Abstract
This systematic review provides a qualitative appraisal of 24 high-quality metabolomics-based studies published over the past decade exploring exercise-induced alterations of the human metabolome. Of these papers, 63% focused on acute metabolite changes following intense and prolonged exercise. The best studies utilized liquid chromatography mass spectrometry (LC-MS/MS) analytical platforms with large chemical standard libraries and strong, multivariate bioinformatics support. These studies reported large-fold changes in diverse lipid-related metabolites, with more than 100 increasing two-fold or greater within a few hours post-exercise. Metabolite shifts, even after strenuous exercise, typically return to near pre-exercise levels after one day of recovery. Few studies investigated metabolite changes following acute exercise bouts of shorter durations (< 60 min) and workload volumes. Plasma metabolite shifts in these types of studies are modest in comparison. More cross-sectional and exercise training studies are needed to improve scientific understanding of the human system’s response to varying, chronic exercise workloads. The findings derived from this review provide direction for future investigations focused on the body’s metabolome response to exercise.
1. Introduction
Acute and chronic physical activity causes extensive adaptations in organs and systems, leading to health benefits [1]. Improvements in technology have allowed investigators to quantify these adaptations using a biological systems approach, overlaying gene information with transcriptomics, proteomics, and metabolomics [1,2,3,4,5,6,7,8,9,10]. Combined data from multi-omics approaches will improve scientific understanding regarding the complex modulating effect that physical activity has on the phenotype at the individual level and related molecular mechanisms.
Metabolomics is defined as the simultaneous measurement of numerous low molecular metabolites that participate as substrates, reactants, signaling agents, intermediates, and products of enzyme-mediated reactions [3,4]. Metabolites are the final endpoints of upstream biochemical processes, and closely reflect the expressed phenotype. With the support of advanced analytical platforms and bioinformatics, metabolomics data can provide valuable insights regarding the biological impact of physical activity, pharmacological treatment, nutritional interventions, and other exposures [3].
Global metabolomics procedures were first performed in the 1960s and 1970s when gas chromatography mass spectrometry (GC-MS) was used to measure human metabolites in blood and urine samples [4]. Despite this, metabolomics was considered an emerging field of scientific endeavor as late as 2010, the year when the earliest studies investigating exercise effects in human athletes were published [3]. Since then, a growing number of research groups have used metabolomics in exercise-based studies. This is due, in large part, to the widespread availability of mass spectrometry platforms, freely accessible online databases of metabolites such as the Human Metabolome Database (HMDB), the expansion of chemical standards libraries, and advanced bioinformatics support to analyze and make sense of the large volumes of data. The net effect has been an improved capacity to accurately detect a greater number of metabolites and then interpret the overall effect on the human metabolome in a wide variety of matrixes.
This systematic review provides a qualitative appraisal of metabolomics-based studies published during the past decade exploring exercise-induced alterations on the human metabolome. The conclusions derived from this review will provide an evidence-based framework for future investigations.
2. Results
A total of 1355 articles were retrieved for this analysis. Of these, 1314 were excluded for not meeting analysis criteria after review of the abstracts. Of the 41 studies selected for full text examination, six were excluded for not meeting analysis criteria. Of the 35 studies included for scoring, 24 achieved a minimum score of 6, and were included in the final analysis (Table 1 and Figure 1).

Table 1.
Studies classification according to score system.
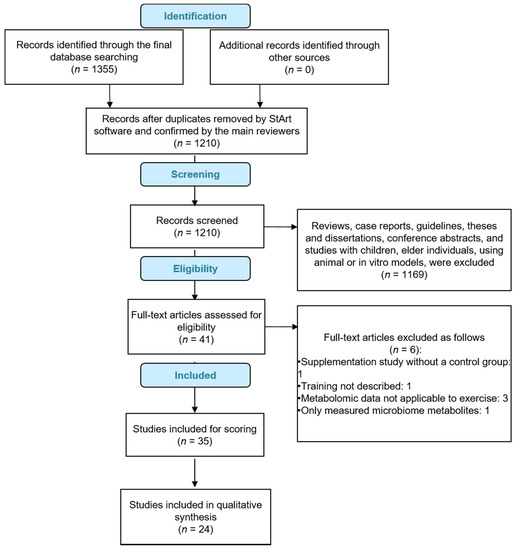
Figure 1.
Outcomes of review flow diagram.
2.1. Exercise Intensity and Duration Effects on Metabolism
Metabolic responses to exercise depend on the intensity and duration of effort. For the purposes of this review, heavy and moderate-intensity were differentiated using an intensity threshold of 60% of the oxygen uptake and heart rate reserve, and long and short-duration using a duration threshold of 60 min [40].
2.2. High-Intensity and Long-Duration
More than half of the studies included in this analysis (62.5%; n = 15) measured metabolite responses to long-duration, high-intensity running (n = 8) [8,9,12,13,14,19,22,24], cycling (n = 5) [5,7,17,18,20], soccer (n = 1), and swimming (n = 1) [16] (Table 2). Liquid chromatography mass spectrometry (LC-MS) with or without GC-MS was used for metabolite identification in 11 [5,7,8,9,12,13,14,17,19,20,24] of these studies, with GC-MS as the primary method in two studies [16,22], capillary electrophoresis time-of-flight mass spectrometry (CE-TOFMS) in one study [21] and nuclear magnetic resonance (NMR) for one study [18]. Large-fold changes in metabolites from the lipid super pathway were reported by most investigators, including increases in plasma medium- and long-chain fatty acids, fatty acid oxidation products (dicarboxylate and monohydroxy fatty acids, acylcarnitines), and ketone bodies, with corresponding decreases in triacylglycerol esters (Table 2). Other metabolite changes included shifts in amino acids and increases in energy tricarboxylic acid (TCA) cycle components.

Table 2.
High-intensity and long-duration studies.
2.3. High-Intensity and Short-Duration, Moderate-Intensity and Short/Long-Duration, Cross-Sectional and Training Studies
(A) High-Intensity, Short-Duration
Two studies measured metabolite responses to high-intensity, short-duration (18 to 30 min) exercise in recreationally active males and soccer athletes [23,28] (Table 3). Metabolite data from these studies were derived from GC-MS and NMR analytical platforms. Relatively small post-exercise changes were reported for metabolites related to the TCA cycle and related bioenergetics pathways.

Table 3.
Summaries of study characteristics and findings from nine [6,10,11,15,23,25,26,27,28] studies using other types of exercise designs.
(B) Moderate-Intensity, Short-Duration
One study reported metabolite responses following moderate-intensity, short-duration (30 min) cycling [6] (Table 3). Metabolites were identified using GC-MS and LC-MS/MS analytical platforms. Small-fold post-exercise changes were reported for metabolites linked to energy metabolism (lipolysis, glycolysis, TCA cycle intermediates, and catecholamines).
(C) Moderate-Intensity, Long-Duration
Two studies investigated metabolite responses to moderate-intensity, long-duration cycling and cross-country skiing [10,11]. Small to moderate post-exercise changes were reported for metabolites related to glycolytic and lipid pathways including free fatty acids, branched chain amino acids, acylcarnitines, mono- and diacylglycerols, and TCA intermediates.
(D) High- and Moderate-Intensity, Long-Duration
One study compared metabolite responses of high-intensity interval training (HIIT) and 60 min of moderate-intensity cycling (MOD) using GC-MS [26]. Small- to moderate-fold changes were reported for metabolites related to energy metabolism, and glycolytic and lipid pathways. HIIT compared to MOD induced higher post-exercise levels for glycolytic-related metabolites, with lower levels of lipid related metabolites.
(E) Cross-Section Elite Athletes
One study compared plasma metabolite levels in athletes from high- (n = 121) and moderate- (n = 70) endurance sports [15] (Table 3). Metabolomics was performed by ultra-performance liquid chromatography mass spectrometry (UPLC-MS/MS). The cross-sectional analysis showed some group differences, including higher levels for metabolites related to oxidative stress, fatty acid metabolism, steroid biosynthesis, and energy metabolism in high power and high endurance athletes. Plasma levels of metabolites related to steroid and polyamine pathways were more prominent in endurance athletes, with sterols, adenine-containing purines, and energy metabolites more evident in power athletes.
(F) Chronic Training, Low-, Moderate-, and High-Intensity
One study compared the chronic effects of cycling training at different intensities [25]. Plasma metabolites were measured with NMR, and only small group differences were reported in a few selected metabolites (hippuric acid, hypoxanthine, creatinine, dimethylamine, 3-methylxanthine).
(G) Chronic Training, High-Intensity
One study investigated the effects of chronic, high-intensity, short-duration running on plasma metabolite levels using NMR [27]. Small changes in selected metabolites were reported including lactate, pyruvate, TCA intermediates, and phospholipids.
3. Discussion
Advances in mass spectrometry since 2010 have led to an increasing number of metabolomics-based studies targeted on whole-body metabolite responses to varying acute and chronic exercise workloads. This systematic review of 24 high-quality papers published during the past decade revealed that the primary focus (63% of studies) has been on acute metabolite perturbations to long-duration, high-intensity aerobic exercise. Little information is available regarding metabolite changes coupled to acute bouts of exercise with lower workload volumes or those linked to long-term exercise training. The best studies utilized LC-MS/MS analytical platforms with large chemical standards libraries to identify and detect exercise-induced shifts in hundreds of metabolites. Strong bioinformatics support has improved predictive and descriptive modelling, discriminative variable selection, and the overall understanding of the body’s metabolome response to exercise.
This review indicates that a bout of prolonged and intensive exercise causes large-fold changes in numerous and diverse lipid-related metabolites [5,7,8,9,12,14,17,19,20,21,22,24]. In a typical study with human athletes exercising intensely for more than two hours, significant increases in at least 300 identified metabolites can be measured by LC-MS/MS analytical platforms, with more than 100 increasing twofold or greater [5,7,14,20,24]. This response includes post-exercise increases in plasma medium- and long-chain fatty acids, ketone bodies, fatty acid oxidation products, and sulfated bile acids. At the same time, related decreases occur in plasma triacylglycerol esters, primary and secondary bile acids, and minor phospholipids such as lysophosphatidylcholines and lysophosphatidylethanolamines [12,19,20,41]. Untargeted metabolomics has revealed post-exercise increases in both common (e.g., oleate/vaccinate, palmitate, linoleate, stearate, palmitoleate, myristate), and atypical fatty acids (adrenate, docosapentaenoate, dihomo-linolenate, dihomolinoleate, docosadienoate, and eicosenoate). The corresponding fatty acid oxidation signature includes acylcarnitines, 3-hydroxybutyrate (BHBA), and dicarboxylate and monohydroxy fatty acids. Other important shifts have been measured for plasma concentrations of tryptophan- and other amino acid-related metabolites, and energy tricarboxylic acid (TCA) cycle components including malate, aconitate, citrate, fumarate, succinate, and alpha-ketoglutarate [13,16,17,18,19,22,41].
Most of the changes in plasma metabolites after prolonged and intensive exercise reach their nadir within a few hours. Plasma deviations in many of these metabolites are still apparent, but largely abated, after one day of recovery [5,7,12,19,20,21]. The large and varied metabolite response to heavy exercise workloads reflects the physiological stress and diminished glycogen stores experienced by the participant [12,21,24,41].
An increasing number of studies are utilizing metabolomics to measure the influence of various nutritional interventions on metabolite perturbations during recovery from prolonged and intensive exercise [5,7,14,16,41,42]. Metabolomics is ideally suited to measuring the impact of nutritional interventions during acute exercise by simultaneously measuring and identifying shifts in hundreds of metabolites from diverse pathways. Emerging data indicate that carbohydrates from both sugar beverages and fruits such as bananas, and flavonoids from food and beverage sources such as blueberries and green tea, have a large effects on the human metabolome response to intense exercise workloads [3,5,14,16,42,43].
Relatively few studies have investigated exercise-induced metabolite changes following acute bouts with lower durations (< 60 min) and workload volumes [23,26]. Half of these studies performed metabolomics using GC-MS or NMR analytical platforms, limiting the number of identified metabolites and the usefulness of these data. As expected, post-exercise shifts in plasma metabolite levels are modest in comparison to high exercise volume workloads due to a moderated reliance on underlying carbohydrate and lipid substrate pathways.
More cross-sectional studies are needed to compare plasma and urine metabolite levels between sedentary and physically active individuals, and athletes from different sports. These studies could provide important information for future randomized, exercise training trials. Using a cross-sectional design, one study showed some metabolite differences between power and endurance athletic groups [15]. The athletes were not tested at the same time or in similar resting states, however, making it difficult to draw definitive conclusions.
Few randomized, exercise training studies have been conducted to investigate potential adaptations in the human metabolome [25,27]. These two studies employed different training protocols and study designs, and performed metabolomics using NMR, limiting the usefulness of these data. Future metabolomics-based randomized exercise training studies, especially when combined with genomics and proteomics outcomes, will improve scientific understanding of the human system’s response to varying exercise workloads [44].
4. Materials and Methods
This systematic review was performed according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [45] and was preregistered in the International Prospective Register of Systematic Review (PROSPERO). To systematize the search and data extraction, a free standardized electronic tool called State of the Art through Systematic Review (StArt) [46] was used. The software StArt tracked duplicated studies during extraction, and this was confirmed with manual examination by the two main reviewers. The studies were selected, extracted and included independently by two researchers (CAS and EFS), and a third independent researcher (RMA) verified the inclusion process in order to solve any disagreement between the two main researchers.
4.1. Search Strategy
The electronic search was performed from inception to November 26th, 2018 and updated on April 10th, 2019. The articles were retrieved from the following electronic databases: PubMed (via National Library of Medicine), Science Direct, SCOPUS (Elsevier) and Web of Science. The MeSH terms were selected and combined according to analysis method (metabolomics) and mandatory activity (sports OR exercise). Moreover, the search strategy was limited to humans (population of interest), English language and clinical trial studies.
4.2. Eligibility Criteria for Inclusion
The abstracts were first examined and evaluated for the listed criteria. Studies were selected if metabolomics were utilized to measure exercise-induced changes in metabolites in healthy study participants using serum, plasma, saliva, or urine samples. Exercise-based studies with nutrition interventions were included, but this review only included data collected from the control groups. Reviews, case reports, guidelines, theses and dissertations, conference abstracts, and studies using animal or in vitro models were not included.
4.3. Data Extraction and Study Inclusion
The following data from the selected studies were extracted: name of the first author, year of publication, characteristics of participants and groups (population, sample size, groups, gender, age, physical activity level), research design elements (type of research, exercise mode, duration, and intensity), metabolomics procedures (analytical platform, metabolite data), and summary comments.
4.4. Studies Quality Assessment
The quality of the studies was assessed by two researchers (CAS and EFS) using a scoring system created for this analysis (see Table 4 and Figure 2).

Table 4.
Score setting for metabolomic studies quality assessment.
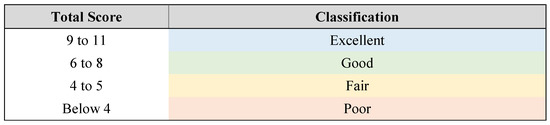
Figure 2.
Classification of studies based on the total score.
5. Conclusions and Future Directions
The first decade of metabolomics-based exercise studies, especially those utilizing sensitive LC-MS/MS analytical platforms with large chemical standards libraries and rigorous bioinformatics support, provided useful systems biology information on the biochemical mechanisms underlying exercise-induced effects on metabolism [13,47]. This area of scientific endeavor is still emerging, and much remains to be discovered, especially in the areas of the metabolite response to acute and chronic moderate exercise workloads. The sensitivity of the analytical platforms will continue to improve, expanding the number of small molecule metabolites that can be detected. These improvements in technology, coupled with improved quality control, bioinformatics support, the expansion of biochemical standards, and an emphasis on larger study groups of both genders, will improve the identification and quantitation of currently known and unknown metabolites in a variety of human matrixes. More emphasis is needed on the influence of activity reduction and physical inactivity on metabolite shifts. These improvements in study design and methodology will broaden our understanding of the influence of acute and chronic exercise on the human metabolome. An increasing number of studies, including the National Institutes of Health project, ’Molecular Transducers of Physical Activity in Humans’, will combine metabolomics with genetics, epigenetics, lipidomics, and proteomics to examine all aspects of the physiological, biochemical, and molecular response to both aerobic- and resistance-based exercise training interventions [44].
Author Contributions
C.A.S., D.C.N. and A.M.C. organized the study and created the scoring and classification system; C.A.S., D.C.N., E.F.S., R.M.A. searched, extracted the studies from the databases and selected for inclusion; D.C.N., C.A.S., E.F.S. wrote; D.C.N., C.A.S., E.F.S., R.M.A., A.M.C. edited and reviewed the study.
Funding
São Paulo Research Fundation—FAPESP (grant #2016/222157).
Acknowledgments
The authors would like to acknowledge the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brazil (CAPES, Postgraduate Program in Physiotherapy, grant: 001).
Conflicts of Interest
Authors declare no conflict of interest.
References
- Grazioli, E.; Dimauro, I.; Mercatelli, N.; Wang, G.; Pitsiladis, Y.; Di Luigi, L.; Caporossi, D. Physical activity in the prevention of human diseases: Role of epigenetic modifications. BMC Genom. 2017, 18, 802. [Google Scholar] [CrossRef] [PubMed]
- Wackerhage, H.; Smith, J.; Wisniewski, D. Molecular Exercise Physiology, 1st ed.; Oxford University Press: Oxford, UK, 2014; p. 1. ISBN 978-0-19-181751-9. [Google Scholar]
- Nieman, D.C.; Mitmesser, S.H. Potential impact of nutrition on immune system recovery from heavy exertion: A metabolomics perspective. Nutrients 2017, 9, 513. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Broadhurst, D.I.; Atherton, H.J.; Goodacre, R.; Griffin, J.L. Systems level studies of mammalian metabolomes: The roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chem. Soc. Rev. 2011, 40, 387–426. [Google Scholar] [CrossRef] [PubMed]
- Nieman, D.C.; Gillitt, N.D.; Sha, W.; Meaney, M.P.; John, C.; Pappan, K.L.; Kinchen, J.M. Metabolomics-based analysis of banana and pear ingestion on exercise performance and recovery. J. Proteome Res. 2015, 14, 5367–5377. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, D.M.; Hodgson, A.B.; Randell, R.K.; Mahabir-Jagessar-T, K.; Garczarek, U.; Jeukendrup, A.E.; Mela, D.J.; Lotito, S. Metabolic response to decaffeinated green tea extract during rest and moderate-intensity exercise. J. Agric. Food Chem. 2014, 62, 9936–9943. [Google Scholar] [CrossRef] [PubMed]
- Nieman, D.C.; Scherr, J.; Luo, B.; Meaney, M.P.; Dréau, D.; Sha, W.; Dew, D.A.; Henson, D.A.; Pappan, K.L. Influence of pistachios on performance and exercise-induced inflammation, oxidative stress, immune dysfunction, and metabolite shifts in cyclists: A randomized, crossover trial. PLoS ONE 2014, 9, e113725. [Google Scholar] [CrossRef] [PubMed]
- Nieman, D.C.; Sha, W.; Pappan, K.L. IL-6 linkage to exercise-induced shifts in lipid-related metabolites: A metabolomics-based analysis. J. Proteome Res. 2017, 16, 970–977. [Google Scholar] [CrossRef] [PubMed]
- Davison, G.; Vinaixa, M.; McGovern, R.; Beltran, A.; Novials, A.; Correig, X.; McClean, C. Metabolomic response to acute hypoxic exercise and recovery in adult males. Front. Physiol. 2018, 9, 1682. [Google Scholar] [CrossRef] [PubMed]
- Hodgson, A.B.; Randell, R.K.; Boon, N.; Garczarek, U.; Mela, D.J.; Jeukendrup, A.E.; Jacobs, D.M. Metabolic response to green tea extract during rest and moderate-intensity exercise. J. Nutr. Biochem. 2013, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Karl, J.P.; Margolis, L.M.; Murphy, N.E.; Carrigan, C.T.; Castellani, J.W.; Madslien, E.H.; Teien, H.-K.; Martini, S.; Montain, S.J.; Pasiakos, S.M. Military training elicits marked increases in plasma metabolomic signatures of energy metabolism, lipolysis, fatty acid oxidation, and ketogenesis. Physiol. Rep. 2017, 5, e13407. [Google Scholar] [CrossRef]
- Lehmann, R.; Zhao, X.; Weigert, C.; Simon, P.; Fehrenbach, E.; Fritsche, J.; Machann, J.; Schick, F.; Wang, J.; Hoene, M.; et al. Medium chain acylcarnitines dominate the metabolite pattern in humans under moderate intensity exercise and support lipid oxidation. PLoS ONE 2010, 5, e11519. [Google Scholar] [CrossRef]
- Lewis, G.D.; Farrell, L.; Wood, M.J.; Martinovic, M.; Arany, Z.; Rowe, G.C.; Souza, A.; Cheng, S.; McCabe, E.L.; Yang, E.; et al. Metabolic signatures of exercise in human plasma. Sci. Transl. Med. 2010, 2, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Nieman, D.C.; Gillitt, N.D.; Knab, A.M.; Shanely, R.A.; Pappan, K.L.; Jin, F.; Lila, M.A. Influence of a polyphenol-enriched protein powder on exercise-induced inflammation and oxidative stress in athletes: A randomized trial using a metabolomics approach. PLoS ONE 2013, 8, e72215. [Google Scholar] [CrossRef] [PubMed]
- Al-Khelaifi, F.; Diboun, I.; Donati, F.; Botrè, F.; Alsayrafi, M.; Georgakopoulos, C.; Suhre, K.; Yousri, N.A.; Elrayess, M.A. A pilot study comparing the metabolic profiles of elite-level athletes from different sporting disciplines. Sports Med. Open 2018, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Knab, A.M.; Nieman, D.C.; Gillitt, N.D.; Shanely, R.A.; Cialdella-Kam, L.; Henson, D.A.; Sha, W. Effects of a flavonoid-rich juice on inflammation, oxidative stress, and immunity in elite swimmers: A metabolomics-based approach. Int. J. Sport Nutr. Exerc. Metab. 2013, 23, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Manaf, F.A.; Lawler, N.; Peiffer, J.J.; Maker, G.L.; Boyce, M.C.; Fairchild, T.J.; Broadhurst, D. Characterizing the plasma metabolome during and following a maximal exercise cycling test. J. Appl. Physiol. 2018, 125, 1193–1203. [Google Scholar] [CrossRef] [PubMed]
- Messier, F.M.; Le Moyec, L.; Santi, C.; Gaston, A.-F.; Triba, M.N.; Roca, E.; Durand, F. The impact of moderate altitude on exercise metabolism in recreational sportsmen: A nuclear magnetic resonance metabolomic approach. Appl. Physiol. Nutr. Metab. Physiol. Appl. Nutr. Metab. 2017, 42, 1135–1141. [Google Scholar] [CrossRef]
- Nieman, D.C.; Shanely, R.A.; Gillitt, N.D.; Pappan, K.L.; Lila, M.A. Serum metabolic signatures induced by a three-day intensified exercise period persist after 14 h of recovery in runners. J. Proteome Res. 2013, 12, 4577–4584. [Google Scholar] [CrossRef]
- Nieman, D.C.; Shanely, R.A.; Luo, B.; Meaney, M.P.; Dew, D.A.; Pappan, K.L. Metabolomics approach to assessing plasma 13- and 9-hydroxy-octadecadienoic acid and linoleic acid metabolite responses to 75-km cycling. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2014, 307, 68–74. [Google Scholar] [CrossRef]
- Ra, S.-G.; Maeda, S.; Higashino, R.; Imai, T.; Miyakawa, S. Metabolomics of salivary fatigue markers in soccer players after consecutive games. Appl. Physiol. Nutr. Metab. Physiol. Appl. Nutr. Metab. 2014, 39, 1120–1126. [Google Scholar] [CrossRef]
- Stander, Z.; Luies, L.; Mienie, L.J.; Keane, K.M.; Howatson, G.; Clifford, T.; Stevenson, E.J.; Loots, D.T. The altered human serum metabolome induced by a marathon. Metabolomics Off. J. Metabolomic Soc. 2018, 14, 150. [Google Scholar] [CrossRef] [PubMed]
- Danaher, J.; Gerber, T.; Wellard, R.M.; Stathis, C.G.; Cooke, M.B. The use of metabolomics to monitor simultaneous changes in metabolic variables following supramaximal low volume high intensity exercise. Metabolomics 2015, 12, 7. [Google Scholar] [CrossRef][Green Version]
- Howe, C.C.F.; Alshehri, A.; Muggeridge, D.; Mullen, A.B.; Boyd, M.; Spendiff, O.; Moir, H.J.; Watson, D.G. Untargeted metabolomics profiling of an 80.5 km simulated treadmill ultramarathon. Metabolites 2018, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Neal, C.M.; Hunter, A.M.; Brennan, L.; O’Sullivan, A.; Hamilton, D.L.; De Vito, G.; Galloway, S.D.R. Six weeks of a polarized training-intensity distribution leads to greater physiological and performance adaptations than a threshold model in trained cyclists. J. Appl. Physiol. 2013, 114, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Peake, J.M.; Tan, S.J.; Markworth, J.F.; Broadbent, J.A.; Skinner, T.L.; Cameron-Smith, D. Metabolic and hormonal responses to isoenergetic high-intensity interval exercise and continuous moderate-intensity exercise. Am. J. Physiol. Endocrinol. Metab. 2014, 307, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Pechlivanis, A.; Kostidis, S.; Saraslanidis, P.; Petridou, A.; Tsalis, G.; Veselkov, K.; Mikros, E.; Mougios, V.; Theodoridis, G.A. 1H NMR study on the short- and long-term impact of two training programs of sprint running on the metabolic fingerprint of human serum. J. Proteome Res. 2013, 12, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Zafeiridis, A.; Chatziioannou, A.C.; Sarivasiliou, H.; Kyparos, A.; Nikolaidis, M.G.; Vrabas, I.S.; Pechlivanis, A.; Zoumpoulakis, P.; Baskakis, C.; Dipla, K.; et al. Global metabolic stress of isoeffort continuous and high intensity interval aerobic exercise: A comparative 1H NMR metabonomic study. J. Proteome Res. 2016, 15, 4452–4463. [Google Scholar] [CrossRef] [PubMed]
- Muhsen Ali, A.; Burleigh, M.; Daskalaki, E.; Zhang, T.; Easton, C.; Watson, D.G. Metabolomic profiling of submaximal exercise at a standardized relative intensity in healthy adults. Metabolites 2016, 6, 9. [Google Scholar] [CrossRef] [PubMed]
- Castro, A.; Duft, R.G.; Ferreira, M.L.V.; de Andrade, A.L.L.; Gáspari, A.F.; de Marchi Silva, L.; de Oliveira-Nunes, S.G.; Cavaglieri, C.R.; Ghosh, S.; Bouchard, C.; et al. Association of skeletal muscle and serum metabolites with maximum power output gains in response to continuous endurance or high-intensity interval training programs: The TIMES study—A randomized controlled trial. PLoS ONE 2019, 14, e0212115. [Google Scholar] [CrossRef]
- Enea, C.; Seguin, F.; Petitpas-Mulliez, J.; Boildieu, N.; Boisseau, N.; Delpech, N.; Diaz, V.; Eugène, M.; Dugué, B. 1H NMR-based metabolomics approach for exploring urinary metabolome modifications after acute and chronic physical exercise. Anal. Bioanal. Chem. 2010, 396, 1167–1176. [Google Scholar] [CrossRef]
- Andersson Hall, U.; Edin, F.; Pedersen, A.; Madsen, K. Whole-body fat oxidation increases more by prior exercise than overnight fasting in elite endurance athletes. Appl. Physiol. Nutr. Metab. Physiol. Appl. Nutr. Metab. 2016, 41, 430–437. [Google Scholar] [CrossRef] [PubMed]
- Pechlivanis, A.; Kostidis, S.; Saraslanidis, P.; Petridou, A.; Tsalis, G.; Mougios, V.; Gika, H.G.; Mikros, E.; Theodoridis, G.A. 1H NMR-based metabonomic investigation of the effect of two different exercise sessions on the metabolic fingerprint of human urine. J. Proteome Res. 2010, 9, 6405–6416. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Han, J.; He, Q.; Geng, Z.; Deng, Z.; Qiao, D. Applying (1)H NMR spectroscopy to detect changes in the urinary metabolite levels of Chinese half-pipe snowboarders after different exercises. J. Anal. Methods Chem. 2015, 2015, 315217. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; A, J.; Wang, G.; Lu, H.; Huang, X.; Liu, Y.; Zha, W.; Hao, H.; Zhang, Y.; Liu, L.; et al. Metabolomic investigation into variation of endogenous metabolites in professional athletes subject to strength-endurance training. J. Appl. Physiol. Bethesda Md 1985 2009, 106, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Prado, E.; Souza, G.H.M.F.; Pegurier, M.; Vieira, C.; Lima-Neto, A.B.M.; Assis, M.; Guedes, M.I.F.; Koblitz, M.G.B.; Ferreira, M.S.L.; Macedo, A.F.; et al. Non-targeted sportomics analyses by mass spectrometry to understand exercise-induced metabolic stress in soccer players. Int. J. Mass Spectrom. 2017, 418, 1–5. [Google Scholar] [CrossRef]
- Sun, T.; Wu, Y.; Wu, X.; Ma, H. Metabolomic profiles investigation on athletes’ urine 35 minutes after an 800-meter race. J. Sports Med. Phys. Fitness 2017, 57, 839–849. [Google Scholar] [CrossRef] [PubMed]
- Berton, R.; Conceição, M.S.; Libardi, C.A.; Canevarolo, R.R.; Gáspari, A.F.; Chacon-Mikahil, M.P.T.; Zeri, A.C.; Cavaglieri, C.R. Metabolic time-course response after resistance exercise: A metabolomics approach. J. Sports Sci. 2017, 35, 1211–1218. [Google Scholar] [CrossRef] [PubMed]
- Valério, D.F.; Berton, R.; Conceição, M.S.; Canevarolo, R.R.; Chacon-Mikahil, M.P.T.; Cavaglieri, C.R.; Meirelles, G.V.; Zeri, A.C.; Libardi, C.A. Early metabolic response after resistance exercise with blood flow restriction in well-trained men: A metabolomics approach. Appl. Physiol. Nutr. Metab. Physiol. Appl. Nutr. Metab. 2018, 43, 240–246. [Google Scholar] [CrossRef]
- Nieman, D.C.; Wentz, L.M. The compelling link between physical activity and the body’s defense system. J. Sport Health Sci. 2019, 8, 201–217. [Google Scholar] [CrossRef]
- Nieman, D.C.; Gillitt, N.D.; Sha, W. Identification of a select metabolite panel for measuring metabolic perturbation in response to heavy exertion. Metabolomics 2018, 14, 147. [Google Scholar] [CrossRef]
- Nieman, D.C.; Gillitt, N.D.; Sha, W.; Esposito, D.; Ramamoorthy, S. Metabolic recovery from heavy exertion following banana compared to sugar beverage or water only ingestion: A randomized, crossover trial. PLoS ONE 2018, 13, e0194843. [Google Scholar] [CrossRef] [PubMed]
- Nieman, D.C.; Gillitt, N.D.; Henson, D.A.; Sha, W.; Shanely, R.A.; Knab, A.M.; Cialdella-Kam, L.; Jin, F. Bananas as an energy source during exercise: A metabolomics approach. PLoS ONE 2012, 7, e37479. [Google Scholar] [CrossRef] [PubMed]
- Sparks, L.M. Exercise training response heterogeneity: Physiological and molecular insights. Diabetologia 2017, 60, 2329–2336. [Google Scholar] [CrossRef] [PubMed]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gøtzsche, P.C.; Ioannidis, J.P.A.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: Explanation and elaboration. PLoS Med. 2009, 6, e1000100. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, S.; Silva, C.; Hernandes, E.; Octaviano, F.; Di Thommazo, A.; Belgamo, A. Improvements in the StArt tool to better support the systematic review process. In Proceedings of the 20th International Conference on Evaluation and Assessment in Software Engineering, Limerick, Ireland, 1–3 June 2016; ACM: New York, NY, USA, 2016. [Google Scholar]
- Heaney, L.M.; Deighton, K.; Suzuki, T. Non-targeted metabolomics in sport and exercise science. J. Sports Sci. 2017, 37, 959–967. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).