. Long and short lipid molecules experience the same interleaflet drag in lipid bilayers. Phys. Rev. Lett. 2013, 110, 268101. [Google Scholar] [CrossRef]
- Saparov, S.M.; Antonenko, Y.N.; Koeppe, R.E.; Pohl, P. Desformylgramicidin: A model channel with an extremely high water permeability. Biophys. J. 2000, 79, 2526–2534. [Google Scholar] [CrossRef][Green Version]
- Kelley, L.A.; Sternberg, M.J. Protein structure prediction on the web: A case study using the phyre server. Nat. Protoc. 2009, 4, 363–371. [Google Scholar] [CrossRef]
- Brooks, B.R.; Bruccoleri, R.E.; Olafson, B.D.; States, D.J.; Swaminathan, S.; Karplus, M. Charmm—A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983, 4, 187–217. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. Vmd: Visual molecular dynamics. J. Mol. Graphics 1996, 14, 33–38. [Google Scholar] [CrossRef]
- MacKerell, A.D.; Bashford, D.; Bellott, M.; Dunbrack, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef] [PubMed]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.; Mittal, J.; Feig, M.; Mackerell, A.D., Jr. Optimization of the additive charmm all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef] [PubMed]
- MacKerell, A.D., Jr.; Feig, M.; Brooks, C.L., 3rd. Improved treatment of the protein backbone in empirical force fields. J. Am. Chem. Soc. 2004, 126, 698–699. [Google Scholar] [CrossRef]
- Woodbury, D.J.; Hall, J.E. Role of channels in the fusion of vesicles with a planar bilayer. Biophys. J. 1988, 54, 1053–1063. [Google Scholar] [CrossRef]
- Zilberstein, D.; Agmon, V.; Schuldiner, S.; Padan, E. Escherichia coli intracellular ph, membrane potential, and cell growth. J. Bacteriol. 1984, 158, 246–252. [Google Scholar] [CrossRef]
- Park, E.; Menetret, J.F.; Gumbart, J.C.; Ludtke, S.J.; Li, W.; Whynot, A.; Rapoport, T.A.; Akey, C.W. Structure of the SecY channel during initiation of protein translocation. Nature 2014, 506, 102–106. [Google Scholar] [CrossRef]
- Voorhees, R.M.; Fernandez, I.S.; Scheres, S.H.; Hegde, R.S. Structure of the mammalian ribosome-Sec61 complex to 3.4 a resolution. Cell 2014, 157, 1632–1643. [Google Scholar] [CrossRef]
- Li, L.; Park, E.; Ling, J.; Ingram, J.; Ploegh, H.; Rapoport, T.A. Crystal structure of a substrate-engaged SecY protein-translocation channel. Nature 2016, 531, 395–399. [Google Scholar] [CrossRef]
- Hannesschlaeger, C.; Horner, A.; Pohl, P. Intrinsic membrane permeability to small molecules. Chem. Rev. 2019, 119, 5922–5953. [Google Scholar] [CrossRef]
- Sengupta, D.; Behera, R.N.; Smith, J.C.; Ullmann, G.M. The α helix dipole: Screened out? Structure 2005, 13, 849–855. [Google Scholar] [CrossRef] [PubMed]
- Bondar, A.N.; del Val, C.; Freites, J.A.; Tobias, D.J.; White, S.H. Dynamics of SecY translocons with translocation-defective mutations. Structure 2010, 18, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Faouri, R.A.; Krueger, E.; Govind Kumar, V.; Fologea, D.; Straub, D.; Alismail, H.; Alfaori, Q.; Kight, A.; Ray, J.; Henry, R.; et al. An effective electric dipole model for voltage-induced gating mechanism of lysenin. Sci. Rep. 2019, 9, 11440. [Google Scholar] [CrossRef] [PubMed]
- Schauble, N.; Lang, S.; Jung, M.; Cappel, S.; Schorr, S.; Ulucan, O.; Linxweiler, J.; Dudek, J.; Blum, R.; Helms, V.; et al. Bip-mediated closing of the Sec61 channel limits Ca2+ leakage from the er. EMBO J. 2012, 31, 3282–3296. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, F.; Schauble, N.; Lang, S.; Jung, M.; Honigmann, A.; Ahmad, M.; Dudek, J.; Benedix, J.; Harsman, A.; Kopp, A.; et al. Interaction of calmodulin with Sec61alpha limits Ca2+ leakage from the endoplasmic reticulum. EMBO J. 2011, 30, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Wirth, A.; Jung, M.; Bies, C.; Frien, M.; Tyedmers, J.; Zimmermann, R.; Wagner, R. The Sec61p complex is a dynamic precursor activated channel. Mol. Cell 2003, 12, 261–268. [Google Scholar] [CrossRef]
- Wonderlin, W.F. Constitutive, translation-independent opening of the protein-conducting channel in the endoplasmic reticulum. Pflügers Arch. 2009, 457, 917–930. [Google Scholar] [CrossRef]
- Ma, C.; Wu, X.; Sun, D.; Park, E.; Catipovic, M.A.; Rapoport, T.A.; Gao, N.; Li, L. Structure of the substrate-engaged SecA-SecY protein translocation machine. Nat. Commun. 2019, 10, 2872. [Google Scholar] [CrossRef]
- Fessl, T.; Watkins, D.; Oatley, P.; Allen, W.J.; Corey, R.A.; Horne, J.; Baldwin, S.A.; Radford, S.E.; Collinson, I.; Tuma, R. Dynamic action of the Sec machinery during initiation, protein translocation and termination. Elife 2018, 7, e35112. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knyazev, D.G.; Kuttner, R.; Bondar, A.-N.; Zimmerman, M.; Siligan, C.; Pohl, P. Voltage Sensing in Bacterial Protein Translocation. Biomolecules 2020, 10, 78. https://doi.org/10.3390/biom10010078
Knyazev DG, Kuttner R, Bondar A-N, Zimmerman M, Siligan C, Pohl P. Voltage Sensing in Bacterial Protein Translocation. Biomolecules. 2020; 10(1):78. https://doi.org/10.3390/biom10010078
Chicago/Turabian StyleKnyazev, Denis G., Roland Kuttner, Ana-Nicoleta Bondar, Mirjam Zimmerman, Christine Siligan, and Peter Pohl. 2020. "Voltage Sensing in Bacterial Protein Translocation" Biomolecules 10, no. 1: 78. https://doi.org/10.3390/biom10010078
APA StyleKnyazev, D. G., Kuttner, R., Bondar, A.-N., Zimmerman, M., Siligan, C., & Pohl, P. (2020). Voltage Sensing in Bacterial Protein Translocation. Biomolecules, 10(1), 78. https://doi.org/10.3390/biom10010078







