Assessing the Pre-Analytical Stability of Small-Molecule Metabolites in Cerebrospinal Fluid Using Direct-Infusion Metabolomics
Abstract
:1. Introduction
2. Results
2.1. Analysis of Variability.
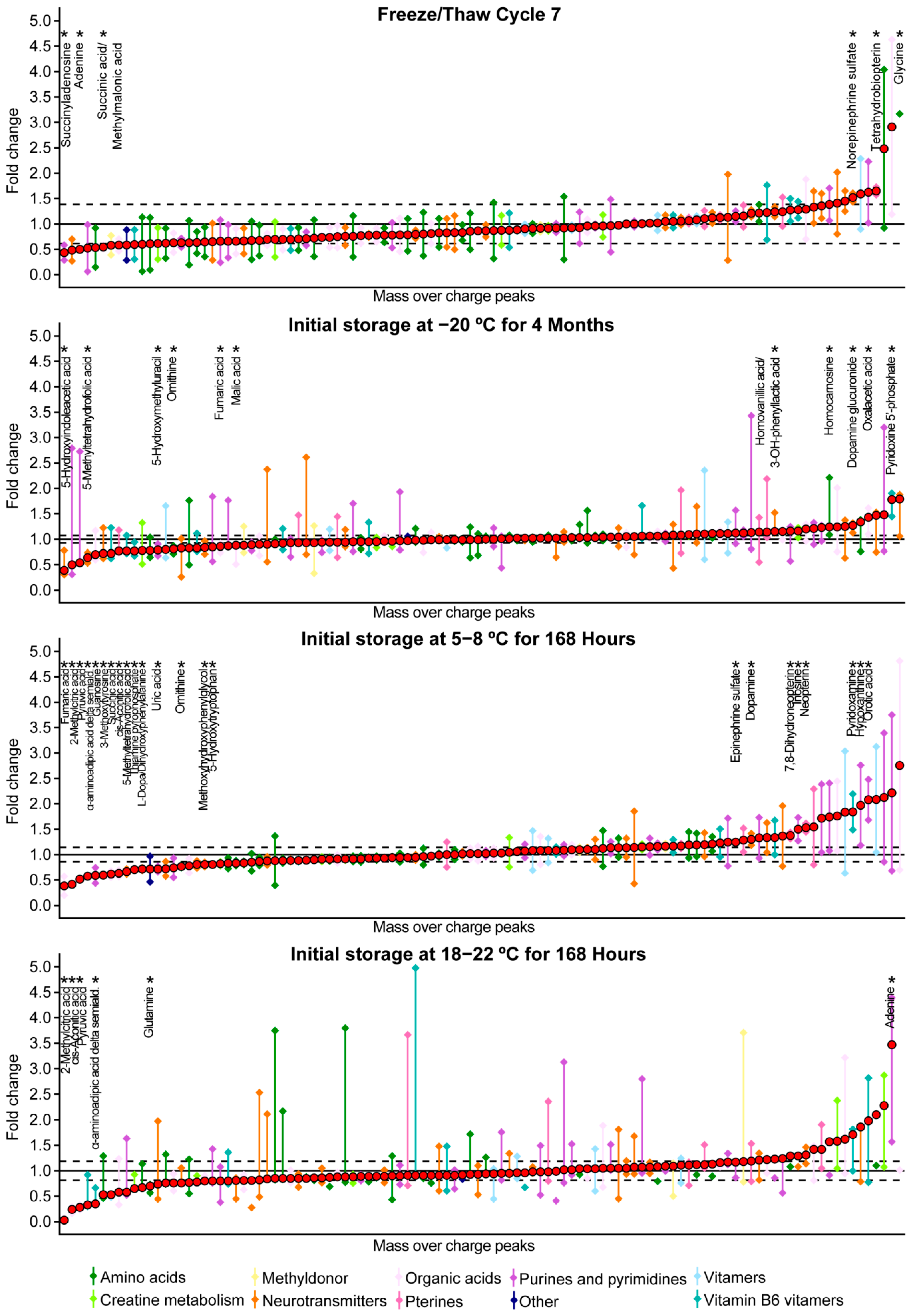
2.2. Freeze/Thaw Stability
2.3. Temperature Stability
3. Discussion
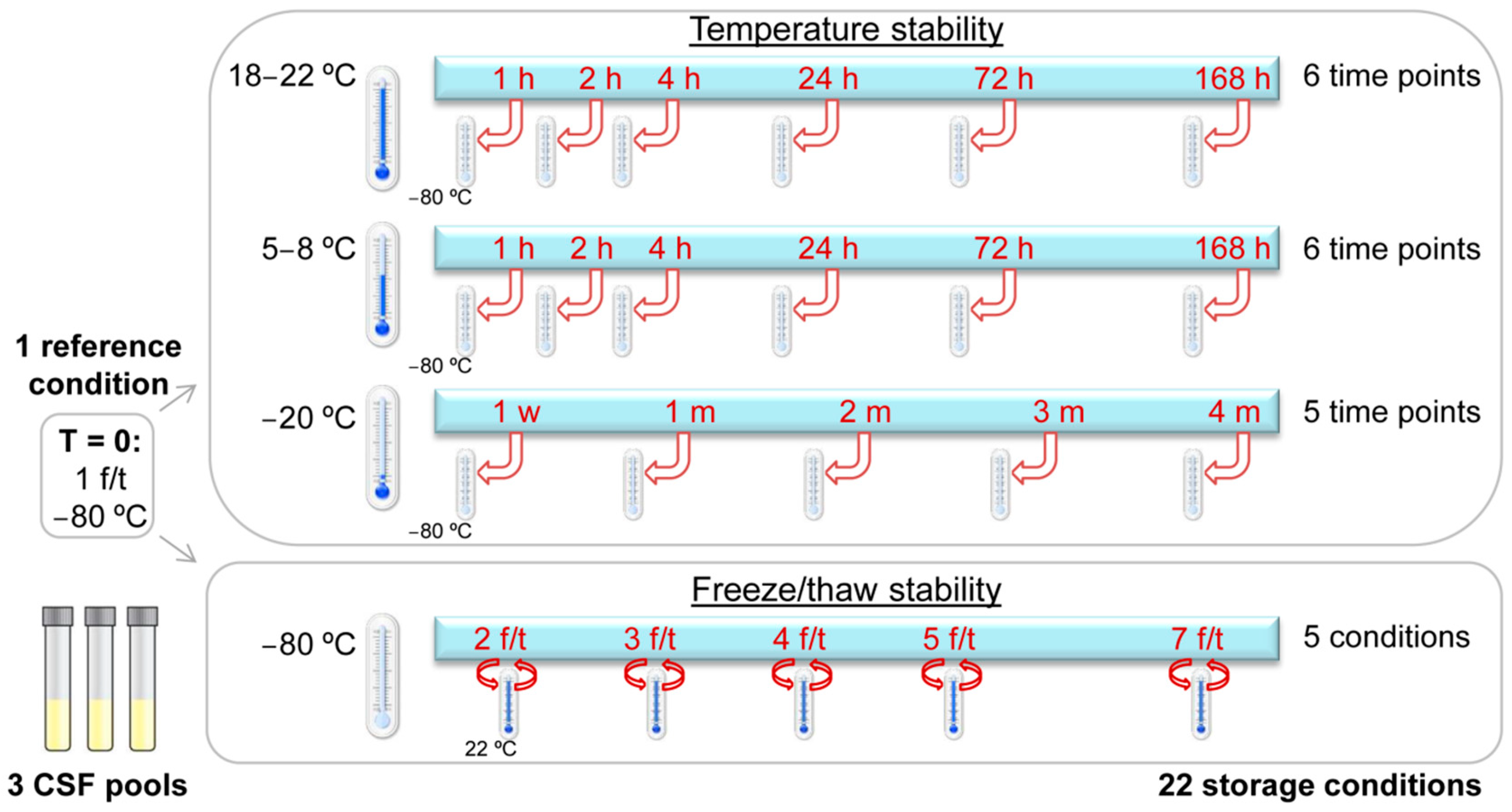
4. Materials and Methods
4.1. Sample Collection and Storage
4.2. Sample Preparation and Analysis
4.3. Data Processing
4.4. Metabolite Groups
4.5. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Cameron, S.; Gillio-Meina, C.; Ranger, A.; Choong, K.; Fraser, D.D. Collection and analyses of cerebrospinal fluid for pediatric translational research. Pediatr. Neurol. 2019, 98, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Anesi, A.; Rondanelli, M.; d’Eril, G.M. Stability of neuroactive amino acids in cerebrospinal fluid under various conditions of processing and storage. Clin. Chem. 1998, 44, 2359–2360. [Google Scholar] [PubMed]
- Levine, J.; Panchalingam, K.; McClure, R.J.; Gershon, S.; Pettegrew, J.W. Stability of CSF metabolites measured by proton NMR. J. Neural Transm. 2000, 107, 843–848. [Google Scholar] [CrossRef] [PubMed]
- Rosenling, T.; Slim, C.L.; Christin, C.; Coulier, L.; Shi, S.; Stoop, M.P.; Bosman, J.; Suits, F.; Horvatovich, P.L.; Stockhofe-Zurwieden, N.; et al. The effect of preanalytical factors on stability of the proteome and selected metabolites in cerebrospinal fluid (CSF). J. Proteome Res. 2009, 8, 5511–5522. [Google Scholar] [CrossRef] [PubMed]
- Haijes, H.A.; Willemsen, M.; van der Ham, M.; Gerrits, J.; Pras-Raves, M.L.; Prinsen, H.C.M.T.; van Hasselt, P.M.; de Sain-van der Velden, M.G.M.; Verhoeven-Duif, N.M.; Jans, J.J.M. Direct infusion based metabolomics identifies metabolic disease in patients’ dried blood spots and plasma. Metabolites. 2019, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Haijes, H.A.; van der Ham, M.; Gerrits, J.; van Hasselt, P.M.; Prinsen, H.C.M.T.; de Sain-van der Velden, M.G.M.; Verhoeven-Duif, N.M.; Jans, J.J.M. Direct-infusion based metabolomics unveils biochemical profiles of inborn errors of metabolism in cerebrospinal fluid. Mol. Genet. Metab. 2019, 127, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.A.; de Sain-van der Velden, M.G.; de Barse, M.M.; Roeleveld, M.W.; Hendriks, M.; Dorland, L.; Klomp, L.W.; Berger, R.; de Koning, T.J. Two mass-spectrometric techniques for quantifying serine enantiomers and glycine in cerebrospinal fluid: Potential confounders and age-dependent ranges. Clin. Chem. 2008, 54, 1443–1450. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.F.; James, C.A.; Zhu, X.; Davis, M.T.; Rose, M.J. Development of a method for the determination of glycine in human cerebrospinal fluid using pre-column derivatization and LC-MS/MS. J. Pharm. Biomed. Anal. 2011, 56, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Willemse, E.A.J.; Vermeiren, Y.; Garcia-Ayllon, M.S.; Bridel, C.; De Deyn, P.P.; Engelborghs, S.; van der Flier, W.M.; Jansen, E.E.W.; Lopez-Font, I.B.; Mendes, V.; et al. Pre-analytical stability of novel cerebrospinal fluid biomarkers. Clin. Chim. Act. 2019, 497, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Van der Flier, W.M.; Pijnenburg, Y.A.; Prins, N.; Lemstra, A.W.; Bouwman, F.H.; Teunissen, C.E.; van Berckel, B.N.; Stam, C.J.; Barkhof, F.; Visser, P.J.; et al. Optimizing patient care and research: The Amsterdam Dementia Cohort. J. Alzheimers Dis. 2014, 41, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Del Campo, M.; Mollenhauer, B.; Bertolotto, A.; Engelborghs, S.; Hampel, H.; Simonsen, A.H.; Kapaki, E.; Kruse, N.; Le Bastard, N.; Lehmann, S.; et al. Recommendations to standardize pre-analytical confounding factors in Alzheimer’s and Parkinson’s disease cerebrospinal fluid biomarkers: An update. Biomark. Med. 2012, 6, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Andreasson, U.; Perret-Liaudet, A.; van Waalwijk van Doorn, L.J.; Blennow, K.; Chiasserini, D.; Engelborghs, S.; Fladby, T.; Genc, S.; Kruse, N.; Kuiperij, H.B.; et al. A practical guide to immunoassay method validation. Front. Neurol. 2015, 19, 179. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res. 2013, 41, 801–807. [Google Scholar] [CrossRef] [PubMed]
- Sumner, I.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, F.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis, Chemical Analysis Working Group (CAWG), Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Batllori, M.; Molero-Luis, M.; Casado, M.; Sierra, C.; Artuch, R.; Armazabal, A. Biochemical analyses of cerebrospinal fluid for the diagnosis of neurometabolic conditions. What can we expect? Semin. Pediatr. Neurol. 2016, 23, 273–284. [Google Scholar] [CrossRef] [PubMed]
| Stable Isotope-Labeled Compound | CV | Median Absolute Variation | |||
|---|---|---|---|---|---|
| Freeze/Thaw Cycle 7 | −20 °C, 4 Months | 5–8 °C, 168 h | 18–22 °C, 168 h | ||
| 15N;2-13C-glycine | 0.580 | 0.975 | 1.732 | 6.999 | 1.855 |
| 2H4-alanine | 0.161 | 0.371 | 0.070 | 0.171 | 0.188 |
| 2H3-leucine | 0.166 | 0.411 | 0.037 | 0.158 | 0.265 |
| 2H3-methionine | 0.775 | 3.388 | 0.188 | 0.607 | 0.359 |
| 13C6-phenylalanine | 0.149 | 0.374 | 0.050 | 0.142 | 0.204 |
| 13C6-tyrosine | 0.150 | 0.349 | 0.043 | 0.098 | 0.206 |
| 2H3-aspartate | 0.160 | 0.349 | 0.137 | 0.110 | 0.155 |
| 2H3-glutamate | 0.126 | 0.247 | 0.099 | 0.110 | 0.074 |
| 2H2-ornithine | 0.183 | 0.540 | 0.014 | 0.108 | 0.168 |
| 2H2-citrulline | 0.134 | 0.231 | 0.070 | 0.131 | 0.038 |
| 2H4;13C-arginine | 0.163 | 0.380 | 0.105 | 0.190 | 0.170 |
| 2H8-valine | 0.155 | 0.377 | 0.042 | 0.124 | 0.222 |
| 2H9-carnitine | 0.202 | 0.580 | 0.072 | 0.179 | 0.162 |
| 2H3-acetylcarnitine | 1.058 | 432.707 | 689.785 | 0.567 | 0.999 |
| 2H3-propionylcarnitine | 0.194 | 0.384 | 0.038 | 0.225 | 0.161 |
| 2H3-butyrylcarnitine | 2.914 | 42.155 | 0.164 | 92.851 | 0.277 |
| 2H9-isovalerylcarnitine | 0.217 | 0.519 | 0.103 | 0.215 | 0.173 |
| 5th percentile | 0.127 | 0.244 | 0.033 | 0.106 | 0.067 |
| Median | 0.166 | 0.384 | 0.072 | 0.171 | 0.188 |
| 95th percentile | 1.565 | 120.266 | 139.343 | 24.170 | 1.170 |
| Amino Acids (24) | Neurotransmitters (21) | Purines, Pyrimidines (20) | Organic Acids (16) |
|---|---|---|---|
| Alanine Arginine | 3-Methoxytyrosine/3-OMD/Methyldopa | 5-Hydroxymethyluracil Adenine | n-Acetylaspartylglut. Acid 2-Methylcitric acid |
| Asparagine | 5-Hydroxyindoleacetic acid | Adenosine/Deoxyguanosine | 3-Hydroxybutyric acid |
| Aspartic acid | 5-Hydroxytryptophan | AICAR | 3-Hydroxyisovaleric acid |
| Cysteine | 5-Methyltetrahydrofolic acid | Deoxyadenosine | 4-Guanidinobutanoic acid |
| Cystine | Dopamine | Deoxyinosine | Acetoacetic acid |
| Glutamic acid | Dopamine 4-sulfate | Dihydrothymine | Cis-Aconitic acid |
| Glutamine | Dopamine glucuronide | Dihydrouracil | Citric acid |
| Glycine | Epinephrine | Guanosine | Fumaric acid |
| Histidine | Epinephrine glucuronide | Hypoxanthine | Lactic acid/ |
| Homoarginine Homocarnosine | Homovanillic acid/3-OH-phenyllactic acid | Inosine Orotic acid | 3-OH-propionic acid Malic acid |
| (Iso)leucine | Epinephrine sulfate | SAICAR | n-Acetylaspartic acid |
| Lysine | Gamma-aminobutyric acid | Succinyladenosine | Oxalacetic acid |
| Methionine | Glutamic acid | Thymidine | Propionic acid |
| Phenylalanine | l-Dopa/Dihydroxyphenylalan. | Thymine | Pyruvic acid |
| Proline | Methoxyhydroxyphenylglycol | Uracil | Succinic acid/ |
| Serine Threonine | n-Acetylserotonin Norepinephrine | Uric acid Uridine | Methylmalonic acid |
| Tryptophan | Norepinephrine sulfate | Xanthine | |
| Tyrosine | Serotonin | ||
| Ornithine | Vanillactic acid | Other (1) | Methyldonor (2) |
| Taurine | Vanillylmandelic acid | Saccharopine | s-Adenosylhomocysteine |
| Valine | s-Adenosylmethionine | ||
| Creatine m. (5) | Vitamers (5) | Pterines (6) | Vitamin B6 vitamers (8) |
| Creatine | Folic acid | 7,8-Dihydroneopterin | Pipecolic acid |
| Creatinine | Thiamine | Dihydrobiopterin | Pyridoxal |
| Guanidoacetic acid Phosphocreatine | Thiamine monophosphate Thiamine pyrophosphate | Neopterin Biopterin/Sepiapterin/Primapterin | Pyridoxal 5′-phosphate Pyridoxamine |
| Phosphocreatinine | Thiamine triphosphate | /6-Pyruvoil-tetrahydropterin | Pyridoxamine 5′-phosph. |
| Tetrahydroneopterin | Pyridoxine | ||
| Tetrahydrobiopterin | Pyridoxine 5′-phosphate | ||
| Alpha-aminoadipic acid delta-semialdehyde |
| Freeze/Thaw Cycle 7 | −20 °C, 4 Months | 5–8 °C, 168 h | 18–22 °C, 168 h | |
|---|---|---|---|---|
| Median absolute variation sILC | 0.384 | 0.072 | 0.171 | 0.188 |
| Range analysis variability | 0.616–1.384 | 0.928–1.072 | 0.829–1.171 | 0.812–1.188 |
| 1852 m/z peaks | ||||
| m/z peaks with decreased intensities | 45 | 142 | 206 | 67 |
| m/z peaks with increased intensities | 74 | 134 | 213 | 54 |
| 106 m/z peaks corresponding to neurometabolic metabolites | ||||
| m/z peaks with decreased intensities | 3 | 5 | 8 | 1 |
| m/z peaks with increased intensities | 3 | 6 | 15 | 5 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haijes, H.A.; Willemse, E.A.J.; Gerrits, J.; van der Flier, W.M.; Teunissen, C.E.; Verhoeven-Duif, N.M.; Jans, J.J.M. Assessing the Pre-Analytical Stability of Small-Molecule Metabolites in Cerebrospinal Fluid Using Direct-Infusion Metabolomics. Metabolites 2019, 9, 236. https://doi.org/10.3390/metabo9100236
Haijes HA, Willemse EAJ, Gerrits J, van der Flier WM, Teunissen CE, Verhoeven-Duif NM, Jans JJM. Assessing the Pre-Analytical Stability of Small-Molecule Metabolites in Cerebrospinal Fluid Using Direct-Infusion Metabolomics. Metabolites. 2019; 9(10):236. https://doi.org/10.3390/metabo9100236
Chicago/Turabian StyleHaijes, Hanneke A., Eline A.J. Willemse, Johan Gerrits, Wiesje M. van der Flier, Charlotte E. Teunissen, Nanda M. Verhoeven-Duif, and Judith J.M. Jans. 2019. "Assessing the Pre-Analytical Stability of Small-Molecule Metabolites in Cerebrospinal Fluid Using Direct-Infusion Metabolomics" Metabolites 9, no. 10: 236. https://doi.org/10.3390/metabo9100236
APA StyleHaijes, H. A., Willemse, E. A. J., Gerrits, J., van der Flier, W. M., Teunissen, C. E., Verhoeven-Duif, N. M., & Jans, J. J. M. (2019). Assessing the Pre-Analytical Stability of Small-Molecule Metabolites in Cerebrospinal Fluid Using Direct-Infusion Metabolomics. Metabolites, 9(10), 236. https://doi.org/10.3390/metabo9100236




_Verhoeven-Duif.png)


