Inhibitory Effects of Green Tea Polyphenols on Microbial Metabolism of Aromatic Amino Acids in Humans Revealed by Metabolomic Analysis
Abstract
1. Introduction
2. Results
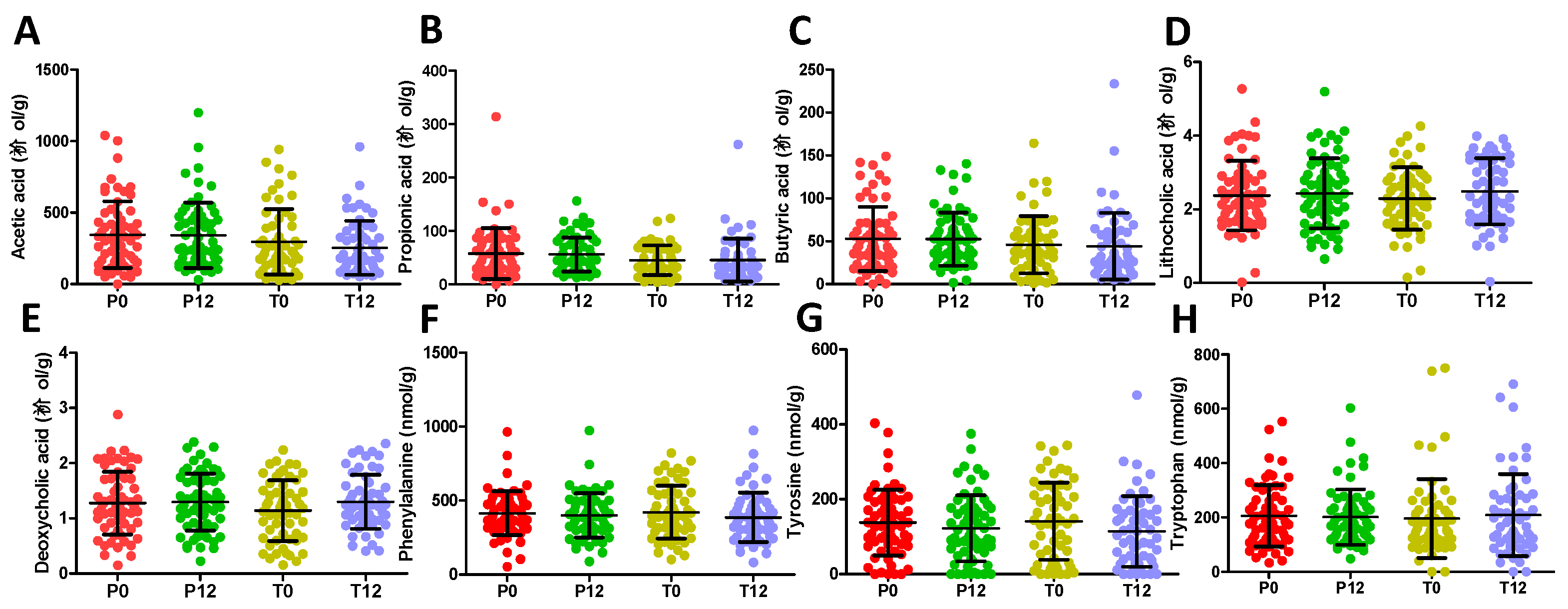
2.1. Influences of GTP on Fecal Metabolome
2.2. Influences of GTP on Urine Metabolome
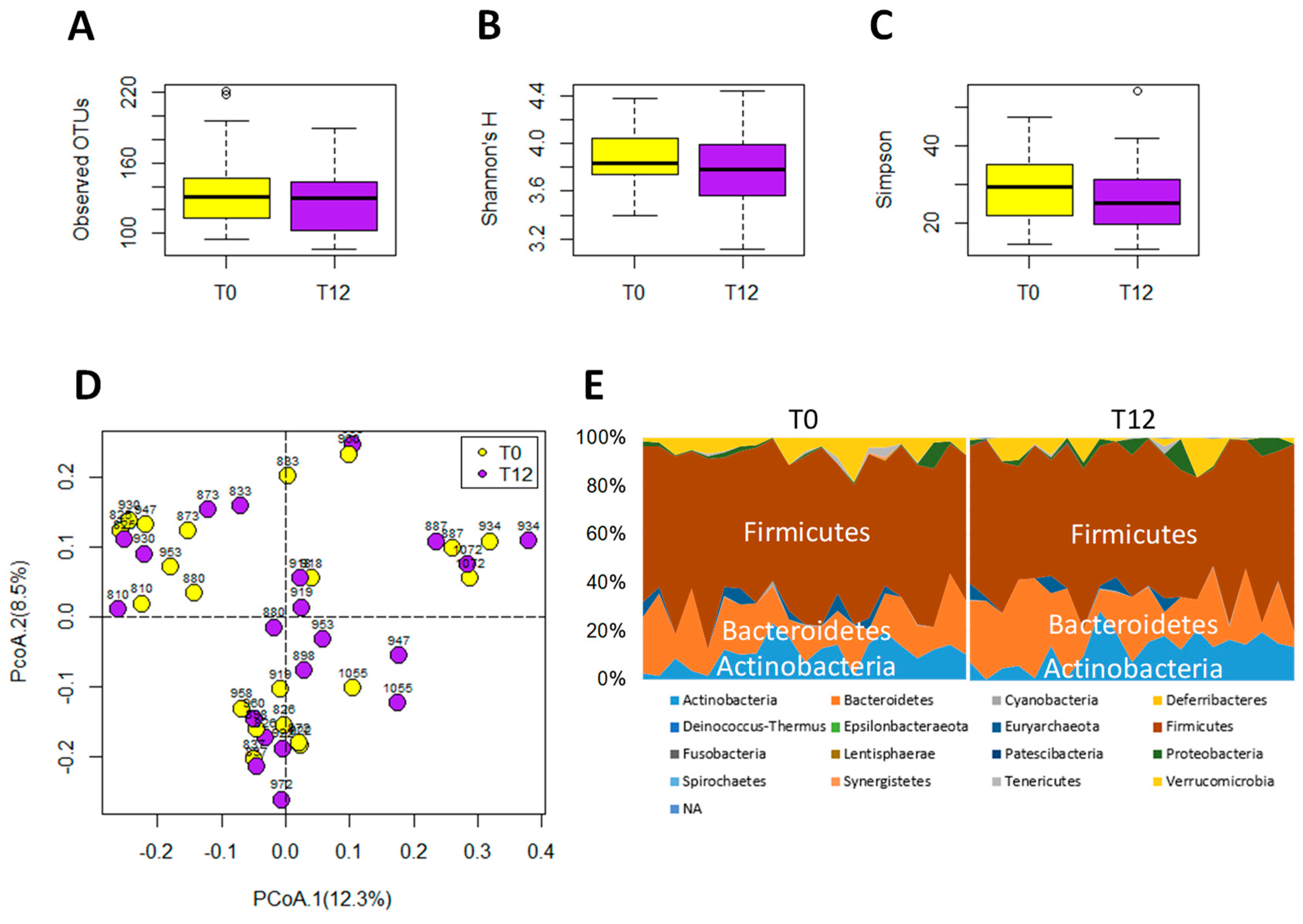
2.3. Influences of GTP on Gut Microbiome
3. Discussion
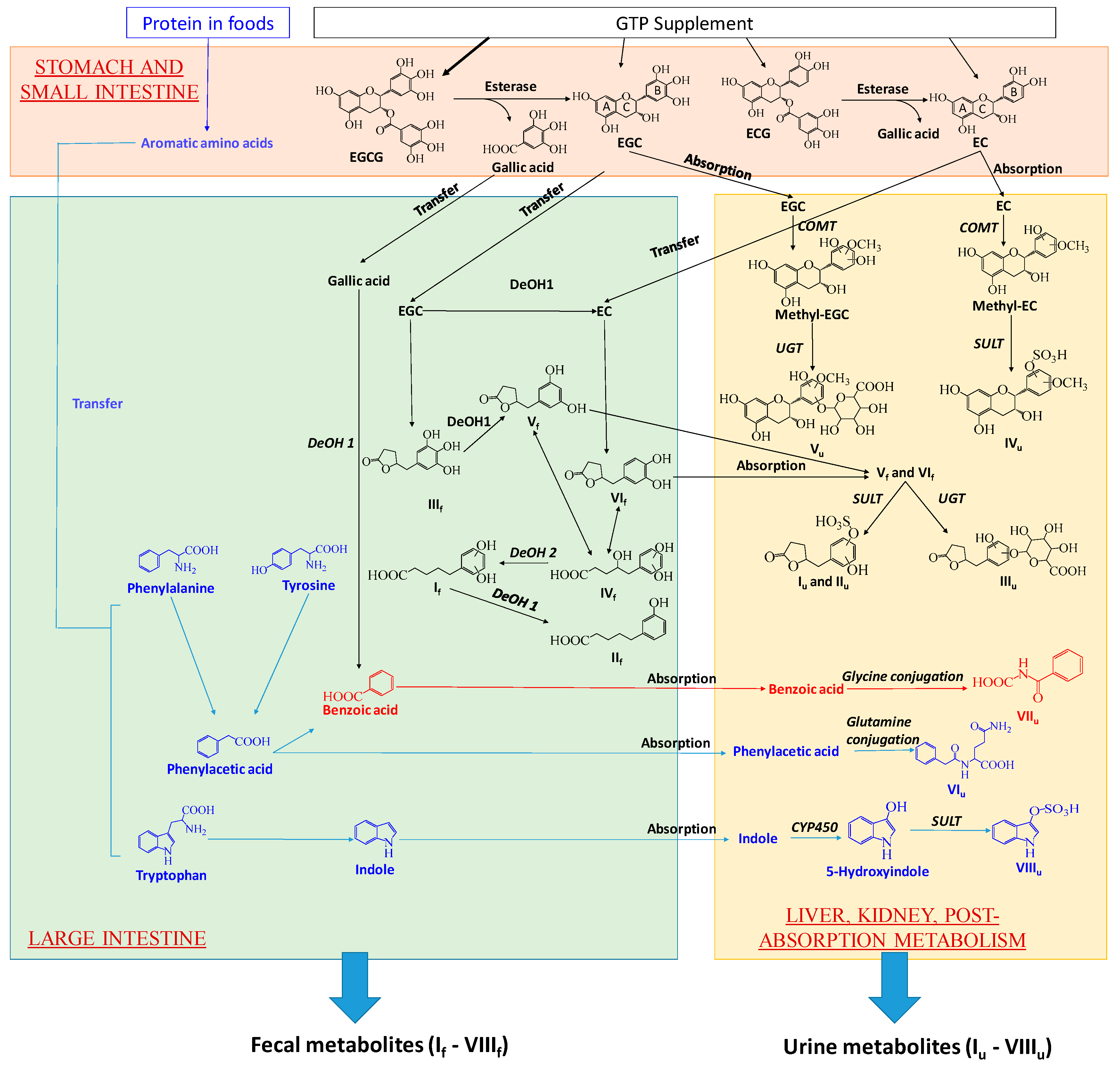
3.1. Formation and Potential Functions of GTP-Derived Metabolites in Human Feces and Urine
3.2. Potential Causes and Consequences of GTP-Elicited Decreases in Microbial Metabolism of AAA
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Green Tea Polyphenols (GTP) Preparation
4.3. Study Design
4.4. Fecal and Urine Samples Collection
4.5. Preparation of Fecal and Urine Samples
4.6. LC−MS Analysis
4.7. Multivariate Analysis and Data Visualization
4.8. Marker Characterization
4.9. Fecal Sample Processing and 16S rRNA Gene Analysis
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Balentine, D.A.; Wiseman, S.A.; Bouwens, L.C. The chemistry of tea flavonoids. Crit. Rev. Food Sci. Nutr. 1997, 37, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ho, C.-T. Polyphenolic chemistry of tea and coffee: A century of progress. J. Agric. Food Chem. 2009, 57, 8109–8114. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.Y.; Zhang, L.; Zuo, Z. Intestinal efflux transport kinetics of green tea catechins in caco-2 monolayer model. J. Pharm. Pharmacol. 2007, 59, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Stalmach, A.; Mullen, W.; Steiling, H.; Williamson, G.; Lean, M.E.; Crozier, A. Absorption, metabolism, and excretion of green tea flavan-3-ols in humans with an ileostomy. Mol. Nutr. Food Res. 2010, 54, 323–334. [Google Scholar] [CrossRef]
- Del Rio, D.; Stalmach, A.; Calani, L.; Crozier, A. Bioavailability of coffee chlorogenic acids and green tea flavan-3-ols. Nutrients 2010, 2, 820–833. [Google Scholar] [CrossRef] [PubMed]
- Monagas, M.; Urpi-Sarda, M.; Sanchez-Patan, F.; Llorach, R.; Garrido, I.; Gomez-Cordoves, C.; Andres-Lacueva, C.; Bartolome, B. Insights into the metabolism and microbial biotransformation of dietary flavan-3-ols and the bioactivity of their metabolites. Food Funct. 2010, 1, 233–253. [Google Scholar] [CrossRef]
- Stoupi, S.; Williamson, G.; Drynan, J.W.; Barron, D.; Clifford, M.N. A comparison of the in vitro biotransformation of (-)-epicatechin and procyanidin b2 by human faecal microbiota. Mol. Nutr. Food Res. 2010, 54, 747–759. [Google Scholar] [CrossRef]
- Kutschera, M.; Engst, W.; Blaut, M.; Braune, A. Isolation of catechin-converting human intestinal bacteria. J. Appl. Microbiol. 2011, 111, 165–175. [Google Scholar] [CrossRef]
- Meng, X.; Sang, S.; Zhu, N.; Lu, H.; Sheng, S.; Lee, M.-J.; Ho, C.-T.; Yang, C.S. Identification and characterization of methylated and ring-fission metabolites of tea catechins formed in humans, mice, and rats. Chem. Res. Toxicol. 2002, 15, 1042–1050. [Google Scholar] [CrossRef] [PubMed]
- Takagaki, A.; Nanjo, F. Metabolism of (-)-epigallocatechin gallate by rat intestinal flora. J. Agric. Food Chem. 2010, 58, 1313–1321. [Google Scholar] [CrossRef]
- van’t Slot, G.; Humpf, H.U. Degradation and metabolism of catechin, epigallocatechin-3-gallate (egcg), and related compounds by the intestinal microbiota in the pig cecum model. J. Agric. Food Chem. 2009, 57, 8041–8048. [Google Scholar] [CrossRef]
- van der Hooft, J.J.; de Vos, R.C.; Mihaleva, V.; Bino, R.J.; Ridder, L.; de Roo, N.; Jacobs, D.M.; van Duynhoven, J.P.; Vervoort, J. Structural elucidation and quantification of phenolic conjugates present in human urine after tea intake. Anal. Chem. 2012, 84, 7263–7271. [Google Scholar] [CrossRef]
- Roowi, S.; Stalmach, A.; Mullen, W.; Lean, M.E.; Edwards, C.A.; Crozier, A. Green tea flavan-3-ols: Colonic degradation and urinary excretion of catabolites by humans. J. Agric. Food Chem. 2010, 58, 1296–1304. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-J.; Maliakal, P.; Chen, L.; Meng, X.; Bondoc, F.Y.; Prabhu, S.; Lambert, G.; Mohr, S.; Yang, C.S. Pharmacokinetics of tea catechins after ingestion of green tea and (−)-epigallocatechin-3-gallate by humans. Cancer Epidemiol. Biomarkers Prev. 2002, 11, 1025–1032. [Google Scholar] [PubMed]
- Lee, H.C.; Jenner, A.M.; Low, C.S.; Lee, Y.K. Effect of tea phenolics and their aromatic fecal bacterial metabolites on intestinal microbiota. Res. Microbiol. 2006, 157, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Okubo, T.; Ishihara, N.; Oura, A.; Serit, M.; Kim, M.; Yamamoto, T.; Mitsuoka, T. In vivo effects of tea polyphenol intake on human intestinal microflora and metabolism. Biosci. Biotechnol. Biochem. 1992, 56, 588–591. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.S.; Touyama, M.; Hisada, T.; Benno, Y. Effects of green tea consumption on human fecal microbiota with special reference to bifidobacterium species. Microbiol. Immunol. 2012, 56, 729–739. [Google Scholar] [CrossRef]
- Janssens, P.L.; Penders, J.; Hursel, R.; Budding, A.E.; Savelkoul, P.H.; Westerterp-Plantenga, M.S. Long-term green tea supplementation does not change the human gut microbiota. PLoS ONE 2016, 11, e0153134. [Google Scholar] [CrossRef]
- Rastmanesh, R. High polyphenol, low probiotic diet for weight loss because of intestinal microbiota interaction. Chem. Biol. Interact. 2011, 189, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Samavat, H.; Dostal, A.M.; Wang, R.; Bedell, S.; Emory, T.H.; Ursin, G.; Torkelson, C.J.; Gross, M.D.; Le, C.T.; Yu, M.C.; et al. The minnesota green tea trial (mgtt), a randomized controlled trial of the efficacy of green tea extract on biomarkers of breast cancer risk: Study rationale, design, methods, and participant characteristics. Cancer Causes Control 2015, 26, 1405–1419. [Google Scholar] [CrossRef]
- Dostal, A.M.; Espejo, L.; Kurzer, M.S.; Arikawa, A. Long-term supplementation of green tea extract does not modify adiposity or bone mineral density in a randomized trial of overweight and obese postmenopausal women. J. Nutr. 2015, 146, 256–264. [Google Scholar] [CrossRef]
- Dostal, A.M.; Samavat, H.; Espejo, L.; Arikawa, A.Y.; Stendell-Hollis, N.R.; Kurzer, M.S. Green tea extract and catechol-o-methyltransferase genotype modify fasting serum insulin and plasma adiponectin concentrations in a randomized controlled trial of overweight and obese postmenopausal women. J. Nutr. 2016, 146, 38–45. [Google Scholar] [CrossRef]
- Dostal, A.M.; Arikawa, A.; Espejo, L.; Bedell, S.; Kurzer, M.S.; Stendell-Hollis, N.R. Green tea extract and catechol-o-methyltransferase genotype modify the post-prandial serum insulin response in a randomised trial of overweight and obese post-menopausal women. J. Hum. Nutr. Diet 2017, 30, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Xu, F.; Zhang, Y.Z.; Zang, X.Y.; Wang, D.; Shang, M.Y.; Wang, X.; Chui, D.H.; Cai, S.Q. The profiling and identification of the metabolites of (+)-catechin and study on their distribution in rats by hplc-dad-esi-it-tof-ms(n) technique. Biomed. Chromatogr. 2014, 28, 401–411. [Google Scholar] [CrossRef]
- Sang, S.; Lee, M.-J.; Yang, I.; Buckley, B.; Yang, C.S. Human urinary metabolite profile of tea polyphenols analyzed by liquid chromatography/electrospray ionization tandem mass spectrometry with data-dependent acquisition. Rapid Commun. Mass Spectrom. 2008, 22, 1567–1578. [Google Scholar] [CrossRef]
- Brindani, N.; Mena, P.; Calani, L.; Benzie, I.; Choi, S.W.; Brighenti, F.; Zanardi, F.; Curti, C.; Del Rio, D. Synthetic and analytical strategies for the quantification of phenyl-gamma-valerolactone conjugated metabolites in human urine. Mol. Nutr. Food Res. 2017, 61. [Google Scholar] [CrossRef]
- Spencer, J.P. Metabolism of tea flavonoids in the gastrointestinal tract. J. Nutr. 2003, 133, 3255s–3261s. [Google Scholar] [CrossRef] [PubMed]
- Schantz, M.; Erk, T.; Richling, E. Metabolism of green tea catechins by the human small intestine. Biotechnol. J. 2010, 5, 1050–1059. [Google Scholar] [CrossRef]
- Del Rio, D.; Costa, L.G.; Lean, M.E.J.; Crozier, A. Polyphenols and health: What compounds are involved? Nutr. Metab. Cardiovasc. Dis. 2010, 20, 1–6. [Google Scholar] [CrossRef]
- Mena, P.; Bresciani, L.; Brindani, N.; Ludwig, I.A.; Pereira-Caro, G.; Angelino, D.; Llorach, R.; Calani, L.; Brighenti, F.; Clifford, M.N.; et al. Phenyl-γ-valerolactones and phenylvaleric acids, the main colonic metabolites of flavan-3-ols: Synthesis, analysis, bioavailability, and bioactivity. Nat. Prod. Rep. 2019. (Epub ahead of print). [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Girón, A.; Muñoz-González, I.; Martínlvarez, P.J.; Moreno-Arribas, M.V.; Bartolomé, B. Towards the fecal metabolome derived from moderate red wine intake. Metabolites 2014, 4, 1101–1118. [Google Scholar] [CrossRef]
- Lu, H.; Meng, X.; Lee, M.-J.; Li, C.; Maliakal, P.; Yang, C.S. Bioavailability and biological activity of tea polyphenols. In Food Factors in Health Promotion and Disease Prevention; American Chemical Society: Washington, DC, USA, 2003; Vol. 851, pp. 9–15. [Google Scholar]
- McKay, D.L.; Blumberg, J.B. The role of tea in human health: An update. J. Am. Coll. Nutr. 2002, 21, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, C.; Artacho, R.; Giménez, R. Beneficial effects of green tea—A review. J. Am. Coll. Nutr. 2006, 25, 79–99. [Google Scholar] [PubMed]
- Lambert, J.D.; Rice, J.E.; Hong, J.; Hou, Z.; Yang, C.S. Synthesis and biological activity of the tea catechin metabolites, m4 and m6 and their methoxy-derivatives. Bioorg. Med. Chem. Lett. 2005, 15, 873–876. [Google Scholar] [CrossRef]
- Uhlenhut, K.; Högger, P. Facilitated cellular uptake and suppression of inducible nitric oxide synthase by a metabolite of maritime pine bark extract (pycnogenol). Free Radic. Biol. Med. 2012, 53, 305–313. [Google Scholar] [CrossRef]
- Sun, Y.N.; Li, W.; Song, S.B.; Yan, X.T.; Zhao, Y.; Jo, A.R.; Kang, J.S.; Young Ho, K. A new phenolic derivative with soluble epoxide hydrolase and nuclear factor-kappab inhibitory activity from the aqueous extract of acacia catechu. Nat. Prod. Res. 2016, 30, 2085–2092. [Google Scholar] [CrossRef]
- Takagaki, A.; Nanjo, F. Effects of metabolites produced from (−)-epigallocatechin gallate by rat intestinal bacteria on angiotensin i-converting enzyme activity and blood pressure in spontaneously hypertensive rats. J. Agric. Food Chem. 2015, 63, 8262–8266. [Google Scholar] [CrossRef]
- Rodriguez-Mateos, A.; Feliciano, R.P.; Boeres, A.; Weber, T.; Dos Santos, C.N.; Ventura, M.R.; Heiss, C. Cranberry (poly)phenol metabolites correlate with improvements in vascular function: A double-blind, randomized, controlled, dose-response, crossover study. Mol. Nutr. Food Res. 2016, 60, 2130–2140. [Google Scholar] [CrossRef]
- Hara-Terawaki, A.; Takagaki, A.; Kobayashi, H.; Nanjo, F. Inhibitory activity of catechin metabolites produced by intestinal microbiota on proliferation of hela cells. Biol. Pharm. Bull. 2017, 40, 1331–1335. [Google Scholar] [PubMed]
- Aronov, P.A.; Luo, F.J.; Plummer, N.S.; Quan, Z.; Holmes, S.; Hostetter, T.H.; Meyer, T.W. Colonic contribution to uremic solutes. J. Am. Soc. Nephrol. 2011, 22, 1769–1776. [Google Scholar] [CrossRef] [PubMed]
- Niwa, T.; Ise, M.; Miyazaki, T. Progression of glomerular sclerosis in experimental uremic rats by administration of indole, a precursor of indoxyl sulfate. Am. J. Nephrol. 1994, 14, 207–212. [Google Scholar] [CrossRef]
- Dazard, J.-E.J.; Sandlers, Y.; Doerner, S.K.; Berger, N.A.; Brunengraber, H. Metabolomics of apc min/+ mice genetically susceptible to intestinal cancer. BMC Syst. Biol. 2014, 8, 72. [Google Scholar] [CrossRef] [PubMed]
- Clayton, T.A.; Baker, D.; Lindon, J.C.; Everett, J.R.; Nicholson, J.K. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl. Acad. Sci. USA 2009, 106, 14728–14733. [Google Scholar] [CrossRef]
- Grümer, H.-D. Formation of hippuric acid from phenylalanine labelled with carbon-14 in phenylketonuric subjects. Nature 1961, 189, 63–64. [Google Scholar] [CrossRef]
- Self, H.L.; Brown, R.R.; Price, J.M. Quantitative studies on the metabolites of tryptophan in the urine of swine. J. Nutr. 1960, 70, 21–25. [Google Scholar] [CrossRef]
- Bridges, J.; French, M.; Smith, R.; Williams, R. The fate of benzoic acid in various species. Biochem. J. 1970, 118, 47–51. [Google Scholar] [CrossRef]
- Dodd, D.; Spitzer, M.H.; Van Treuren, W.; Merrill, B.D.; Hryckowian, A.J.; Higginbottom, S.K.; Le, A.; Cowan, T.M.; Nolan, G.P.; Fischbach, M.A.; et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 2017, 551, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Agus, A.; Planchais, J.; Sokol, H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef] [PubMed]
- Stevens, J.F.; Maier, C.S. The chemistry of gut microbial metabolism of polyphenols. Phytochem. Rev. 2016, 15, 425–444. [Google Scholar] [CrossRef]
- Pournourmohammadi, S.; Grimaldi, M.; Stridh, M.H.; Lavallard, V.; Waagepetersen, H.S.; Wollheim, C.B.; Maechler, P. Epigallocatechin-3-gallate (egcg) activates ampk through the inhibition of glutamate dehydrogenase in muscle and pancreatic ss-cells: A potential beneficial effect in the pre-diabetic state? Int. J. Biochem. Cell Biol. 2017, 88, 220–225. [Google Scholar] [CrossRef]
- Rodriguez-Caso, C.; Rodriguez-Agudo, D.; Sanchez-Jimenez, F.; Medina, M.A. Green tea epigallocatechin-3-gallate is an inhibitor of mammalian histidine decarboxylase. Cell Mol. Life Sci. 2003, 60, 1760–1763. [Google Scholar] [PubMed]
- Li, C.; Li, M.; Chen, P.; Narayan, S.; Matschinsky, F.M.; Bennett, M.J.; Stanley, C.A.; Smith, T.J. Green tea polyphenols control dysregulated glutamate dehydrogenase in transgenic mice by hijacking the adp activation site. J. Biol. Chem. 2011, 286, 34164–34174. [Google Scholar] [CrossRef]
- Mulder, T.P.; Rietveld, A.G.; van Amelsvoort, J.M. Consumption of both black tea and green tea results in an increase in the excretion of hippuric acid into urine. Am. J. Clin. Nutr. 2005, 81, 256s–260s. [Google Scholar] [CrossRef] [PubMed]
- Gholson, R.K.; Sanders, D.C.; Henderson, L.M. Glutaric acid: A product of tryptophan metabolism. Biochem. Biophys. Res. Commun. 1959, 1, 98–100. [Google Scholar] [CrossRef]
- Fothergill, J.C.; Guest, J.R. Catabolism of l-lysine by pseudomonas aeruginosa. J. Gen. Microbiol. 1977, 99, 139–155. [Google Scholar] [CrossRef] [PubMed]
- Hung, S.-C.; Kuo, K.-L.; Wu, C.-C.; Tarng, D.-C. Indoxyl sulfate: A novel cardiovascular risk factor in chronic kidney disease. J. Am. Heart Assoc. 2017, 6, e005022. [Google Scholar] [CrossRef] [PubMed]
- Sallee, M.; Dou, L.; Cerini, C.; Poitevin, S.; Brunet, P.; Burtey, S. The aryl hydrocarbon receptor-activating effect of uremic toxins from tryptophan metabolism: A new concept to understand cardiovascular complications of chronic kidney disease. Toxins 2014, 6, 934–949. [Google Scholar] [CrossRef]
- Bao, H.; Peng, A. The green tea polyphenol(-)-epigallocatechin-3-gallate and its beneficial roles in chronic kidney disease. J. Transl. Int. Med. 2016, 4, 99–103. [Google Scholar] [CrossRef]
- Yang, C.S.; Zhang, J. Studies on the prevention of cancer and cardiometabolic diseases by tea: Issues on mechanisms, effective doses and toxicities. J. Agric. Food Chem. 2018. (Epub ahead of print). [Google Scholar] [CrossRef]
- Lu, Y.; Yao, D.; Chen, C. 2-hydrazinoquinoline as a derivatization agent for lc-ms-based metabolomic investigation of diabetic ketoacidosis. Metabolites 2013, 3, 993–1010. [Google Scholar] [CrossRef]
- Gohl, D.M.; Vangay, P.; Garbe, J.; MacLean, A.; Hauge, A.; Becker, A.; Gould, T.J.; Clayton, J.B.; Johnson, T.J.; Hunter, R.; et al. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat. Biotechnol. 2016, 34, 942. [Google Scholar] [CrossRef] [PubMed]
- Burns, M.B.; Lynch, J.; Starr, T.K.; Knights, D.; Blekhman, R. Virulence genes are a signature of the microbiome in the colorectal tumor microenvironment. Genome Med. 2015, 7, 55. [Google Scholar] [CrossRef] [PubMed]
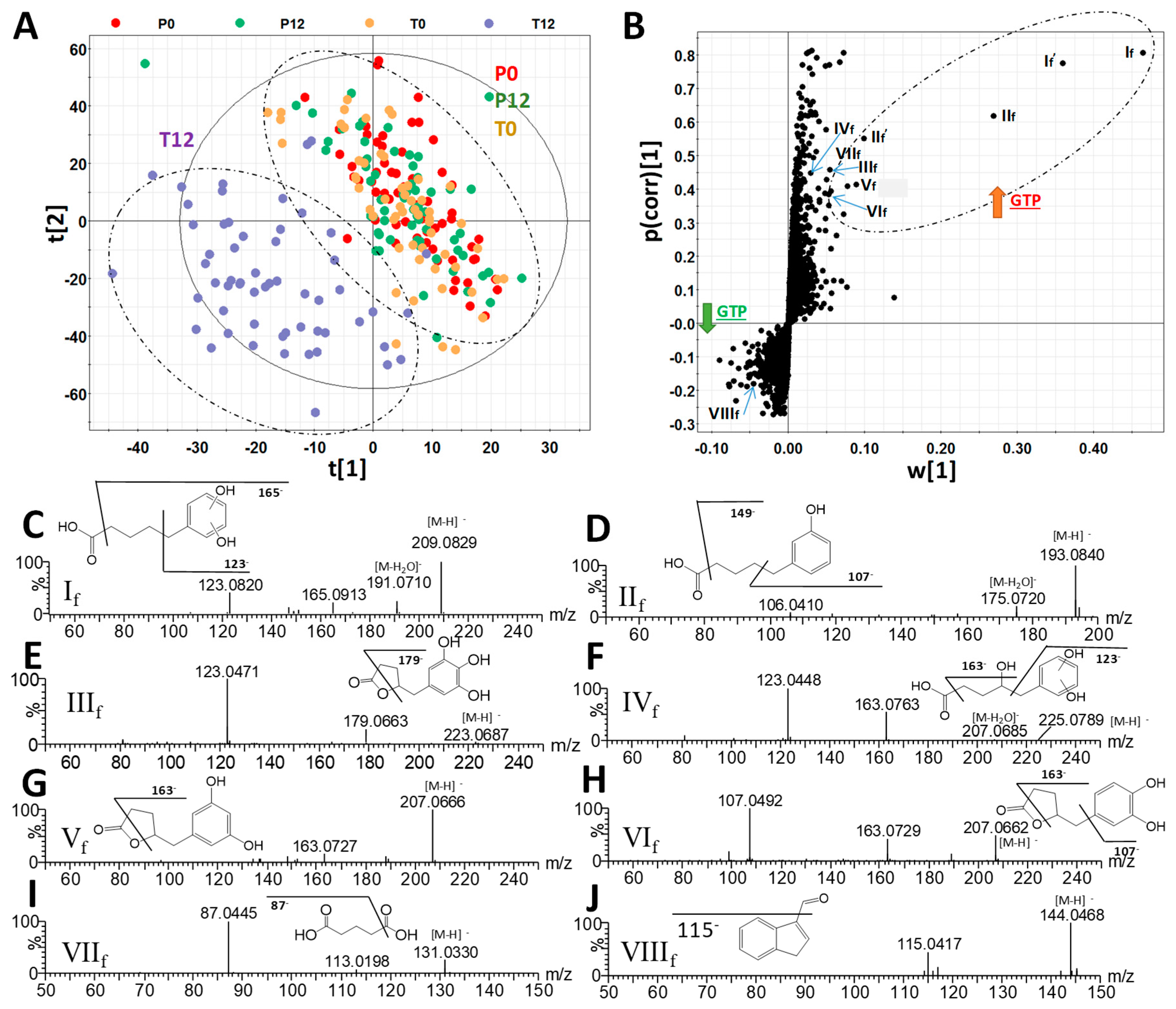
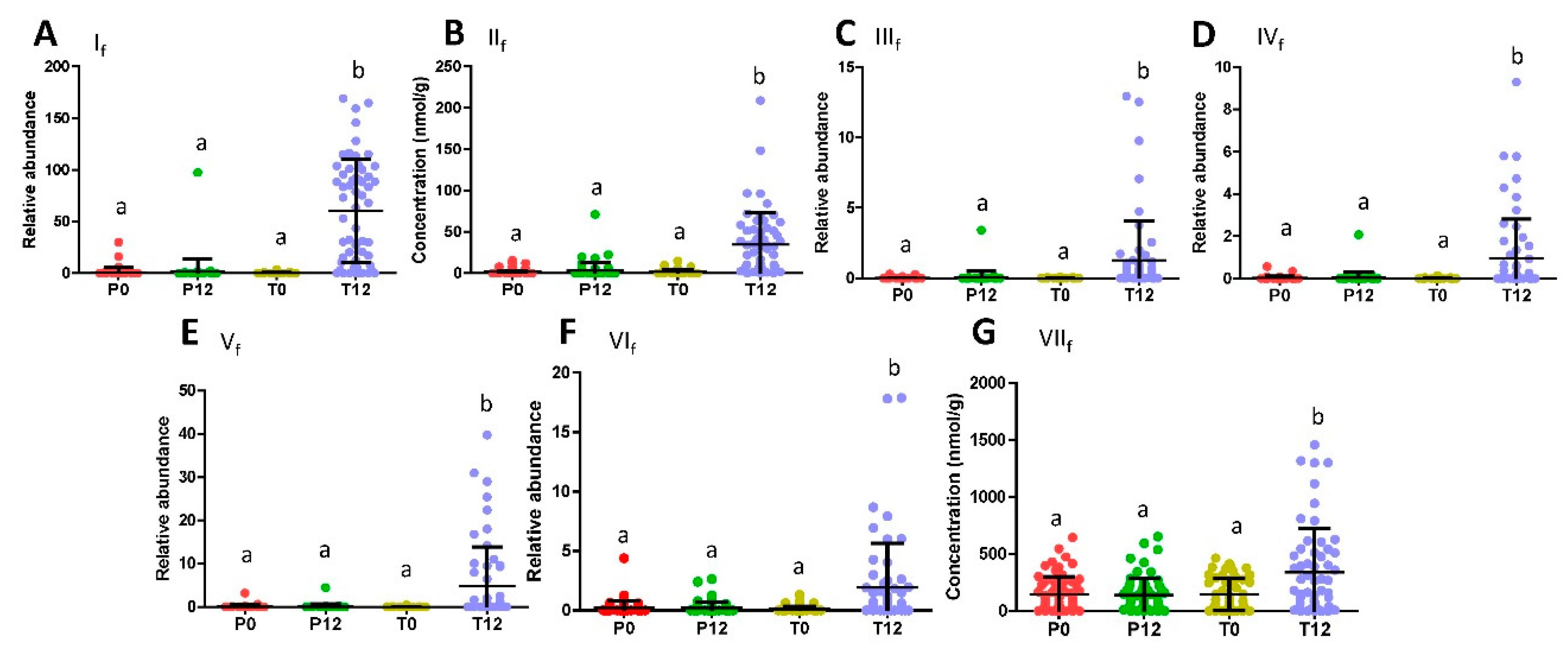
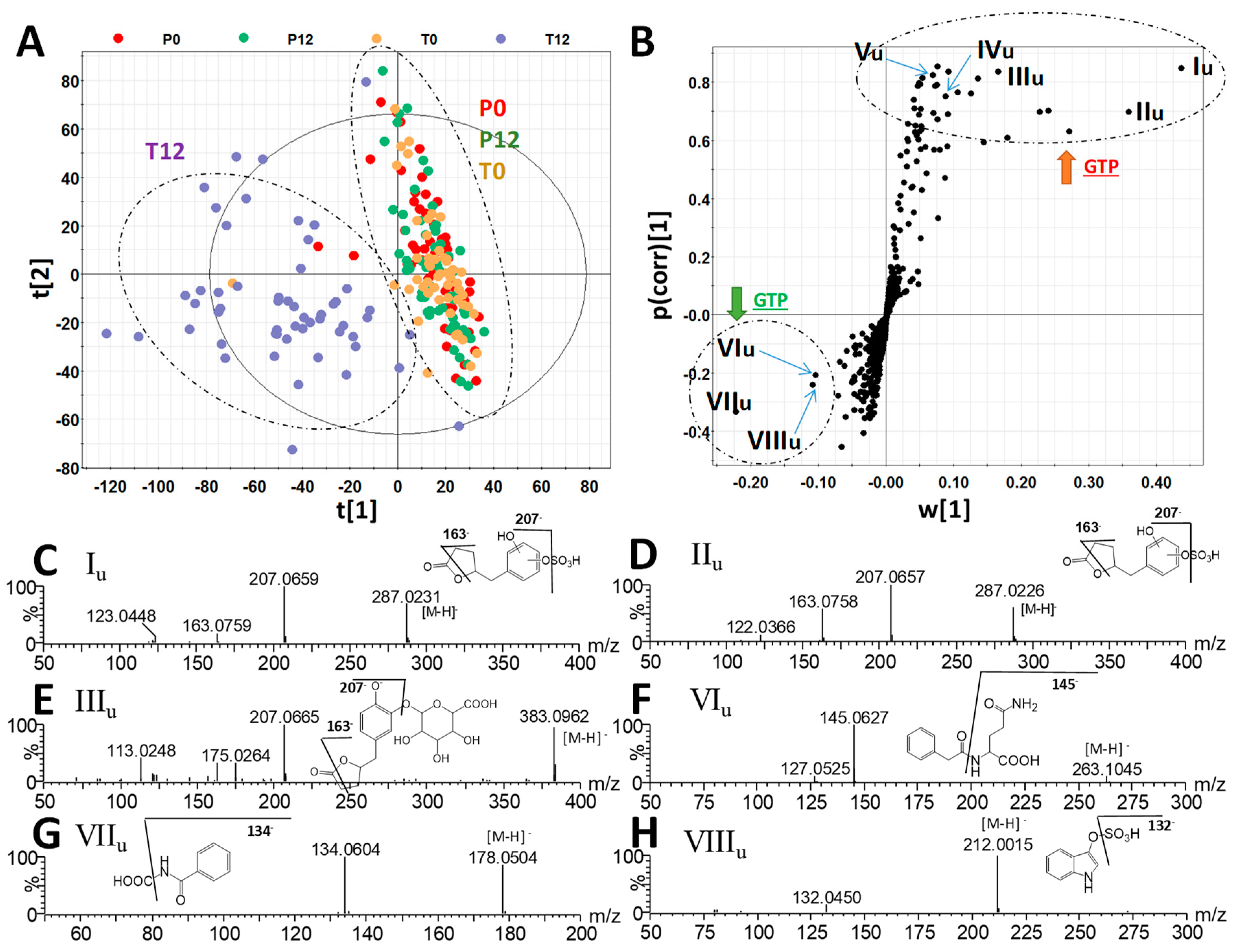
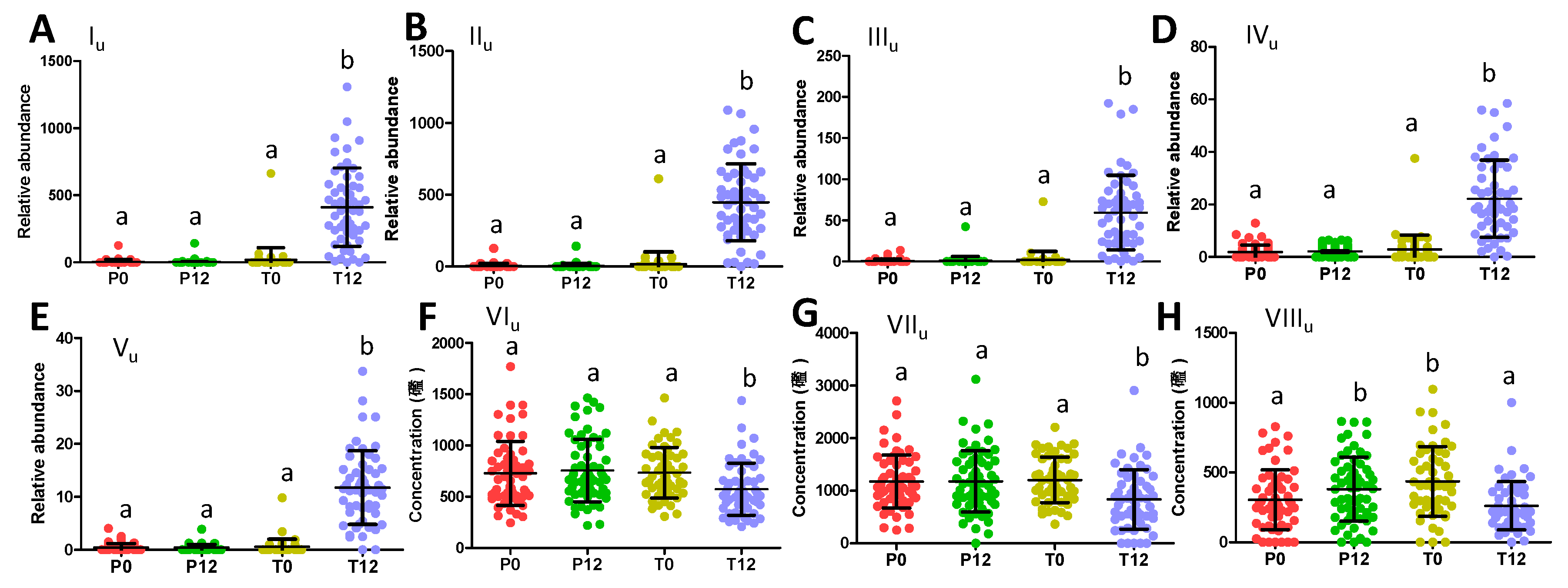
| Ions | Retention Time (min) | m/z of [M–H]- | Δppm | Formula | Identity |
|---|---|---|---|---|---|
| If | 3.40 | 209.0819 | 2 | C11H14O4 | 5-(Dihydroxyphenyl)-valeric acid |
| If’ | 3.40 | 419.1704 | 0 | C22H28O8 | 5-(Dihydroxyphenyl)-valeric acid in-source dimer |
| IIf | 4.79 | 193.0867 | 1 | C11H14O3 | 3-Hydroxyphenyl-valeric acid |
| IIf’ | 4.79 | 387.1799 | 2 | C22H28O6 | 3-Hydroxyphenyl-valeric acid in-source dimer |
| IIIf | 2.09 | 223.0609 | 1 | C11H12O5 | 5-(3′,4′,5′-Trihydroxyphenyl)-γ-valerolactone |
| IVf | 2.01 | 225.0763 | 2 | C11H14O5 | 4-Hydroxy-5-(dihydroxyphenyl)-valeric acid |
| Vf | 2.72 | 207.0661 | 1 | C11H12O4 | 5-(3′,5′-Dihydroxyphenyl)-γ-valerolactone |
| VIf | 3.05 | 207.0664 | 3 | C11H12O4 | 5-(3′,4′-Dihydroxyphenyl)-γ-valerolactone |
| VIIf | 0.68 | 131.0341 | 6 | C5H8O4 | Glutaric acid |
| VIIIf | 3.81 | 144.0449 | 4 | C9H7NO | Indole-3-carboxaldehyde |
| Ions | RT (min) | m/z of [M – H]- | Δppm | Formula | Identity |
|---|---|---|---|---|---|
| Iu | 2.41 | 287.0225 | 2 | C11H12O7S | 5-(Dihydroxyphenyl)-γ-valerolactone sulfate |
| IIu | 2.93 | 287.0225 | 2 | C11H12O7S | 5-(Dihydroxyphenyl)-γ-valerolactone sulfate |
| IIIu | 2.10 | 383.0971 | 3 | C17H20O10 | 5-(Dihydroxyphenyl)-γ-valerolactone glucuronide |
| IVu | 3.29 | 383.0420 | 4 | C16H16O9S | Methyl epicatechin sulfate |
| Vu | 2.42 | 495.1121 | 4 | C22H24O13 | Methyl epigallocatechin glucuronide |
| VIu | 2.59 | 263.1031 | 2 | C13H16N2O4 | Phenylacetylglutamine |
| VIIu | 2.39 | 178.0503 | 3 | C9H8NO3 | Hippuric acid |
| VIIIu | 2.42 | 212.0017 | 2 | C8H7NO4S | Indoxyl sulfate |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Zhang, N.; Arikawa, A.Y.; Chen, C. Inhibitory Effects of Green Tea Polyphenols on Microbial Metabolism of Aromatic Amino Acids in Humans Revealed by Metabolomic Analysis. Metabolites 2019, 9, 96. https://doi.org/10.3390/metabo9050096
Zhou Y, Zhang N, Arikawa AY, Chen C. Inhibitory Effects of Green Tea Polyphenols on Microbial Metabolism of Aromatic Amino Acids in Humans Revealed by Metabolomic Analysis. Metabolites. 2019; 9(5):96. https://doi.org/10.3390/metabo9050096
Chicago/Turabian StyleZhou, Yuyin, Ningning Zhang, Andrea Y. Arikawa, and Chi Chen. 2019. "Inhibitory Effects of Green Tea Polyphenols on Microbial Metabolism of Aromatic Amino Acids in Humans Revealed by Metabolomic Analysis" Metabolites 9, no. 5: 96. https://doi.org/10.3390/metabo9050096
APA StyleZhou, Y., Zhang, N., Arikawa, A. Y., & Chen, C. (2019). Inhibitory Effects of Green Tea Polyphenols on Microbial Metabolism of Aromatic Amino Acids in Humans Revealed by Metabolomic Analysis. Metabolites, 9(5), 96. https://doi.org/10.3390/metabo9050096






