Comparison of Bi- and Tri-Linear PLS Models for Variable Selection in Metabolomic Time-Series Experiments
Abstract
1. Introduction
2. Materials and Methods
2.1. PLS and NPLS
2.2. PLS-DA and Dummy Y
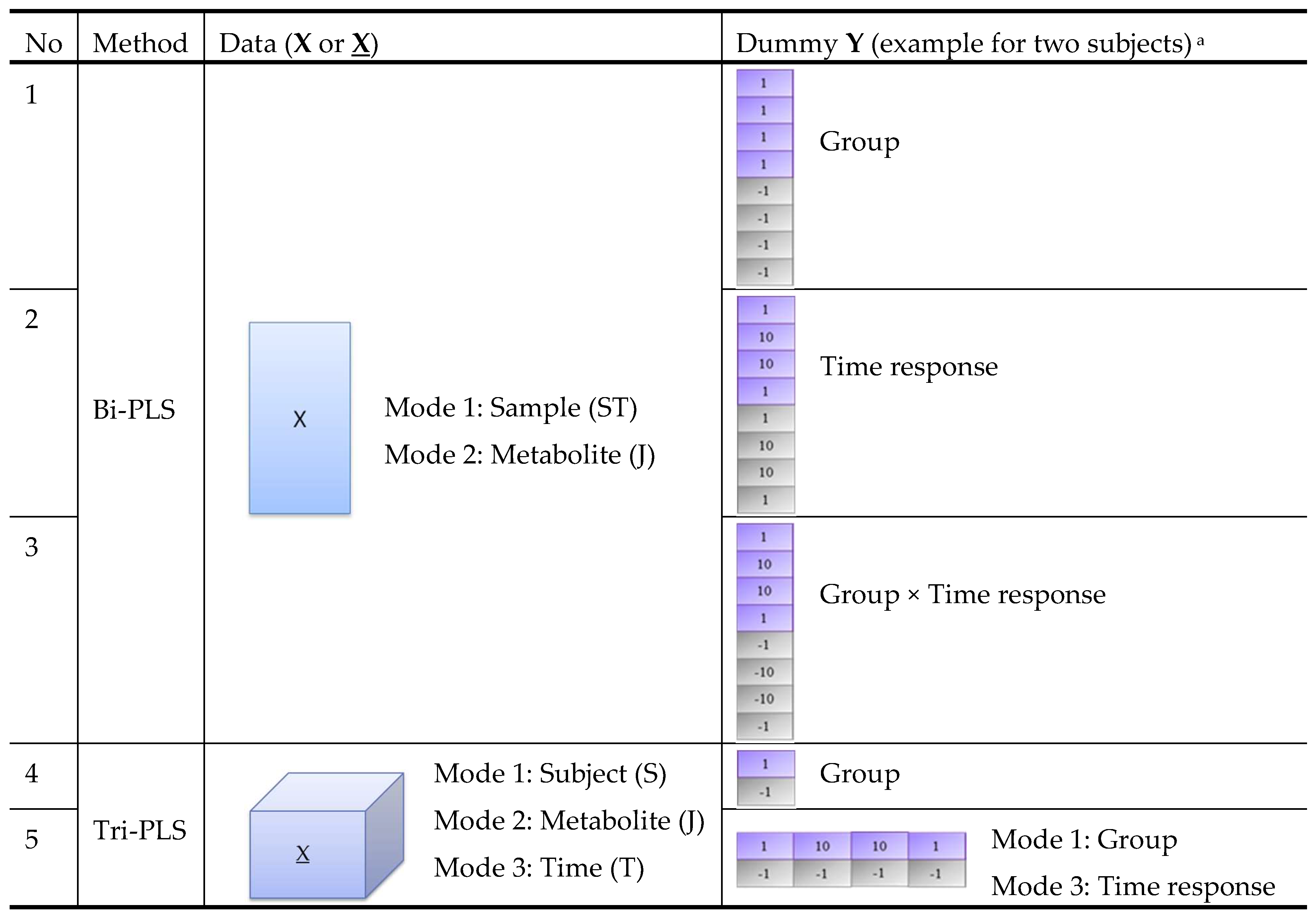
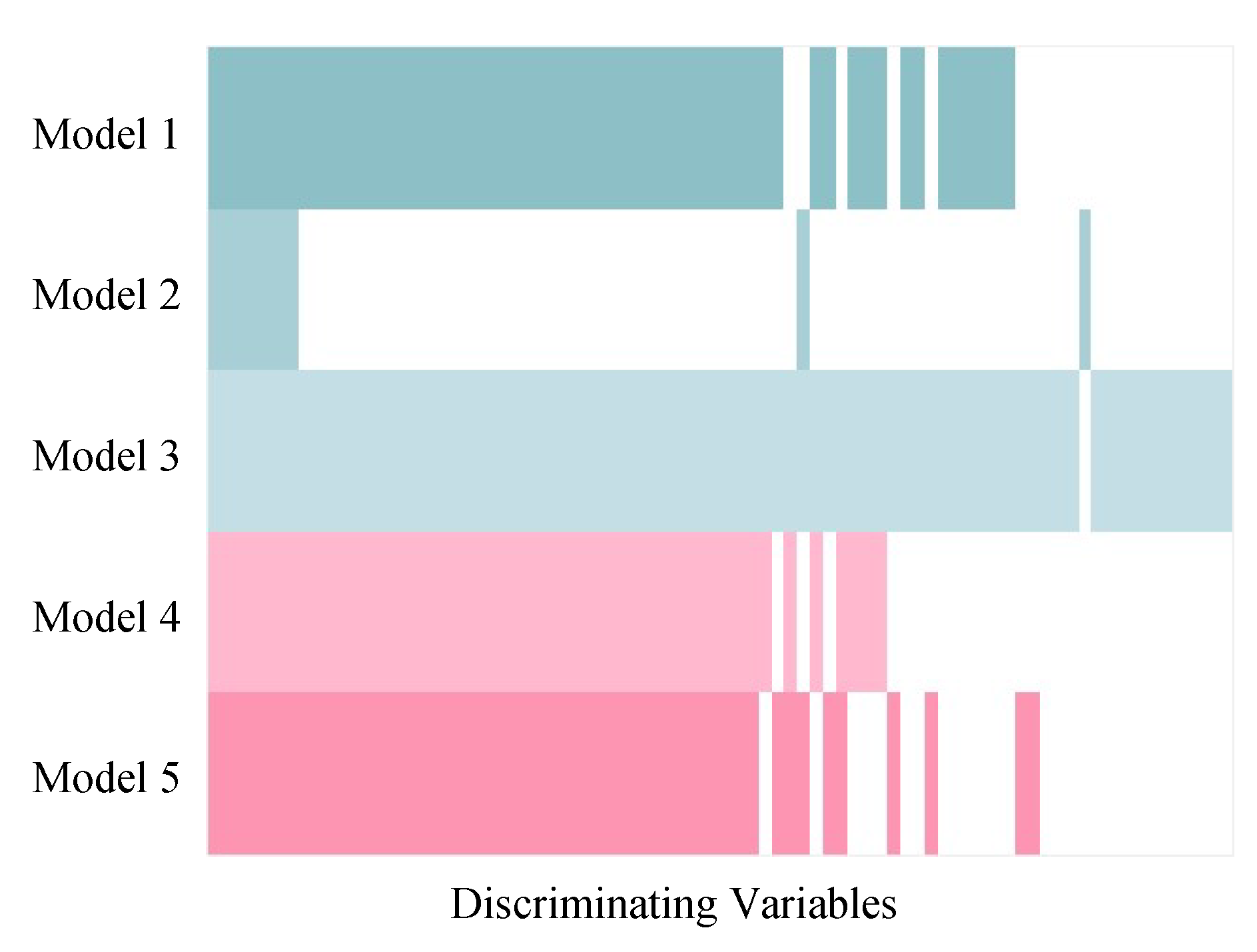
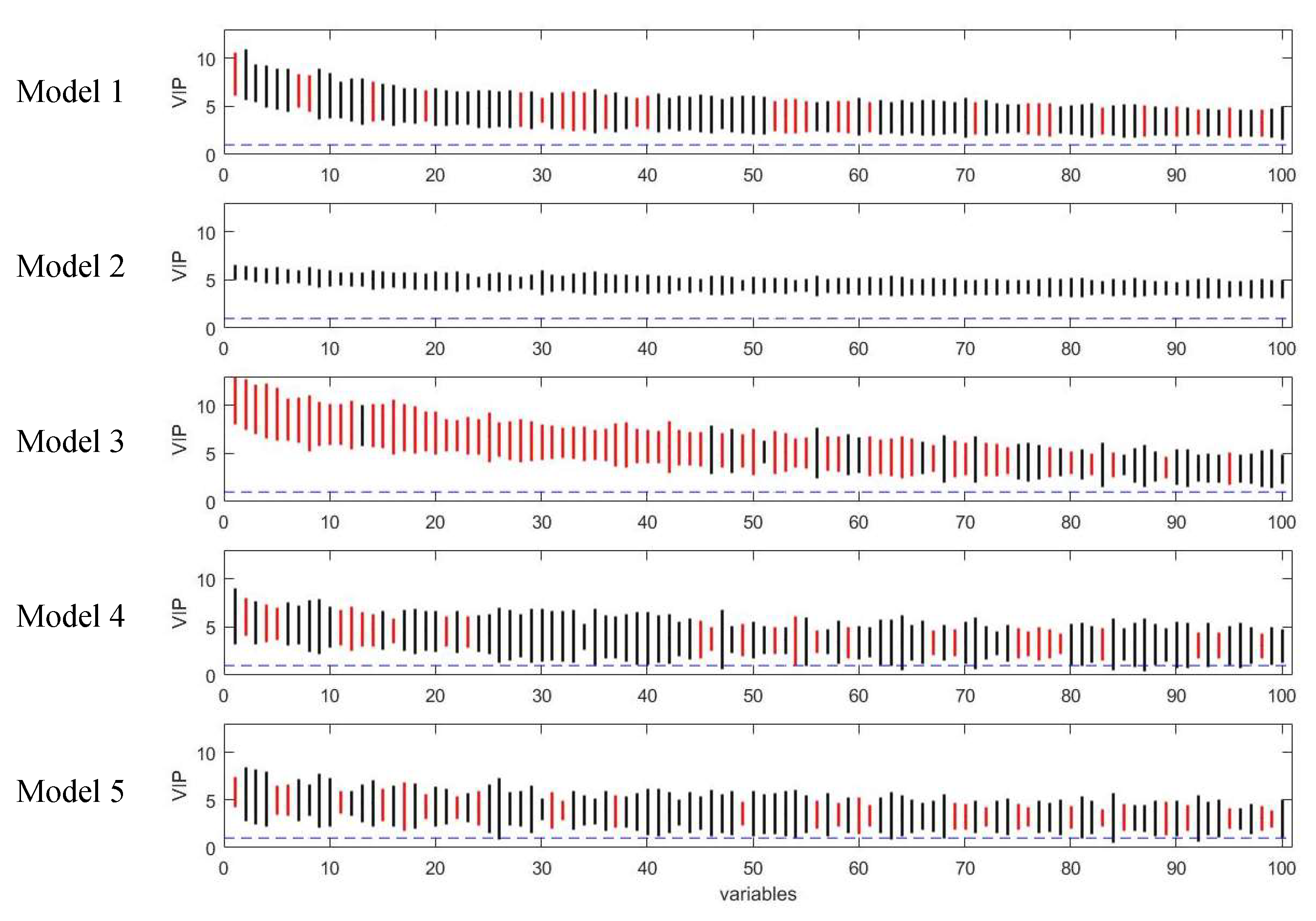
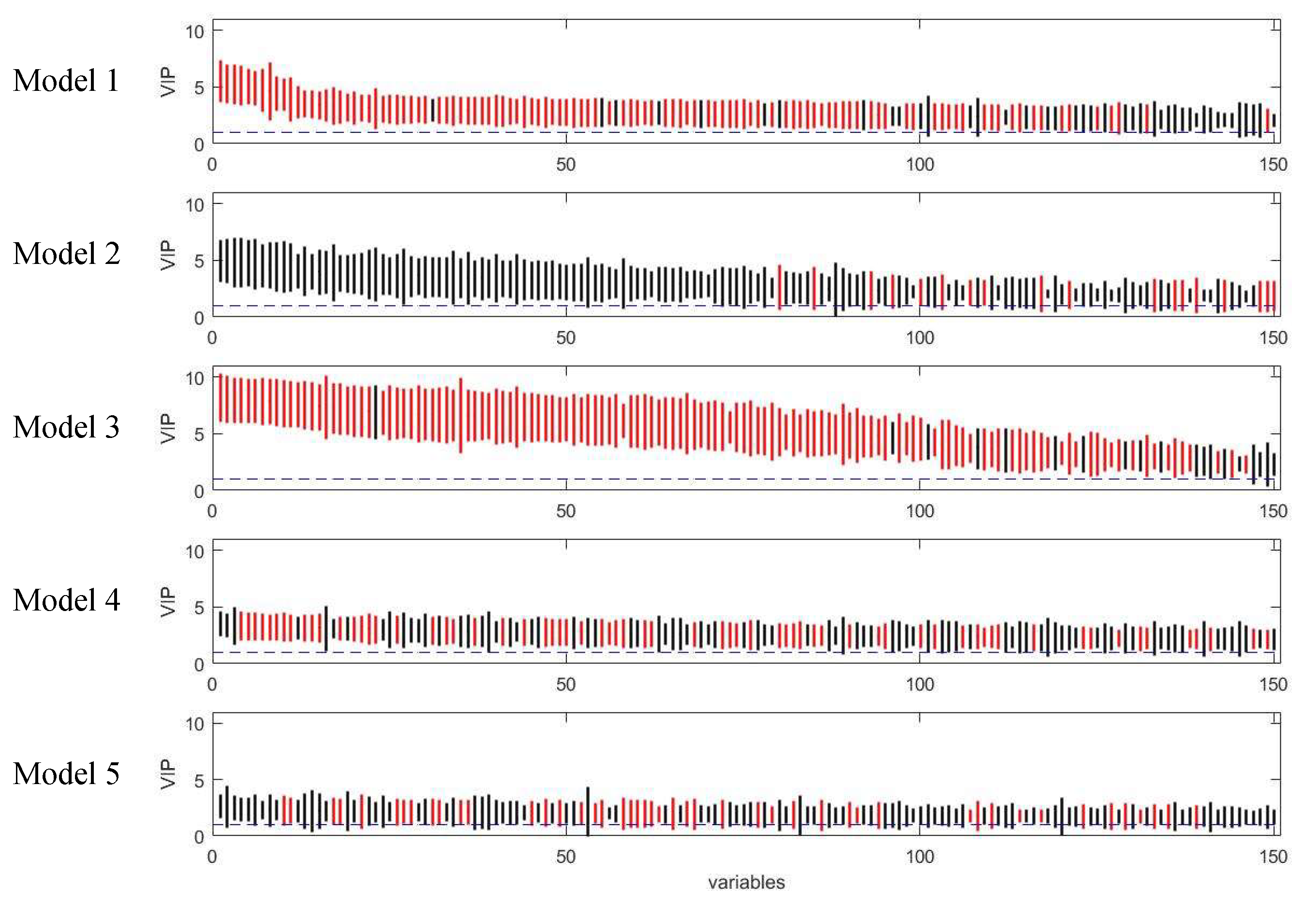
2.3. Comparison of Variable Selection by Five PLS Models
2.4. Datasets
2.4.1. Simulated Datasets
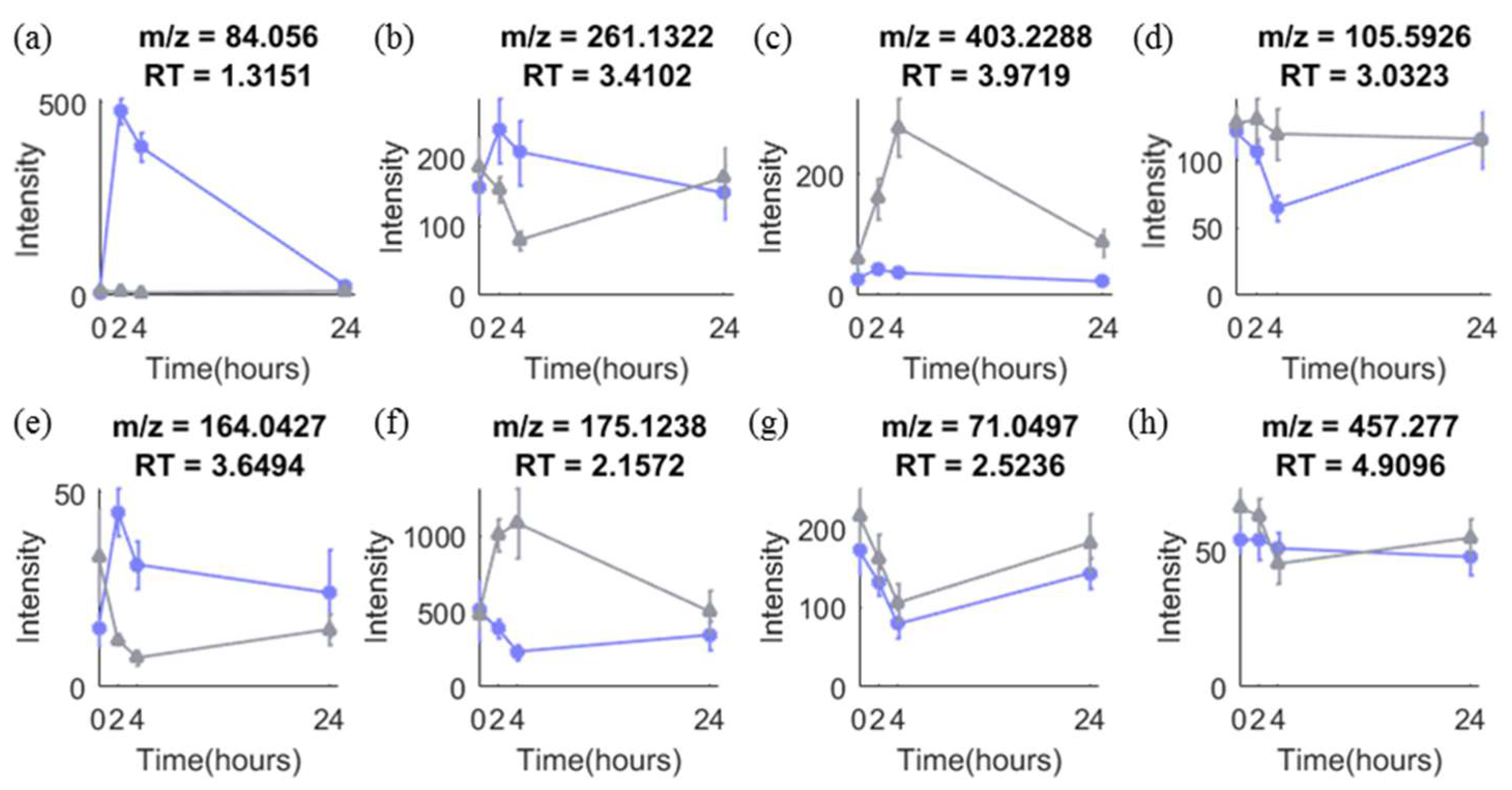
2.4.2. Onion Intervention Data
2.4.3. Coffee Intervention Data
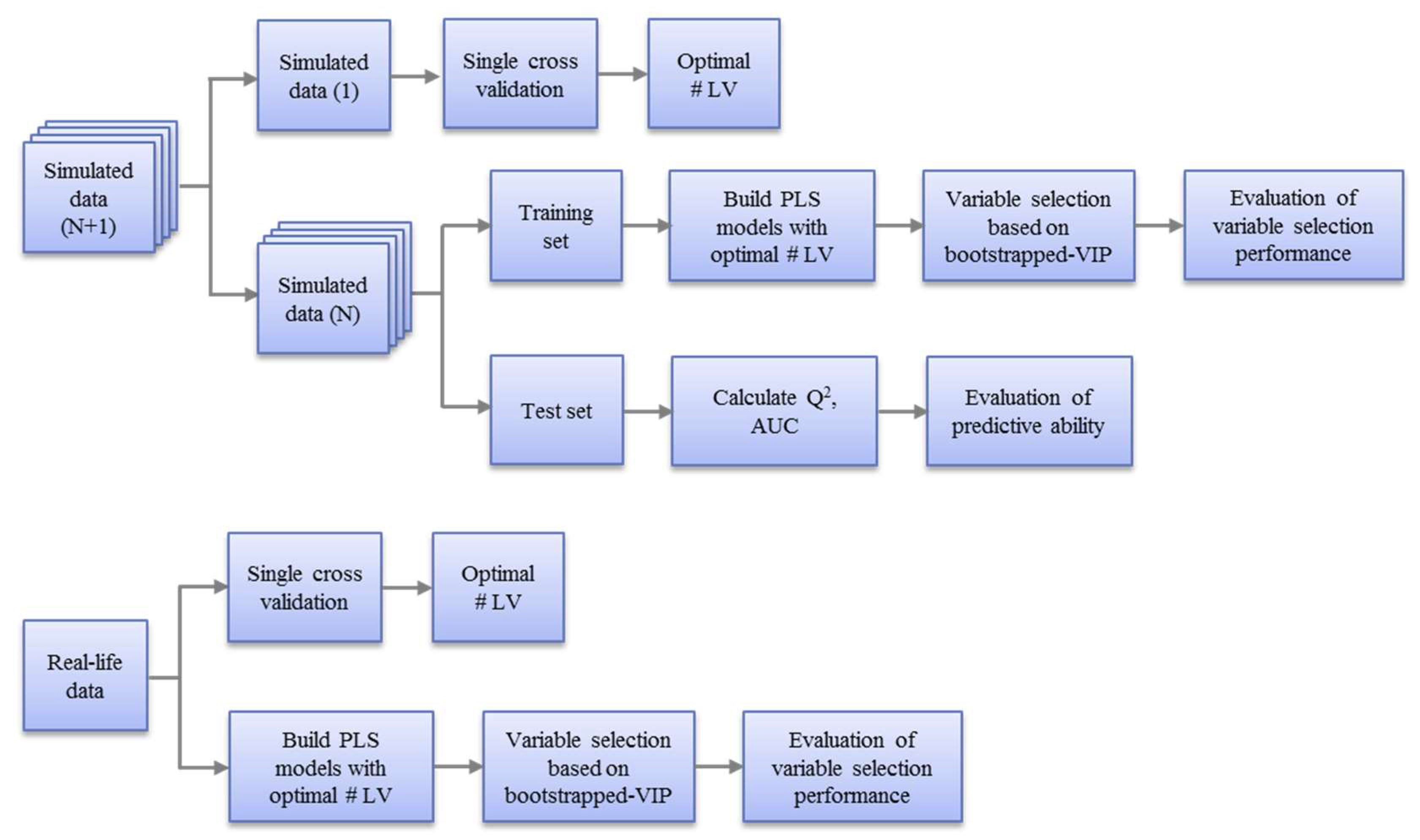
2.5. Workflow
2.5.1. Pre-Processing of Data
2.5.2. Model Optimization and Evaluation
(1). Evaluation of Variable Selection Performance with Training Sets
- Recall = TP/(TP + FN)
- Precision = TP/(TP + FP)
- F1-score = (β2+1)× Precision × Recall/β2× (Precision + Recall)
(2). Evaluation of predictive ability with test sets
2.6. Evaluation of the Influence of Characteristics of the Dataset on the Model Performance
3. Results
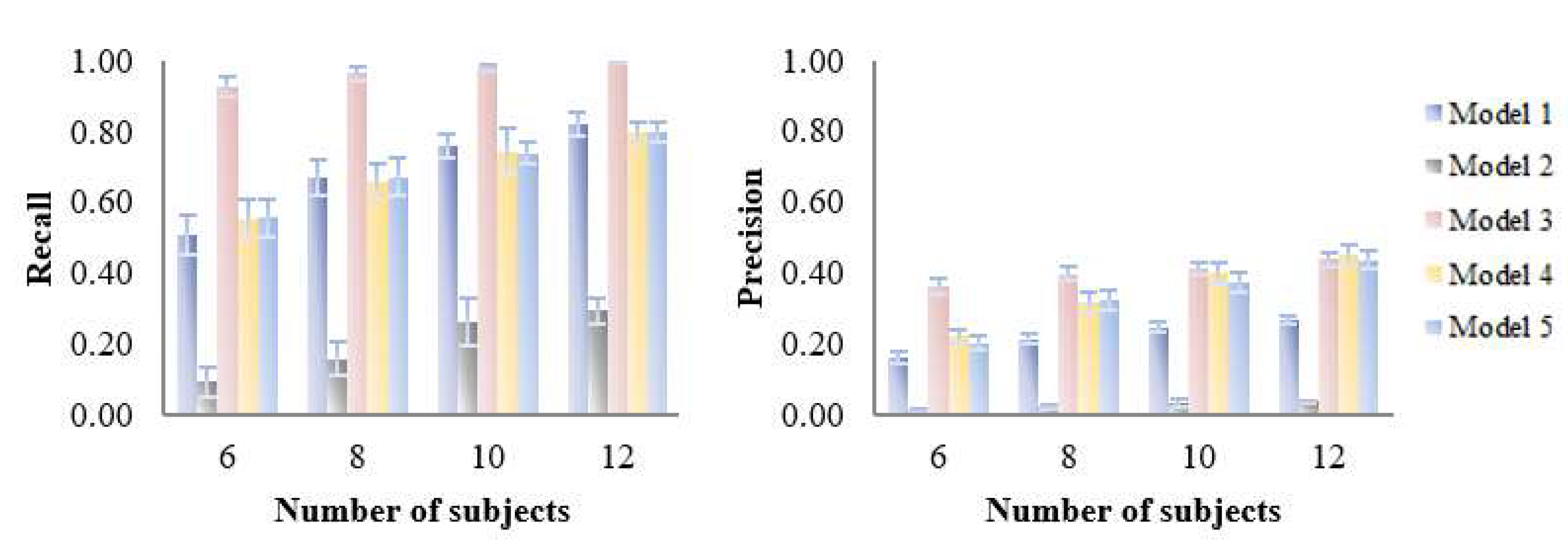
3.1. Assessment of Variable Selection Performance on Simulated Data
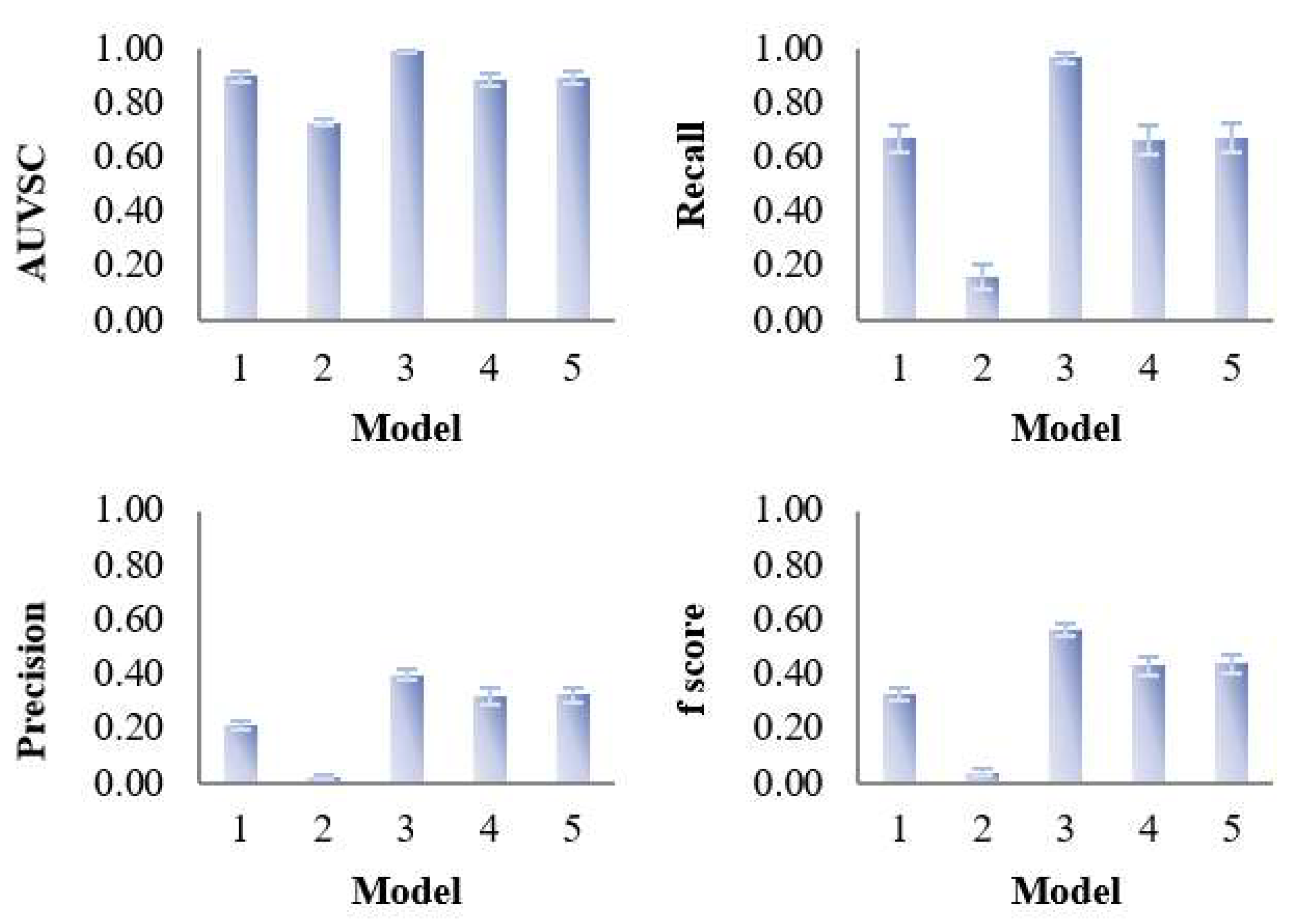
3.1.1. Overall Evaluation
3.1.2. Influence of Characteristics of the Dataset on the Performance of the Five PLS Models
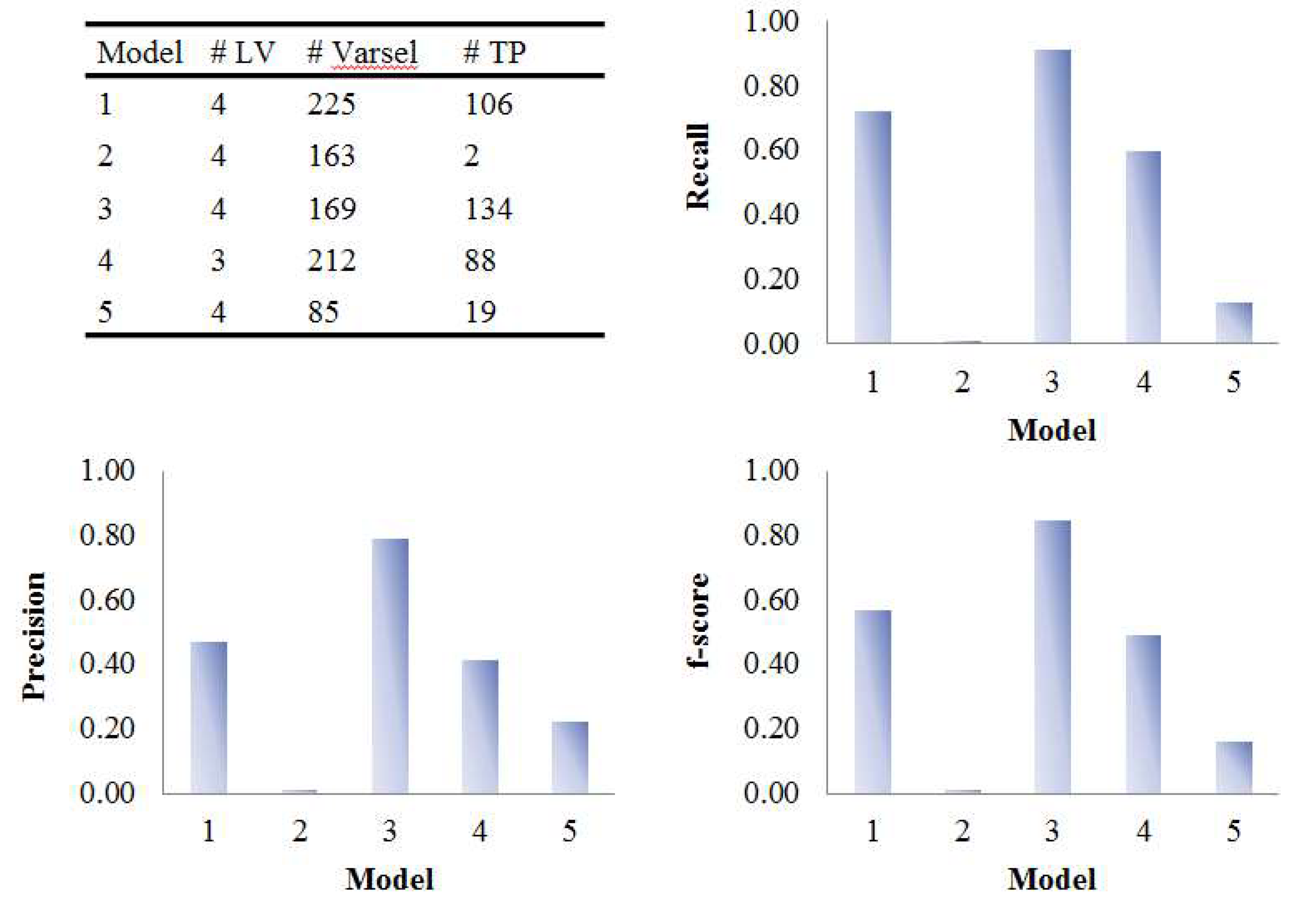
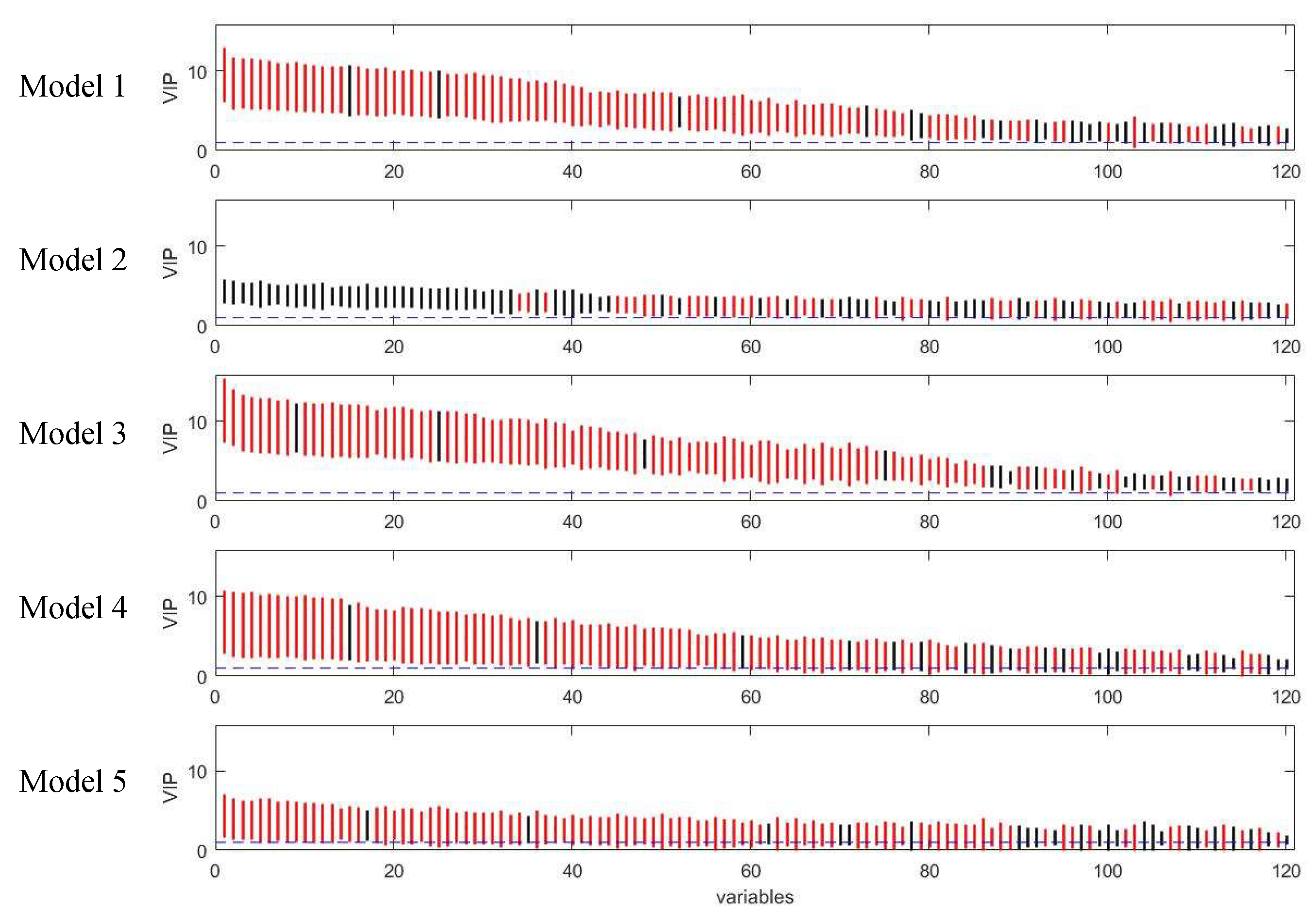
3.2. Assessment of Variable Selection Performance on Real Data
3.2.1. Coffee Intervention Study
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rezzi, S.; Ramadan, Z.; Fay, L.B.; Kochhar, S. Nutritional metabonomics: applications and perspectives. J. Proteome Res. 2007, 6, 513–525. [Google Scholar] [CrossRef]
- Broadhurst, D.I.; Kella, D.B. Statistical strategies for avoiding false discoveries in metabolomics and related experiments. Metabolomics 2008, 4, 81–89. [Google Scholar] [CrossRef]
- Brennan, L. Metabolomics in nutrition research: current status and perspectives. Biochem. Soc. Trans. 2013, 41, 670–673. [Google Scholar] [CrossRef]
- Christin, C.; Hoefsloot, H.C.J.; Smilde, A.K.; Hoekman, B.; Suits, F.; Bischoff, R.; Horvatovich, P. A critical assessment of feature selection methods for biomarker discovery in clinical proteomics. Mol. Cell. Proteom. 2013, 12, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Dragsted, L.O.; Gao, Q.; Scalbert, A.; Vergères, G.; Kolehmainen, M.; Manach, C.; Brennan, L.; Afman, L.A.; Wishart, D.S.; Andres-Lacueva, C.; Garcia-Aloy, M.; Verhagen, H.; Feskens, E.J.M.; Praticò, G. Validation of biomarkers of food intake: critical assessment of candidate biomarkers. Genes Nutr. 2018, 13, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Szymańska, E.; Saccenti, E.; Smilde, A.K.; Westerhuis, J.A. Double-check: validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics 2012, 8, 3–16. [Google Scholar] [CrossRef]
- Barker, M.; Rayens, W. Partial least squares for discrimination. J. Chemom. 2003, 17, 166–173. [Google Scholar] [CrossRef]
- Wold, S.; Kettaneh, N.; Fridén, H.; Holmberg, A. Modelling and diagnostics of batch processes and analogous kinetic experiments. Chemom. Intell. Lab. Syst. 1998, 44, 331–340. [Google Scholar] [CrossRef]
- Antti, H.; Bollard, M.E.; Ebbels, T.; Keun, H.; Lindon, J.C.; Nicholson, J.K.; Holmes, E. Batch statistical processing of1H NMR-derived urinary spectral data. J. Chemom. 2002, 16, 461–468. [Google Scholar] [CrossRef]
- Jonsson, P.; Stenlund, H.; Moritz, T.; Trygg, J.; Sjöström, M.; Verheij, E.R.; Lindberg, J.; Schuppe-Koistinen, I.; Antti, H. A strategy for modelling dynamic responses in metabolic samples characterized by GC/MS. Metabolomics 2006, 2, 135–143. [Google Scholar] [CrossRef]
- Rantalainen, M.; Cloarec, O.; Ebbels, T.M.D.; Lundstedt, T.; Nicholson, J.K.; Holmes, E.; Trygg, J. Piecewise multivariate modelling of sequential metabolic profiling data. BMC Bioinform. 2008, 9, 1–13. [Google Scholar] [CrossRef]
- Kusalik, A.J. State-space model with time delays for gene regulatory networks. J. Biol. Syst. 2004, 12, 483–500. [Google Scholar]
- Smilde, A.K.; Westerhuis, J.A.; Hoefsloot, H.C.J.; Bijlsma, S.; Rubingh, C.M.; Vis, D.J.; Jellema, R.H.; Pijl, H.; Roelfsema, F.; van der Greef, J. Dynamic metabolomic data analysis: a tutorial review. Metabolomics 2010, 6, 3–17. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-regression: a basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Bro, R. Multiway calibration. Multilinear PLS. J. Chemom. 1996, 10, 47–61. [Google Scholar] [CrossRef]
- Rubingh, C.M.; Bijlsma, S.; Jellema, R.H.; Overkamp, K.M.; Van Der Werf, M.J.; Smilde, A.K. Analyzing longitudinal microbial metabolomics data. J. Proteome Res. 2009, 8, 4319–4327. [Google Scholar] [CrossRef]
- Andersen, C.M.; Bro, R. Variable selection in regression-a tutorial. J. Chemom. 2010, 24, 728–737. [Google Scholar] [CrossRef]
- Barri, T.; Holmer-Jensen, J.; Hermansen, K.; Dragsted, L.O. Metabolic fingerprinting of high-fat plasma samples processed by centrifugation-and filtration-based protein precipitation delineates significant differences in metabolite information coverage. Anal. Chim. Acta 2012, 718, 47–57. [Google Scholar] [CrossRef]
- Gürdeniz, G.; Kristensen, M.; Skov, T.; Dragsted, L.O. The effect of LC-MS data preprocessing methods on the selection of plasma biomarkers in fed vs. fasted rats. Metabolites 2012, 2, 77–99. [Google Scholar] [CrossRef]
- Gürdeniz, G.; Jensen, M.G.; Meier, S.; Bech, L.; Lund, E.; Dragsted, L.O. Detecting beer intake by unique metabolite patterns. J. Proteome Res. 2016, 15, 4544–4556. [Google Scholar] [CrossRef]
- Smilde, A.; Bro, R.; Geladi, P. Multi-way Analysis: Applications in the Chemical Sciences; John Wiley & Sons: Hoboken, NJ, USA, 2005; ISBN 0470012102. [Google Scholar]
- Kiers, H.A.L.; Van Mechelen, I. Three-way component analysis: Principles and illustrative application. Psychol. Methods 2001, 6, 84–110. [Google Scholar] [CrossRef] [PubMed]
- Gurden, S.P.; Westerhuis, J.A.; Bro, R.; Smilde, A.K. A comparison of multiway regression and scaling methods. Chemom. Intell. Lab. Syst. 2001, 59, 121–136. [Google Scholar] [CrossRef]
- Gosselin, R.; Rodrigue, D.; Duchesne, C. A Bootstrap-VIP approach for selecting wavelength intervals in spectral imaging applications. Chemom. Intell. Lab. Syst. 2010, 100, 12–21. [Google Scholar] [CrossRef]
- Gleason, J.R. Algorithms for balanced bootstrap simulations. Am. Stat. 1988, 42, 263–266. [Google Scholar] [CrossRef]
- Wold, S.; Johansson, E.; Cocchi, M. 3D QSAR in Drug Design: Theory, Methods and Applications; ESCOM: Leiden, The Netherlands, 1993; pp. 523–550. [Google Scholar]
- Favilla, S.; Durante, C.; Vigni, M.L.; Cocchi, M. Assessing feature relevance in NPLS models by VIP. Chemom. Intell. Lab. Syst. 2013, 129, 76–86. [Google Scholar] [CrossRef]
- Andersson, C.A.; Bro, R. The N-way Toolbox for MATLAB. Chemom. Intell. Lab. Syst. 2000, 52, 1–4. [Google Scholar] [CrossRef]
- Chong, I.-G.; Jun, C.-H. Performance of some variable selection methods when multicollinearity is present. Chemom. Intell. Lab. Syst. 2005, 78, 103–112. [Google Scholar] [CrossRef]
- Alves, A.C.; Li, J.V.; Garcia-Perez, I.; Sands, C.; Barbas, C.; Holmes, E.; Ebbels, T.M.D. Characterization of data analysis methods for information recovery from metabolic 1H NMR spectra using artificial complex mixtures. Metabolomics 2012, 8, 1170–1180. [Google Scholar] [CrossRef]
- Gidskehaug, L.; Anderssen, E.; Flatberg, A.; Alsberg, B.K. A framework for significance analysis of gene expression data using dimension reduction methods. BMC Bioinform. 2007, 8, 346. [Google Scholar] [CrossRef]
- Bar-Joseph, Z.; Gerber, G.; Simon, I.; Gifford, D.K.; Jaakkola, T.S. Comparing the continuous representation of time-series expression profiles to identify differentially expressed genes. Proc. Natl. Acad. Sci. USA 2003, 100, 10146–10151. [Google Scholar] [CrossRef]
- Berk, M.; Ebbels, T.; Montana, G. A statistical framework for biomarker discovery in metabolomic time course data. Bioinformatics 2011, 27, 1979–1985. [Google Scholar] [CrossRef] [PubMed]
- Smilde, A.K.; Jansen, J.J.; Hoefsloot, H.C.J.; Lamers, R.J.A.N.; van der Greef, J.; Timmerman, M.E. ANOVA-simultaneous component analysis (ASCA): A new tool for analyzing designed metabolomics data. Bioinformatics 2005, 21, 3043–3048. [Google Scholar] [CrossRef] [PubMed]
| True Condition | |||
|---|---|---|---|
| Total Variables | Discriminating Variables | Non-Discriminating Variables | |
| Predicted Condition | Selected variables | True positive (TP) | False positive (FP) |
| Unselected variables | False negative (FN) | True negative (TN) | |
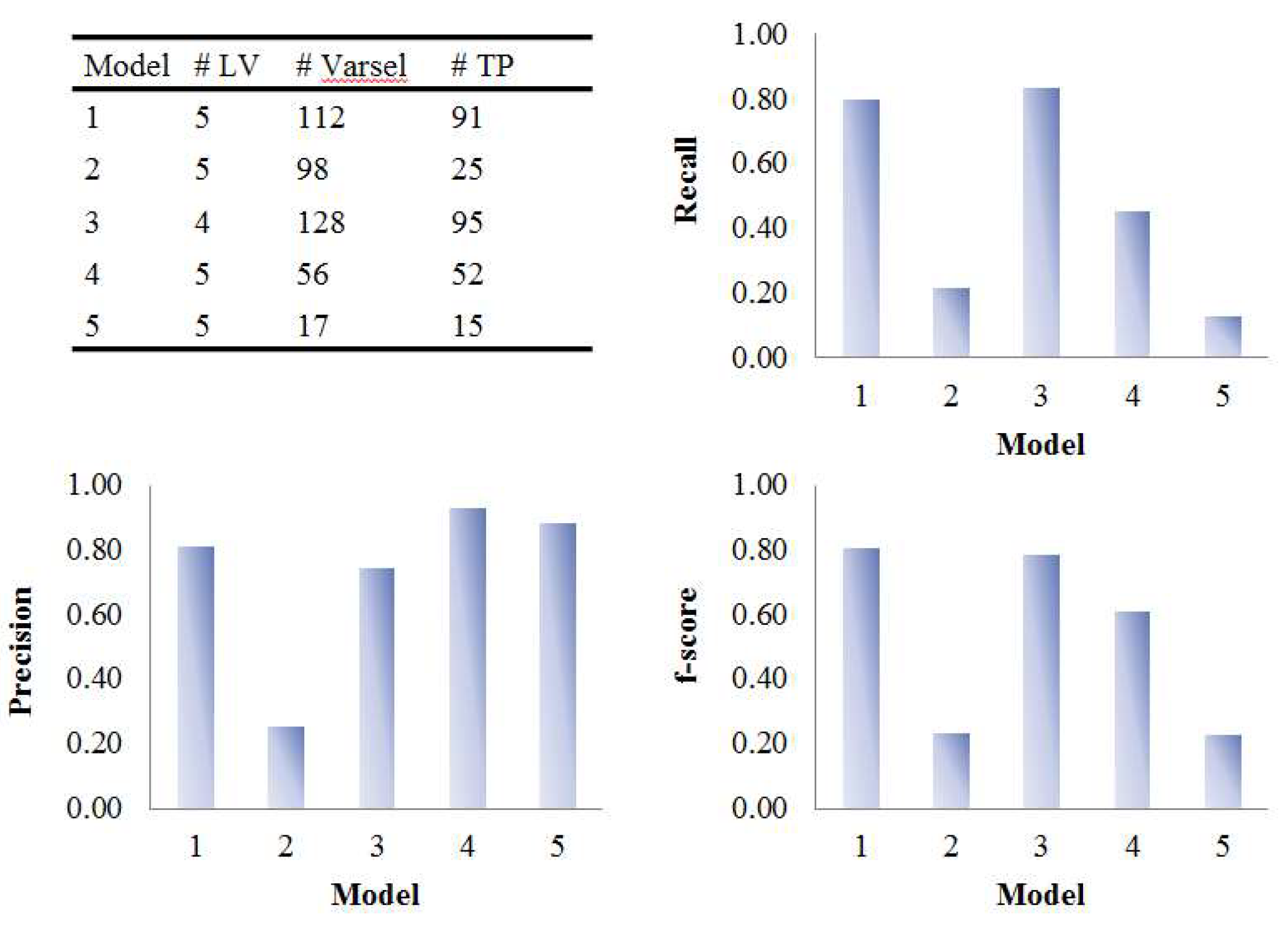
| Model | Data (X or X) | Dummy Y | # LVa | Predictive Ability | Variable Selection Performance | ||
|---|---|---|---|---|---|---|---|
| Q2 | AUC | # Varselb | # TPc | ||||
| 1 | Mode 1: Sample Mode 2: Metabolite | Group | 3 | 0.57 (0.02) | 0.93 (0.06) | 249.3 (11.2) | 53.6 (4) |
| 2 | Time response | 5 | 1 (0) | 0.58 (0.08) | 591.1 (16) | 12.8 (3.9) | |
| 3 | Group × Time response | 5 | 0.83 (0.01) | 0.75 (0) | 194.5 (9.1) | 77.3 (1.6) | |
| 4 | Mode 1: Subject Mode 2: Metabolite Mode 3: Time | Group | 1 | 0.6 (0.01) | 1 (0) | 165.9 (11.4) | 53 (4.1) |
| 5 | Mode 1: Group Mode 3: Time | 1 | 0.6 (0.02) | 0.75 (0) | 166.7 (13.5) | 53.9 (4.3) | |
| Model. | Number of Variables (No. Discriminating Variables Kept at 80 in All Cases). | |||||||
|---|---|---|---|---|---|---|---|---|
| 1000 | 3000 | 5000 | 7000 | |||||
| # Varsela | # TPb | # Varsel | # TP | # Varsel | # TP | # Varsel | # TP | |
| 1 | 105 (5.2) | 51.7 (3.1) | 249.3 (11.2) | 53.6 (4) | 396.3 (12.2) | 53.9 (4) | 540.4 (20.3) | 53.3 (4.4) |
| 2 | 189.8 (6.9) | 15.1 (3.9) | 591.1 (16) | 12.8 (3.9) | 983.1 (19.8) | 15.8 (3.7) | 1379.1 (22.1) | 13.7 (2.1) |
| 3 | 95.7 (4.6) | 74.3 (1.8) | 194.5 (9.1) | 77.3 (1.6) | 304.7 (14.8) | 77.1 (2.2) | 409.3 (18.2) | 77.5 (1.4) |
| 4 | 86.2 (7.1) | 58 (3.6) | 165.9 (11.4) | 53 (4.1) | 243.8 (18.8) | 50.2 (4.1) | 325.8 (21.7) | 49.6 (2.9) |
| 5 | 89 (5.2) | 58.4 (3.8) | 166.7 (13.5) | 53.9 (4.3) | 247.4 (14.8) | 51.7 (2.8) | 325.2 (15.6) | 51.3 (3.3) |
| Model | Inter-Individual Variability | |||||||
|---|---|---|---|---|---|---|---|---|
| 0.1 | 0.3 | 0.5 | 0.7 | |||||
| # Varsela | # TPb | # Varsel | # TP | # Varsel | # TP | # Varsel | # TP | |
| 1 | 153.5 (8.2) | 75.1 (1.9) | 249.3 (11.2) | 53.6 (4) | 301 (11.5) | 39.2 (3.8) | 323 (11.1) | 34.2 (3.3) |
| 2 | 681.5 (17.6) | 2.7 (1.4) | 591.1 (16) | 12.8 (3.9) | 513.5 (15.3) | 19.4 (3) | 493 (18.3) | 19.2 (3.6) |
| 3 | 143.5 (4.5) | 79.9 (0.3) | 194.5 (9.1) | 77.3 (1.6) | 216.5 (14.4) | 72.2 (2.5) | 243.1 (13.3) | 68.3 (3.5) |
| 4 | 173.9 (12.1) | 78.7 (1.2) | 165.9 (11.4) | 53 (4.1) | 155.2 (11.6) | 35.9 (4.3) | 165.9 (15.8) | 27 (2.5) |
| 5 | 177.5 (12.5) | 79.2 (0.9) | 166.7 (13.5) | 53.9 (4.3) | 158.8 (12.7) | 35.4 (3.4) | 170.1 (13.4) | 28.1 (5) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, Q.; Dragsted, L.O.; Ebbels, T. Comparison of Bi- and Tri-Linear PLS Models for Variable Selection in Metabolomic Time-Series Experiments. Metabolites 2019, 9, 92. https://doi.org/10.3390/metabo9050092
Gao Q, Dragsted LO, Ebbels T. Comparison of Bi- and Tri-Linear PLS Models for Variable Selection in Metabolomic Time-Series Experiments. Metabolites. 2019; 9(5):92. https://doi.org/10.3390/metabo9050092
Chicago/Turabian StyleGao, Qian, Lars O. Dragsted, and Timothy Ebbels. 2019. "Comparison of Bi- and Tri-Linear PLS Models for Variable Selection in Metabolomic Time-Series Experiments" Metabolites 9, no. 5: 92. https://doi.org/10.3390/metabo9050092
APA StyleGao, Q., Dragsted, L. O., & Ebbels, T. (2019). Comparison of Bi- and Tri-Linear PLS Models for Variable Selection in Metabolomic Time-Series Experiments. Metabolites, 9(5), 92. https://doi.org/10.3390/metabo9050092






