Single Spheroid Metabolomics: Optimizing Sample Preparation of Three-Dimensional Multicellular Tumor Spheroids
Abstract
1. Introduction
2. Results
2.1. Establishment and Validation of Sample Preparation Procedures Suitable for Probing the Metabolome of Spheroids
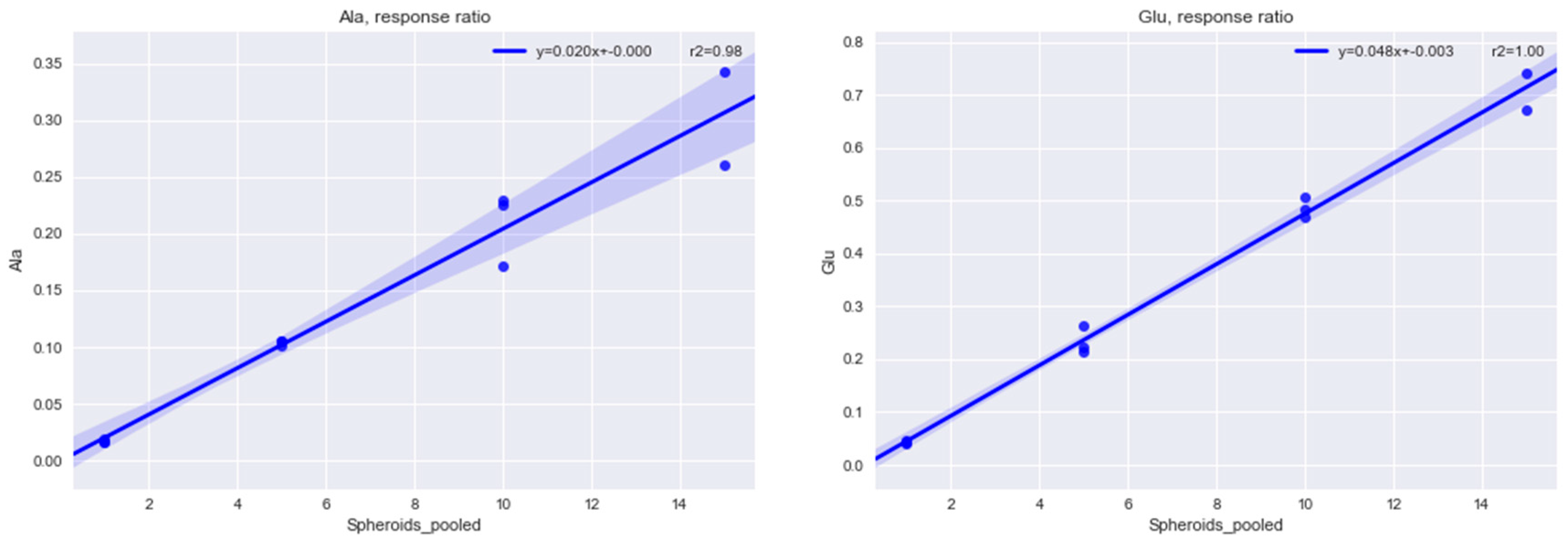
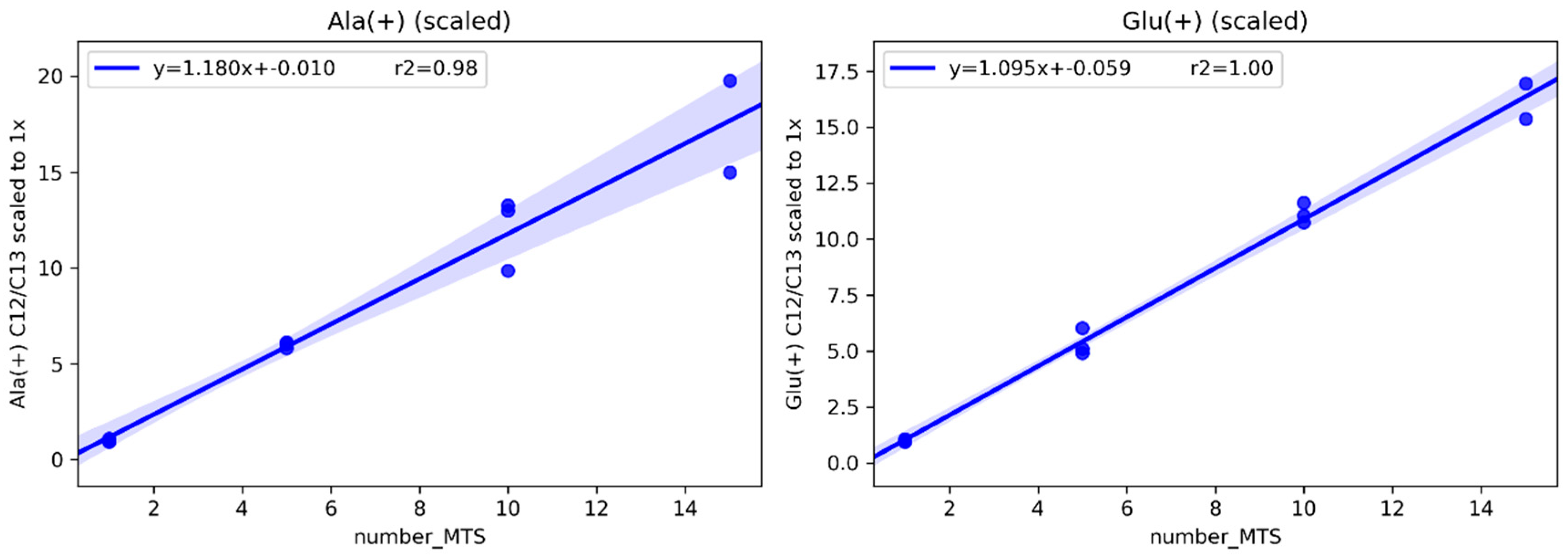
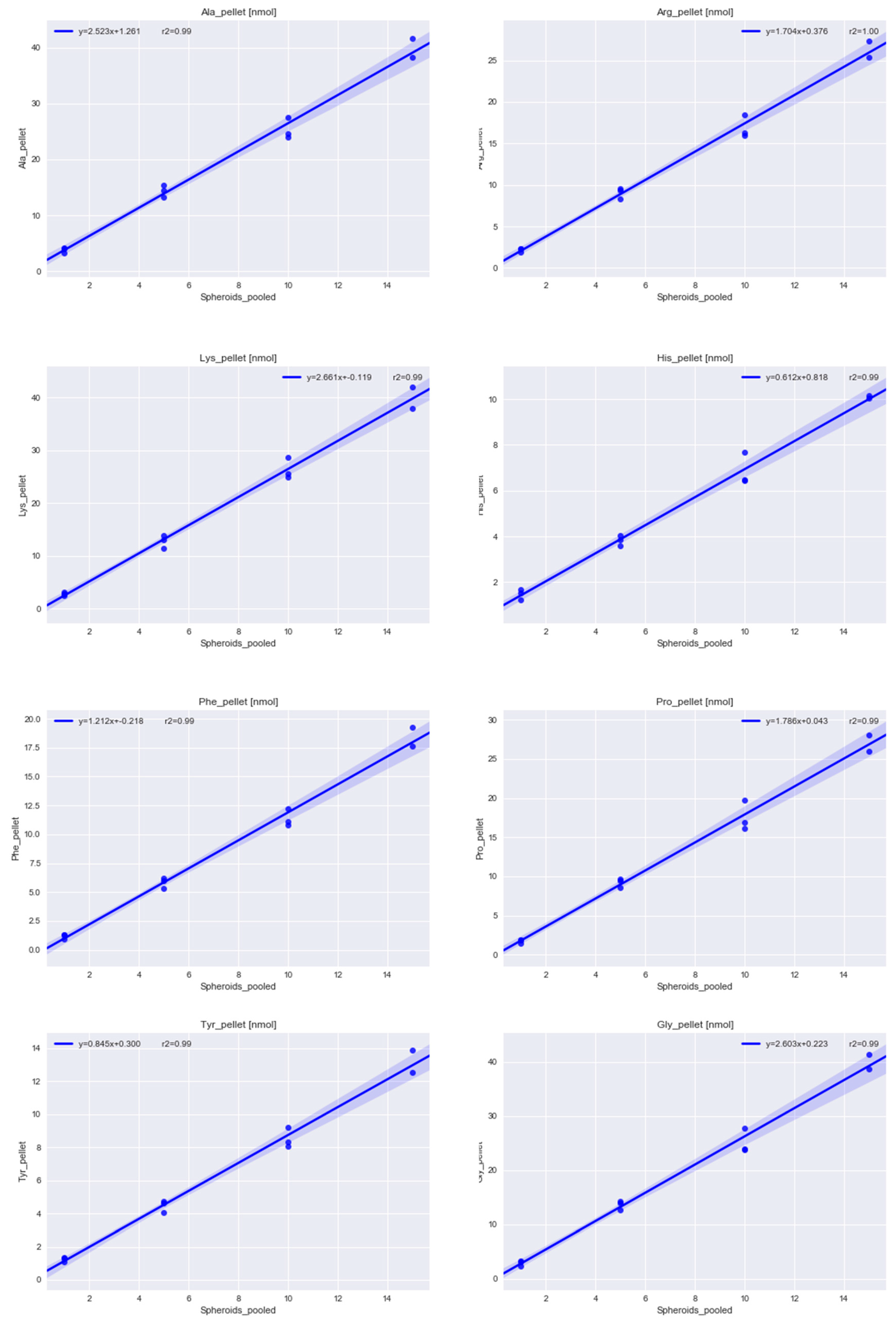
2.1.1. Determining the Minimal Number of MTS Required for Metabolomics Experiment
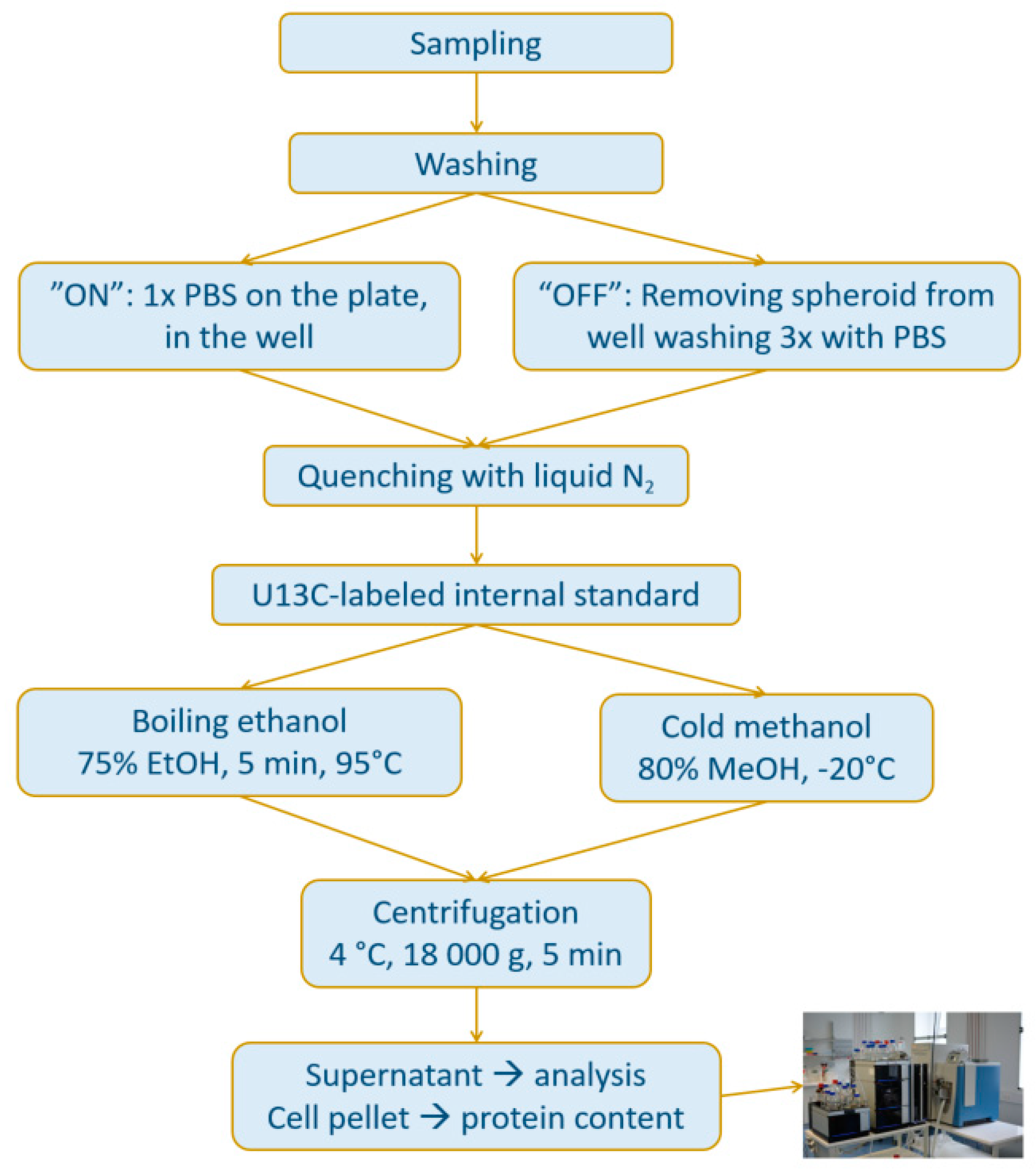
2.1.2. Speeding Up the Sample Preparation
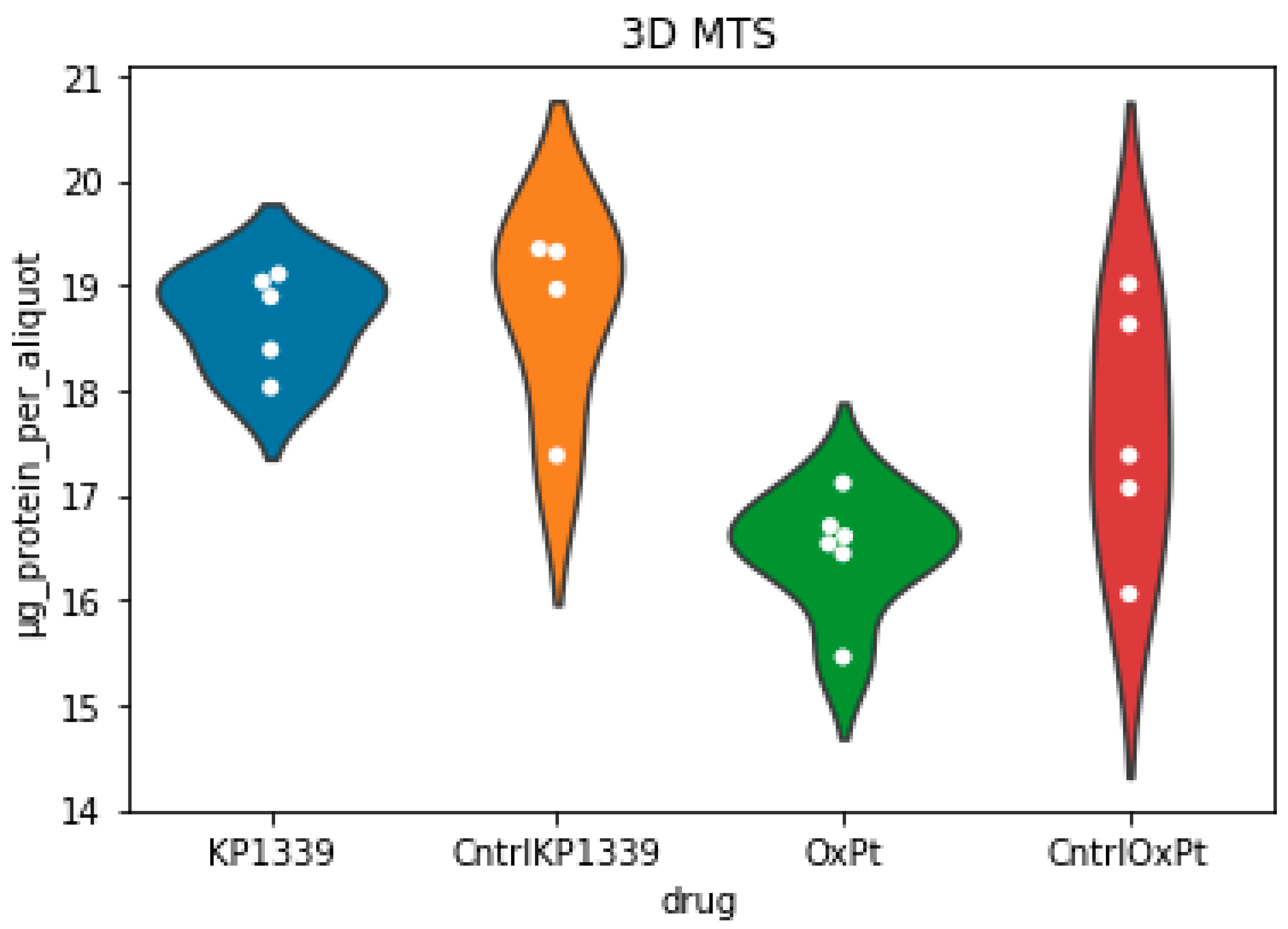
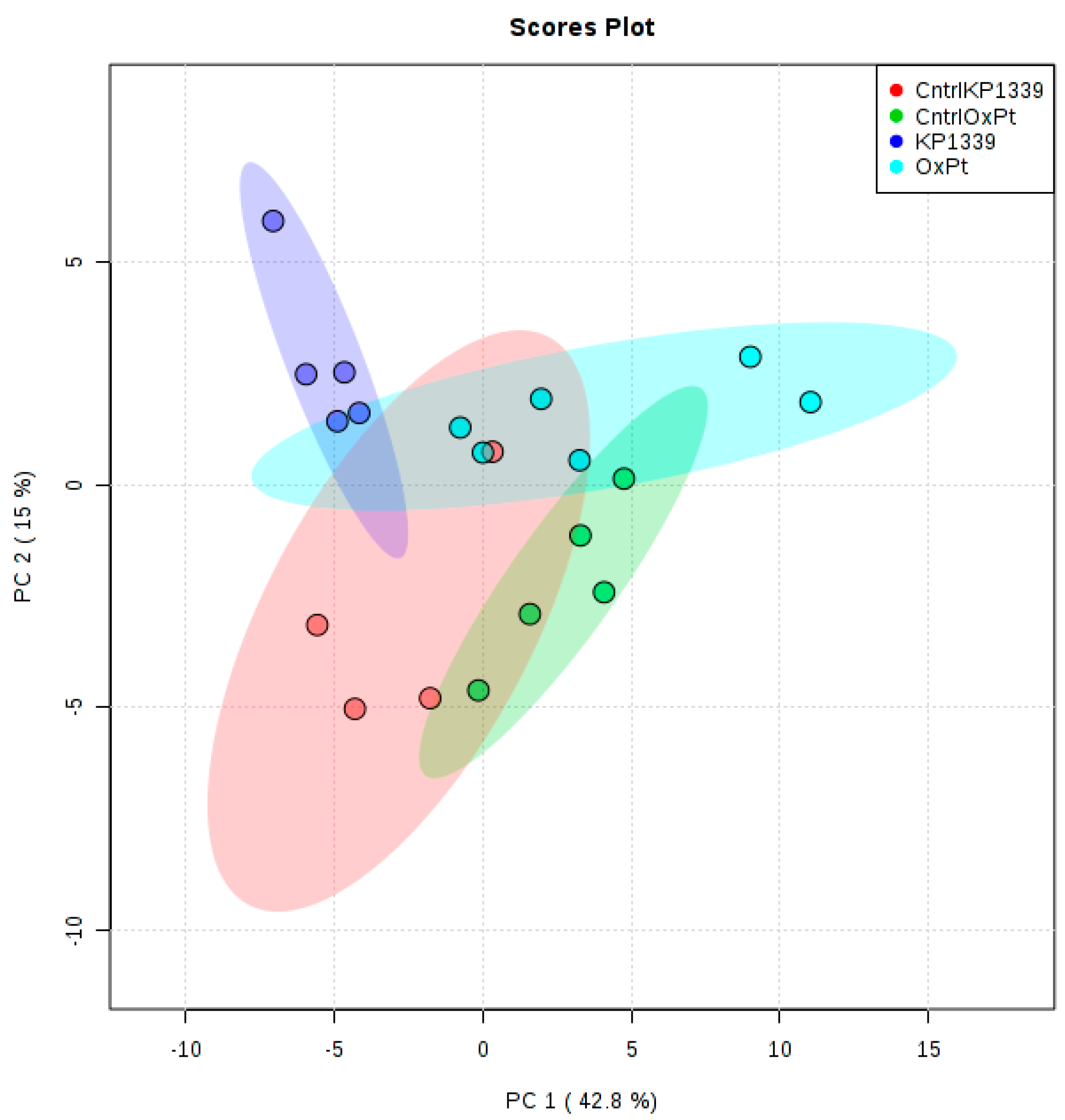
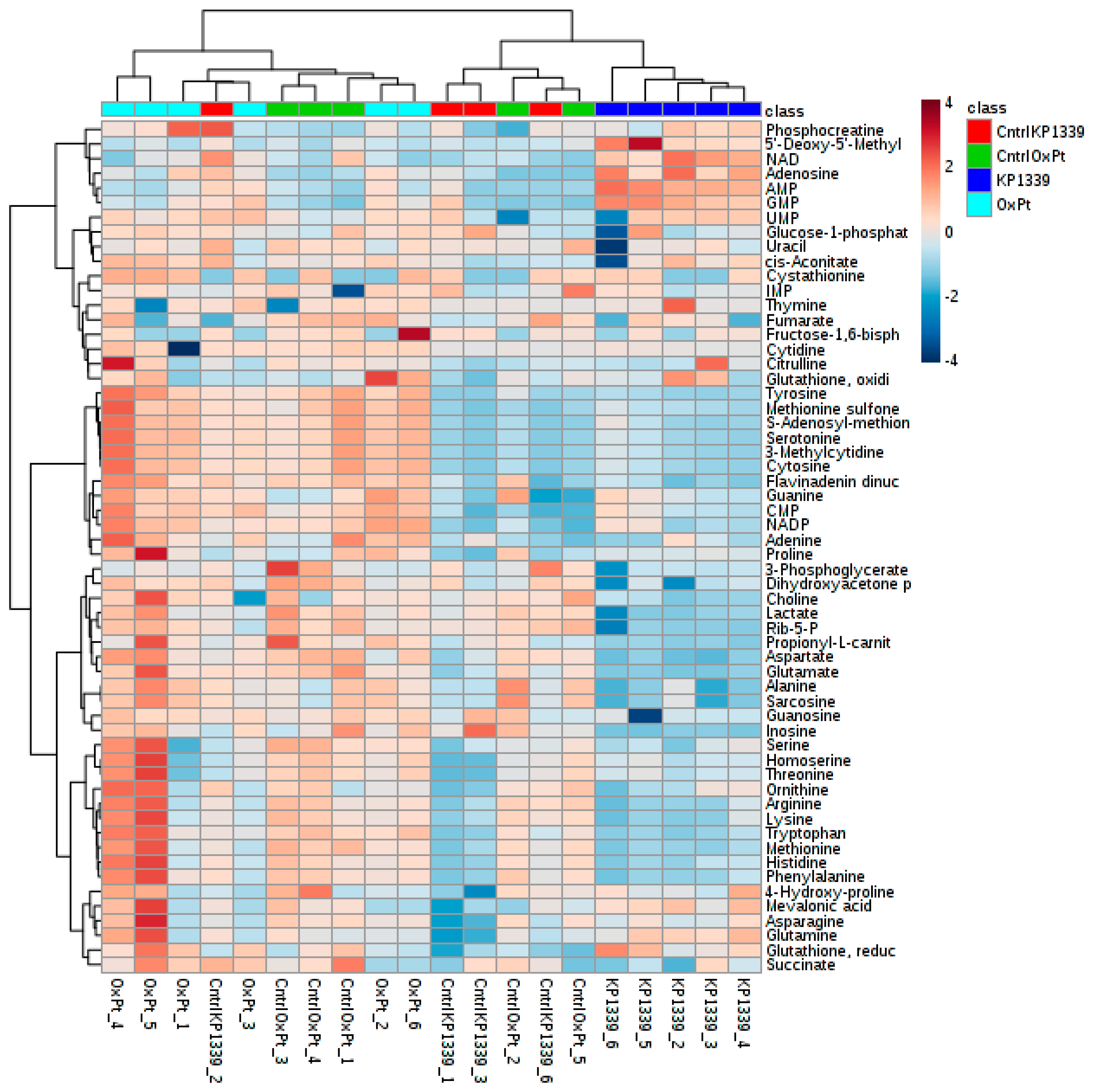
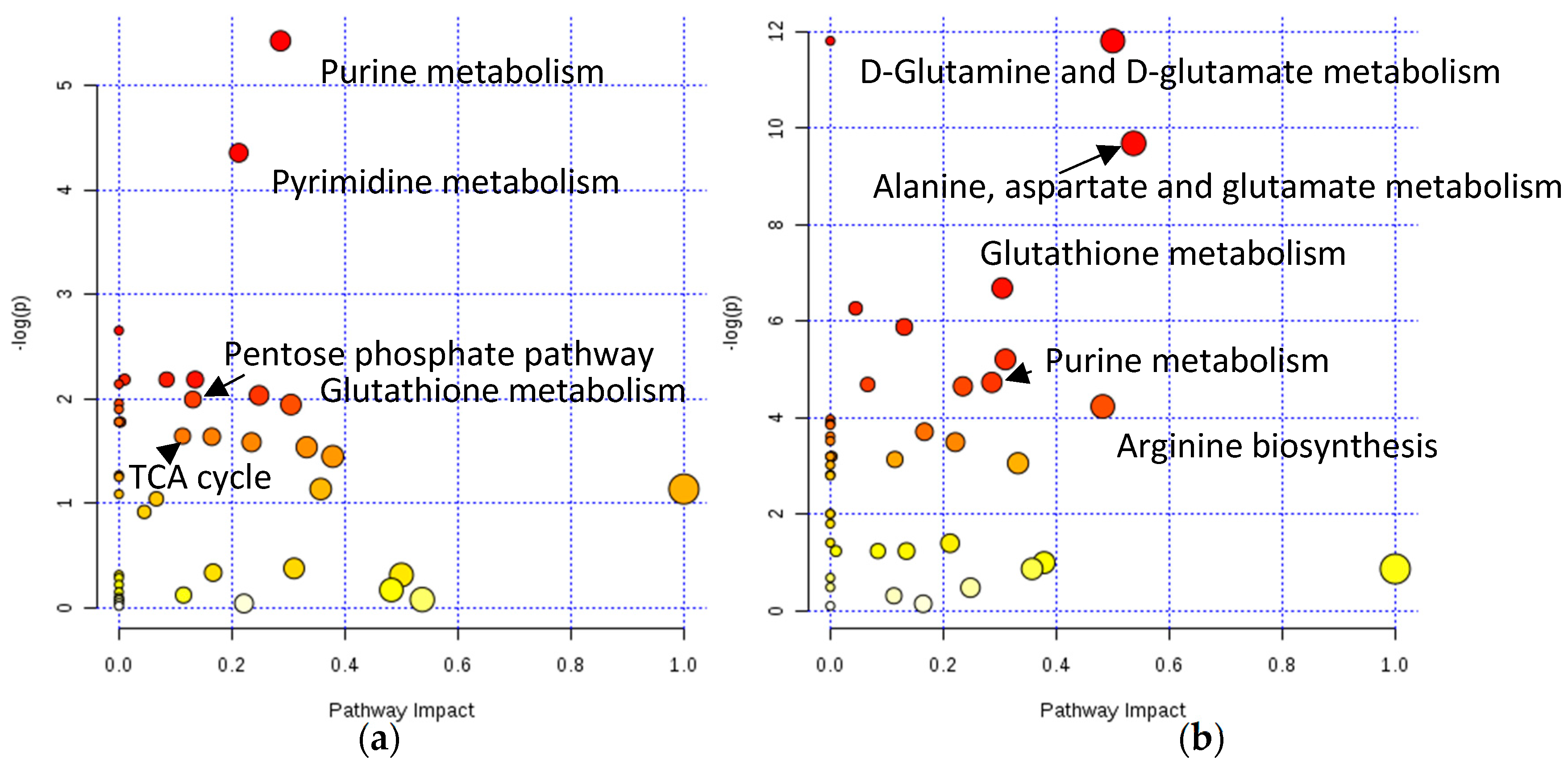
2.2. Assessing Metabolic Shifts in Single MTS Exposed to Metal-Based Anticancer Drugs
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.1.1. Cultivation of Spheroids
4.1.2. Microscopy
4.1.3. Cell Number Estimation
4.2. Methods for Determining the Minimal Number of MTS Required for a Metabolomics Experiment
4.2.1. Sampling 1, 5, 10, 15 Multicellular Tumor Spheroids
4.2.2. Internal Standardization
4.2.3. Boiling Ethanol Extraction for 1×-5×-10×-15× MTSs
4.2.4. Metabolomics Measurement of Extracts 1×-5×-10×-15× MTSs
4.2.5. Acidic Hydrolysis of the Pellet and Amino Acid Analysis
4.3. Methods for Comparison Boiling Ethanol (BE) and Cold Methanol (CM) Extraction and Washing Procedures (OFF vs. ON) of 3D MTS
4.3.1. Transfer of Spheroids
4.3.2. “BE-OFF” Sample Preparation
4.3.3. “CM-OFF” Sample Preparation
4.3.4. “BE-ON” Sample Preparation
4.3.5. “CM-ON” Sample Preparation
4.3.6. LC-MS Method Applied for Proof of Principle Experiment with Metallodrugs
4.3.7. Total Protein Content Determination
4.4. Data Analysis
4.4.1. Targeted Metabolomics Data Treatment and Normalization
4.4.2. Exploratory Data Analysis
4.4.3. Pathway Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Abbreviation | Explanation |
| 5AMP, AMP | adenosine monophosphate |
| 5CMP, CMP | cytidine monophosphate |
| 5GMP, UMP | guanosine monophosphate |
| 5UMP | uridine monophosphate |
| AAA | alpha aminoadipic acid |
| Ade | adenine |
| ADP | adenosine diphosphate |
| Ala | alanine |
| Arg | arginine |
| Asin | adenosine |
| Asn | aspragine |
| Asp | aspartic acid |
| DHAP | dihydroxyacetone phosphate |
| Gln | glutamine |
| Glu | glutamate |
| Gly | glycine |
| GMP | guanosine monophosphate |
| GSH | glutathione (reduced) |
| Gsin | guanosine |
| GSSG | Glutathione (oxidized) |
| His | histidine |
| H-Ser | homoserine |
| IMP | inosine monophosphate |
| Isin | inosine |
| K-Ile | ketoisoleucine |
| Lys | lysine |
| Met | methionine |
| NAD+ | nicotinamide adenine dinucleotide (oxidized) |
| NADP+ | nicotinamide adenine dinucleotide phosphate (oxidized) |
| OAS | O-acetylserine |
| Phe | phenylalanine |
| Pro | proline |
| Pyr | pyruvate |
| Rib-5-P | ribose 5-phosphate/ribulose 5-phosphate |
| Ser | serine |
| Thr | threonine |
| Trp | tryptophan |
| Tyr | tyrosine |
Appendix A
References
- Stock, K.; Estrada, M.F.; Vidic, S.; Gjerde, K.; Rudisch, A.; Santo, V.E.; Barbier, M.; Blom, S.; Arundkar, S.C.; Selvam, I.; et al. Capturing tumor complexity in vitro: Comparative analysis of 2D and 3D tumor models for drug discovery. Sci. Rep. 2016, 6, 28951. [Google Scholar] [CrossRef] [PubMed]
- Hirschhaeuser, F.; Menne, H.; Dittfeld, C.; West, J.; Mueller-Klieser, W.; Kunz-Schughart, L.A. Multicellular tumor spheroids: An underestimated tool is catching up again. J. Biotechnol. 2010, 148, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Mehta, G.; Hsiao, A.Y.; Ingram, M.; Luker, G.D.; Takayama, S. Opportunities and Challenges for use of Tumor Spheroids as Models to Test Drug Delivery and Efficacy. J. Control. Release 2012, 164, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Costa, E.C.; Moreira, A.F.; de Melo-Diogo, D.; Gaspar, V.M.; Carvalho, M.P.; Correia, I.J. 3D tumor spheroids: An overview on the tools and techniques used for their analysis. Biotechnol. Adv. 2016, 34, 1427–1441. [Google Scholar] [CrossRef] [PubMed]
- Stewart, D.J. Mechanisms of resistance to cisplatin and carboplatin. Crit. Rev. Oncol. Hematol. 2007, 63, 12–31. [Google Scholar] [CrossRef]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-Dimensional Cell Culture Systems and Their Applications in Drug Discovery and Cell-Based Biosensors. Assay Drug Dev. Technol. 2014, 12, 207–218. [Google Scholar] [CrossRef]
- Tafani, M.; Sansone, L.; Limana, F.; Arcangeli, T.; De Santis, E.; Polese, M.; Fini, M.; Russo, M.A. The Interplay of Reactive Oxygen Species, Hypoxia, Inflammation, and Sirtuins in Cancer Initiation and Progression. Oxid. Med. Cell. Longev. 2016, 2016, 18. [Google Scholar] [CrossRef]
- Kondoh, M.; Ohga, N.; Akiyama, K.; Hida, Y.; Maishi, N.; Towfik, A.M.; Inoue, N.; Shindoh, M.; Hida, K. Hypoxia-Induced Reactive Oxygen Species Cause Chromosomal Abnormalities in Endothelial Cells in the Tumor Microenvironment. PLoS ONE 2013, 8, e80349. [Google Scholar] [CrossRef]
- Wang, Q.-S.; Zheng, Y.-M.; Dong, L.; Ho, Y.-S.; Guo, Z.; Wang, Y.-X. Role of mitochondrial reactive oxygen species in hypoxia-dependent increase in intracellular calcium in pulmonary artery myocytes. Free Radic. Biol. Med. 2007, 42, 642–653. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Armitage, E.G.; Southam, A.D. Monitoring cancer prognosis, diagnosis and treatment efficacy using metabolomics and lipidomics. Metabolomics 2016, 12, 146. [Google Scholar] [CrossRef] [PubMed]
- do Valle, Í.F.; Menichetti, G.; Simonetti, G.; Bruno, S.; Zironi, I.; Durso, D.F.; Mombach, J.C.M.; Martinelli, G.; Castellani, G.; Remondini, D. Network integration of multi-tumour omics data suggests novel targeting strategies. Nat. Commun. 2018, 9, 4514. [Google Scholar] [CrossRef] [PubMed]
- Beger, R.D. A Review of Applications of Metabolomics in Cancer. Metabolites 2013, 3, 552–574. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Metabolomics: The apogee of the omic triology. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Armitage, E.G.; Barbas, C. Metabolomics in cancer biomarker discovery: Current trends and future perspectives. J. Pharm. Biomed. Anal. 2014, 87, 1–11. [Google Scholar] [CrossRef]
- Beger, R.D. Interest is high in improving quality control for clinical metabolomics: Setting the path forward for community harmonization of quality control standards. Metabolomics 2018, 15, 1. [Google Scholar] [CrossRef]
- Cui, L.; Lu, H.; Lee, Y.H. Challenges and emergent solutions for LC-MS/MS based untargeted metabolomics in diseases. Mass Spectrom. Rev. 2018, 37, 772–792. [Google Scholar] [CrossRef]
- Schwaiger, M.; Schoeny, H.; Abiead, Y.E.; Hermann, G.; Rampler, E.; Koellensperger, G. Merging metabolomics and lipidomics into one analytical run. Analyst 2018, 144, 220–229. [Google Scholar] [CrossRef]
- León, Z.; García-Cañaveras, J.C.; Donato, M.T.; Lahoz, A. Mammalian cell metabolomics: Experimental design and sample preparation. Electrophoresis 2013, 34, 2762–2775. [Google Scholar] [CrossRef]
- Coloff, J.L.; Murphy, J.P.; Braun, C.R.; Harris, I.S.; Shelton, L.M.; Kami, K.; Gygi, S.P.; Selfors, L.M.; Brugge, J.S. Differential Glutamate Metabolism in Proliferating and Quiescent Mammary Epithelial Cells. Cell Metab. 2016, 23, 867–880. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.W.-M.; El-Amouri, S.S.; Macedo, J.K.A.; Wang, Q.J.; Song, H.; Cassel, T.; Lane, A.N. Stable Isotope-Resolved Metabolomics Shows Metabolic Resistance to Anti-Cancer Selenite in 3D Spheroids versus 2D Cell Cultures. Metabolites 2018, 8, 40. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Valli, A.; Haider, S.; Zhang, Q.; Smethurst, E.A.; Schug, Z.T.; Peck, B.; Aboagye, E.O.; Critchlow, S.E.; Schulze, A.; et al. 3D Growth of Cancer Cells Elicits Sensitivity to Kinase Inhibitors but Not Lipid Metabolism Modifiers. Mol. Cancer Ther. 2019, 18, 376–388. [Google Scholar] [CrossRef] [PubMed]
- Vorrink, S.U.; Ullah, S.; Schmidt, S.; Nandania, J.; Velagapudi, V.; Beck, O.; Ingelman-Sundberg, M.; Lauschke, V.M. Endogenous and xenobiotic metabolic stability of primary human hepatocytes in long-term 3D spheroid cultures revealed by a combination of targeted and untargeted metabolomics. FASEB J. 2017, 31, 2696–2708. [Google Scholar] [CrossRef]
- Sato, M.; Kawana, K.; Adachi, K.; Fujimoto, A.; Yoshida, M.; Nakamura, H.; Nishida, H.; Inoue, T.; Taguchi, A.; Takahashi, J.; et al. Spheroid cancer stem cells display reprogrammed metabolism and obtain energy by actively running the tricarboxylic acid (TCA) cycle. Oncotarget 2016, 7, 33297–33305. [Google Scholar] [CrossRef]
- Eljack, N.D.; Ma, H.-Y.M.; Drucker, J.; Shen, C.; Hambley, T.W.; New, E.J.; Friedrich, T.; Clarke, R.J. Mechanisms of cell uptake and toxicity of the anticancer drug cisplatin. Metallomics 2014, 6, 2126–2133. [Google Scholar] [CrossRef]
- Gibson, D. The mechanism of action of platinum anticancer agents—What do we really know about it? Dalton Trans. 2009, 48, 10681–10689. [Google Scholar] [CrossRef]
- Johnstone, T.C.; Suntharalingam, K.; Lippard, S.J. The Next Generation of Platinum Drugs: Targeted Pt (II) Agents, Nanoparticle Delivery and Pt (IV) Prodrugs. Chem. Rev. 2016, 116, 3436–3486. [Google Scholar] [CrossRef]
- Mucaki, E.J.; Zhao, J.Z.L.; Lizotte, D.J.; Rogan, P.K. Predicting responses to platin chemotherapy agents with biochemically-inspired machine learning. Signal Transduct. Target. Ther. 2019, 4, 1. [Google Scholar] [CrossRef]
- Wang, D.; Lippard, S.J. Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov. 2005, 4, 307–320. [Google Scholar] [CrossRef]
- Mjos, K.D.; Orvig, C. Metallodrugs in Medicinal Inorganic Chemistry. Chem. Rev. 2014, 114, 4540–4563. [Google Scholar] [CrossRef] [PubMed]
- Canelas, A.B.; ten Pierick, A.; Ras, C.; Seifar, R.M.; van Dam, J.C.; van Gulik, W.M.; Heijnen, J.J. Quantitative evaluation of intracellular metabolite extraction techniques for yeast metabolomics. Anal. Chem. 2009, 81, 7379–7389. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, B.; François, J.; Renaud, M. A rapid and reliable method for metabolite extraction in yeast using boiling buffered ethanol. Yeast 1997, 13, 1347–1355. [Google Scholar] [CrossRef]
- Dettmer, K.; Nürnberger, N.; Kaspar, H.; Gruber, M.A.; Almstetter, M.F.; Oefner, P.J. Metabolite extraction from adherently growing mammalian cells for metabolomics studies: Optimization of harvesting and extraction protocols. Anal. Bioanal. Chem. 2011, 399, 1127–1139. [Google Scholar] [CrossRef] [PubMed]
- Hermann, G.; Schwaiger, M.; Volejnik, P.; Koellensperger, G. 13C-labelled yeast as internal standard for LC–MS/MS and LC high resolution MS based amino acid quantification in human plasma. J. Pharm. Biomed. Anal. 2018, 155, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Hoofnagle, A.N.; Whiteaker, J.R.; Carr, S.A.; Kuhn, E.; Liu, T.; Massoni, S.A.; Thomas, S.N.; Townsend, R.R.; Zimmerman, L.J.; Boja, E.; et al. Recommendations for the generation, quantification, storage and handling of peptides used for mass spectrometry-based assays. Clin. Chem. 2016, 62, 48–69. [Google Scholar] [CrossRef]
- Galvez, L.; Rusz, M.; Schwaiger, M.; Abiead, Y.E.; Hermann, G.; Jungwirth, U.; Berger, W.; Keppler, B.K.; Jakupec, M.; Koellensperger, G. Preclinical studies on metal based anticancer drugs as enabled by integrated metallomics and metabolomics. Metallomics 2019, 11, 1716–1728. [Google Scholar] [CrossRef]
- Bruno, P.M.; Liu, Y.; Park, G.Y.; Murai, J.; Koch, C.E.; Eisen, T.J.; Pritchard, J.R.; Pommier, Y.; Lippard, S.J.; Hemann, M.T. A subset of platinum-containing chemotherapeutic agents kill cells by inducing ribosome biogenesis stress rather than by engaging a DNA damage response. Nat. Med. 2017, 23, 461–471. [Google Scholar] [CrossRef]
- Schoenhacker-Alte, B.; Mohr, T.; Pirker, C.; Kryeziu, K.; Kuhn, P.-S.; Buck, A.; Hofmann, T.; Gerner, C.; Hermann, G.; Koellensperger, G.; et al. Sensitivity towards the GRP78 inhibitor KP1339/IT-139 is characterized by apoptosis induction via caspase 8 upon disruption of ER homeostasis. Cancer Lett. 2017, 404, 79–88. [Google Scholar] [CrossRef]
- Terenzi, A.; Pirker, C.; Keppler, B.K.; Berger, W. Anticancer metal drugs and immunogenic cell death. J. Inorg. Biochem. 2016, 165, 71–79. [Google Scholar] [CrossRef]
- Kepp, O.; Senovilla, L.; Vitale, I.; Vacchelli, E.; Adjemian, S.; Agostinis, P.; Apetoh, L.; Aranda, F.; Barnaba, V.; Bloy, N.; et al. Consensus guidelines for the detection of immunogenic cell death. Oncoimmunology 2014, 3, e955691. [Google Scholar] [CrossRef] [PubMed]
- Wernitznig, D.; Kiakos, K.; Del Favero, G.; Harrer, N.; Machat, H.; Osswald, A.; Jakupec, M.A.; Wernitznig, A.; Sommergruber, W.; Keppler, B.K. First-in-class ruthenium anticancer drug (KP1339/IT-139) induces an immunogenic cell death signature in colorectal spheroids in vitro. Met. Integr. Biomet. Sci. 2019, 11, 1044–1048. [Google Scholar] [CrossRef] [PubMed]
- Broadhurst, D.; Goodacre, R.; Reinke, S.N.; Kuligowski, J.; Wilson, I.D.; Lewis, M.R.; Dunn, W.B. Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics 2018, 14, 72. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Broadhurst, D.I.; Edison, A.; Guillou, C.; Viant, M.R.; Bearden, D.W.; Beger, R.D. Quality assurance and quality control processes: Summary of a metabolomics community questionnaire. Metabolomics 2017, 13, 50. [Google Scholar] [CrossRef]
- Alcindor, T.; Beauger, N. Oxaliplatin: A review in the era of molecularly targeted therapy. Curr. Oncol. 2011, 18, 18–25. [Google Scholar] [CrossRef]
- Chong, J.; Xia, J. MetaboAnalystR: An R package for flexible and reproducible analysis of metabolomics data. Bioinformatics 2018, 34, 4313–4314. [Google Scholar] [CrossRef]
- Cavill, R.; Kamburov, A.; Ellis, J.K.; Athersuch, T.J.; Blagrove, M.S.C.; Herwig, R.; Ebbels, T.M.D.; Keun, H.C. Consensus-Phenotype Integration of Transcriptomic and Metabolomic Data Implies a Role for Metabolism in the Chemosensitivity of Tumour Cells. PLoS Comput. Biol. 2011, 7, e1001113. [Google Scholar] [CrossRef]
- Von Stechow, L.; Ruiz-Aracama, A.; van de Water, B.; Peijnenburg, A.; Danen, E.; Lommen, A. Identification of Cisplatin-Regulated Metabolic Pathways in Pluripotent Stem Cells. PLoS ONE 2013, 8, e76476. [Google Scholar] [CrossRef][Green Version]
- Gifford, J.B.; Huang, W.; Zeleniak, A.E.; Hindoyan, A.; Wu, H.; Donahue, T.R.; Hill, R. Expression of GRP78, Master Regulator of the Unfolded Protein Response, increases chemoresistance in pancreatic ductal adenocarcinoma. Mol. Cancer Ther. 2016, 15, 1043–1052. [Google Scholar] [CrossRef]
- Kreutz, D.; Bileck, A.; Plessl, K.; Wolrab, D.; Groessl, M.; Keppler, B.K.; Meier, S.M.; Gerner, C. Response Profiling Using Shotgun Proteomics Enables Global Metallodrug Mechanisms of Action To Be Established. Chemistry 2017, 23, 1881–1890. [Google Scholar] [CrossRef]
- Gonen, N.; Meller, A.; Sabath, N.; Shalgi, R. Amino Acid Biosynthesis Regulation during Endoplasmic Reticulum Stress Is Coupled to Protein Expression Demands. IScience 2019, 19, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Scannell, J.W.; Blanckley, A.; Boldon, H.; Warrington, B. Diagnosing the decline in pharmaceutical R & D efficiency. Nat. Rev. Drug Discov. 2012, 11, 191–200. [Google Scholar] [PubMed]
- Hermann, G.; Møller, L.H.; Gammelgaard, B.; Hohlweg, J.; Mattanovich, D.; Hann, S.; Koellensperger, G. In vivo synthesized 34 S enriched amino acid standards for species specific isotope dilution of proteins. J. Anal. At. Spectrom. 2016, 31, 1830–1835. [Google Scholar] [CrossRef][Green Version]
- Rampler, E.; Dalik, T.; Stingeder, G.; Hann, S.; Koellensperger, G. Sulfur containing amino acids—Challenge of accurate quantification. J. Anal. At. Spectrom. 2012, 27, 1018–1023. [Google Scholar] [CrossRef]
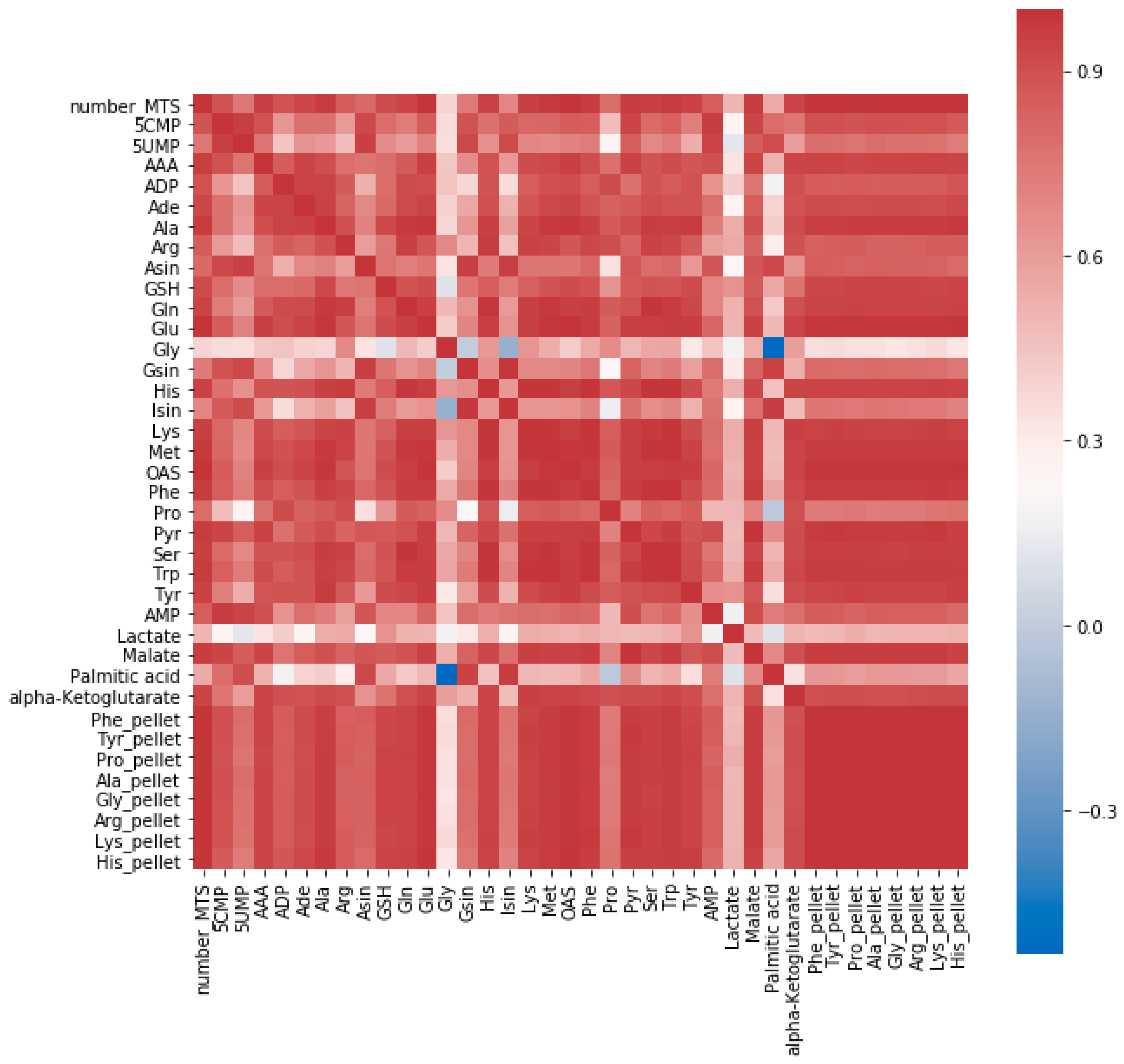
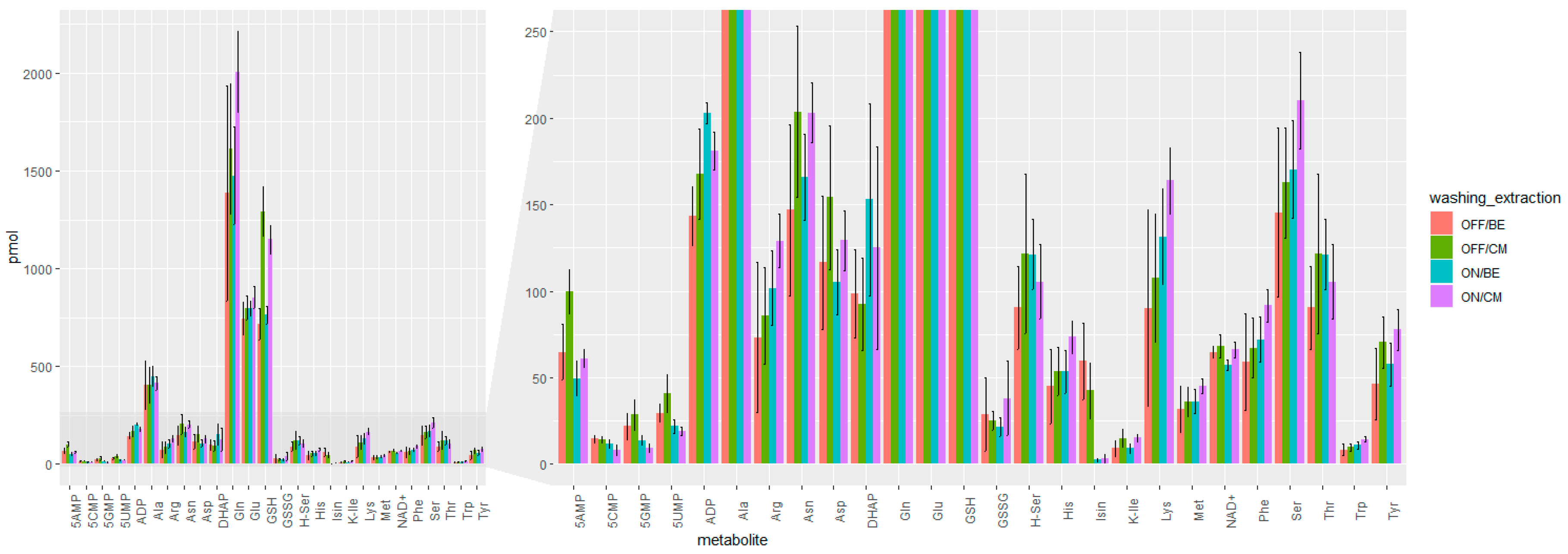
| Sample Preparation | Average RSD [%] |
|---|---|
| OFF/BE | 34% |
| ON/BE | 19% |
| OFF/CM | 24% |
| ON/CM | 18% |
| QC | 7% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rusz, M.; Rampler, E.; Keppler, B.K.; Jakupec, M.A.; Koellensperger, G. Single Spheroid Metabolomics: Optimizing Sample Preparation of Three-Dimensional Multicellular Tumor Spheroids. Metabolites 2019, 9, 304. https://doi.org/10.3390/metabo9120304
Rusz M, Rampler E, Keppler BK, Jakupec MA, Koellensperger G. Single Spheroid Metabolomics: Optimizing Sample Preparation of Three-Dimensional Multicellular Tumor Spheroids. Metabolites. 2019; 9(12):304. https://doi.org/10.3390/metabo9120304
Chicago/Turabian StyleRusz, Mate, Evelyn Rampler, Bernhard K. Keppler, Michael A. Jakupec, and Gunda Koellensperger. 2019. "Single Spheroid Metabolomics: Optimizing Sample Preparation of Three-Dimensional Multicellular Tumor Spheroids" Metabolites 9, no. 12: 304. https://doi.org/10.3390/metabo9120304
APA StyleRusz, M., Rampler, E., Keppler, B. K., Jakupec, M. A., & Koellensperger, G. (2019). Single Spheroid Metabolomics: Optimizing Sample Preparation of Three-Dimensional Multicellular Tumor Spheroids. Metabolites, 9(12), 304. https://doi.org/10.3390/metabo9120304







