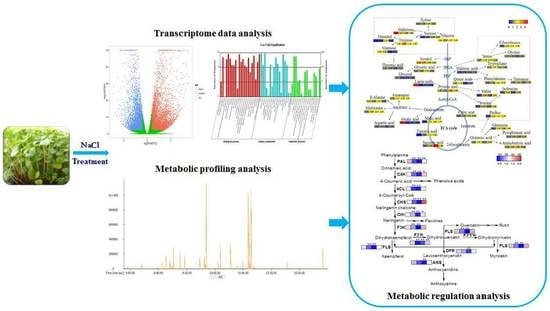
Comparative Transcriptome and Metabolic Profiling Analysis of Buckwheat (Fagopyrum Tataricum (L.) Gaertn.) under Salinity Stress
Abstract
1. Introduction
2. Results
2.1. De Novo Transcriptome Assembly of F. Tataricum and Sequence Analysis
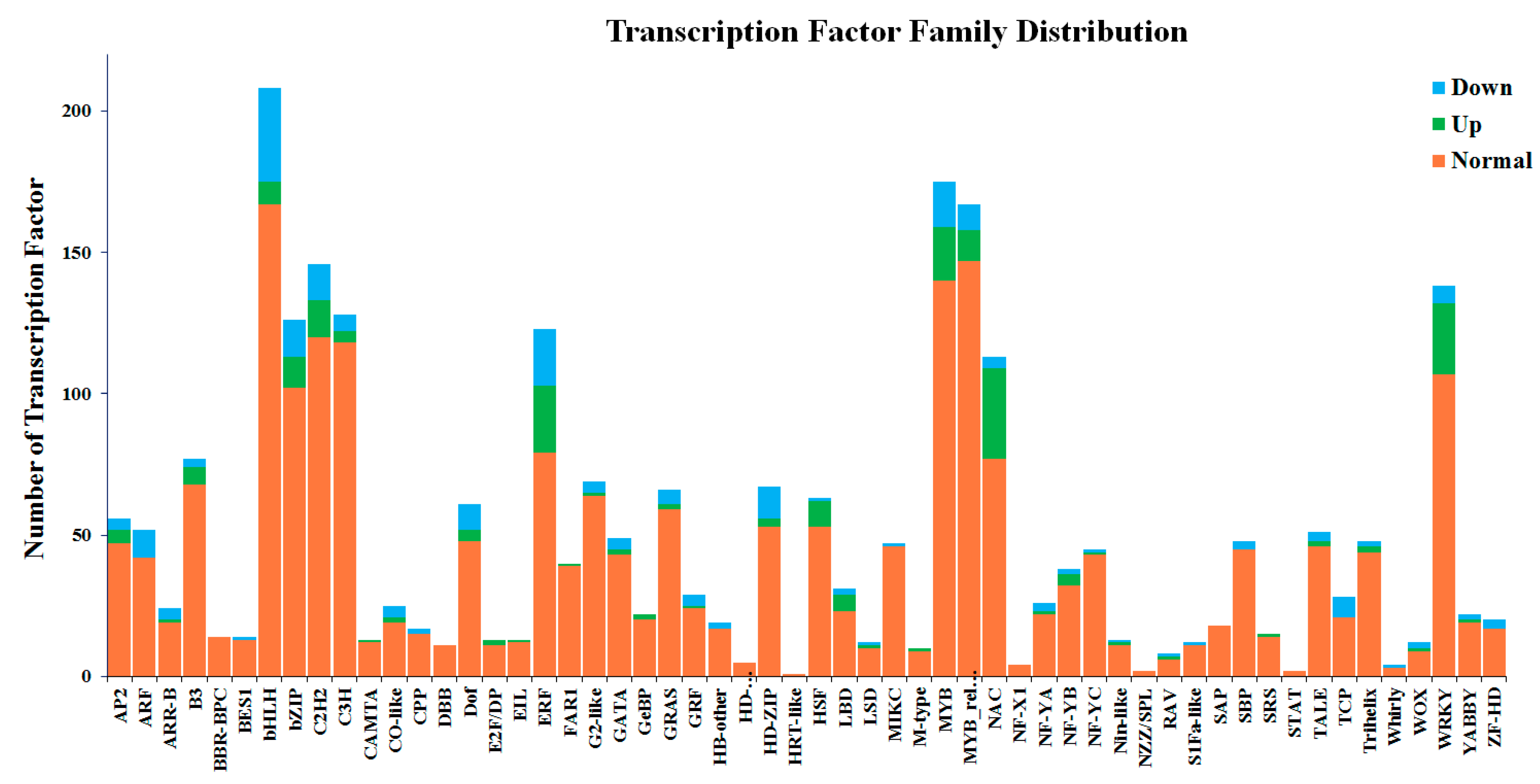
2.2. Functional Annotation and Classification of the Buckwheat Transcriptome
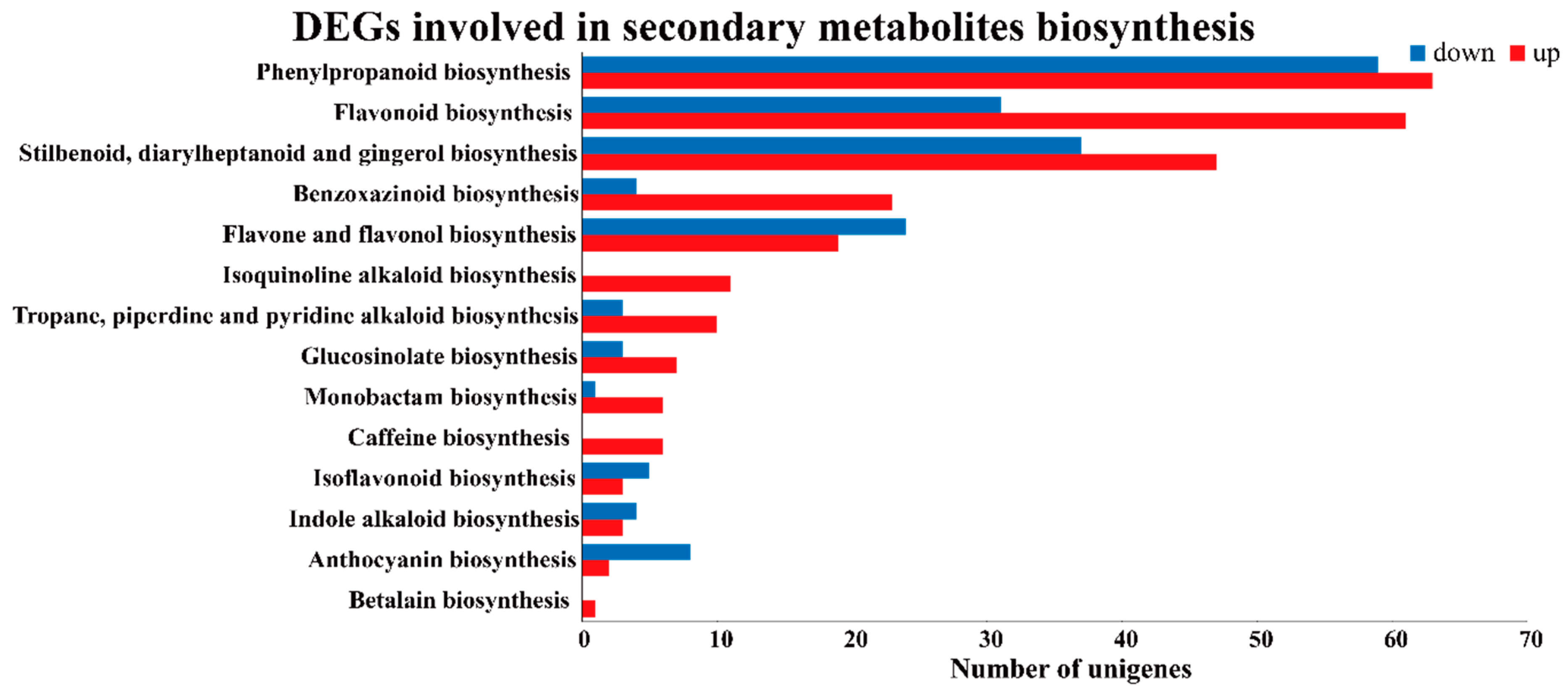
2.3. Differentially Expressed Genes (DEGs) of Buckwheat in Response to Salt Stress
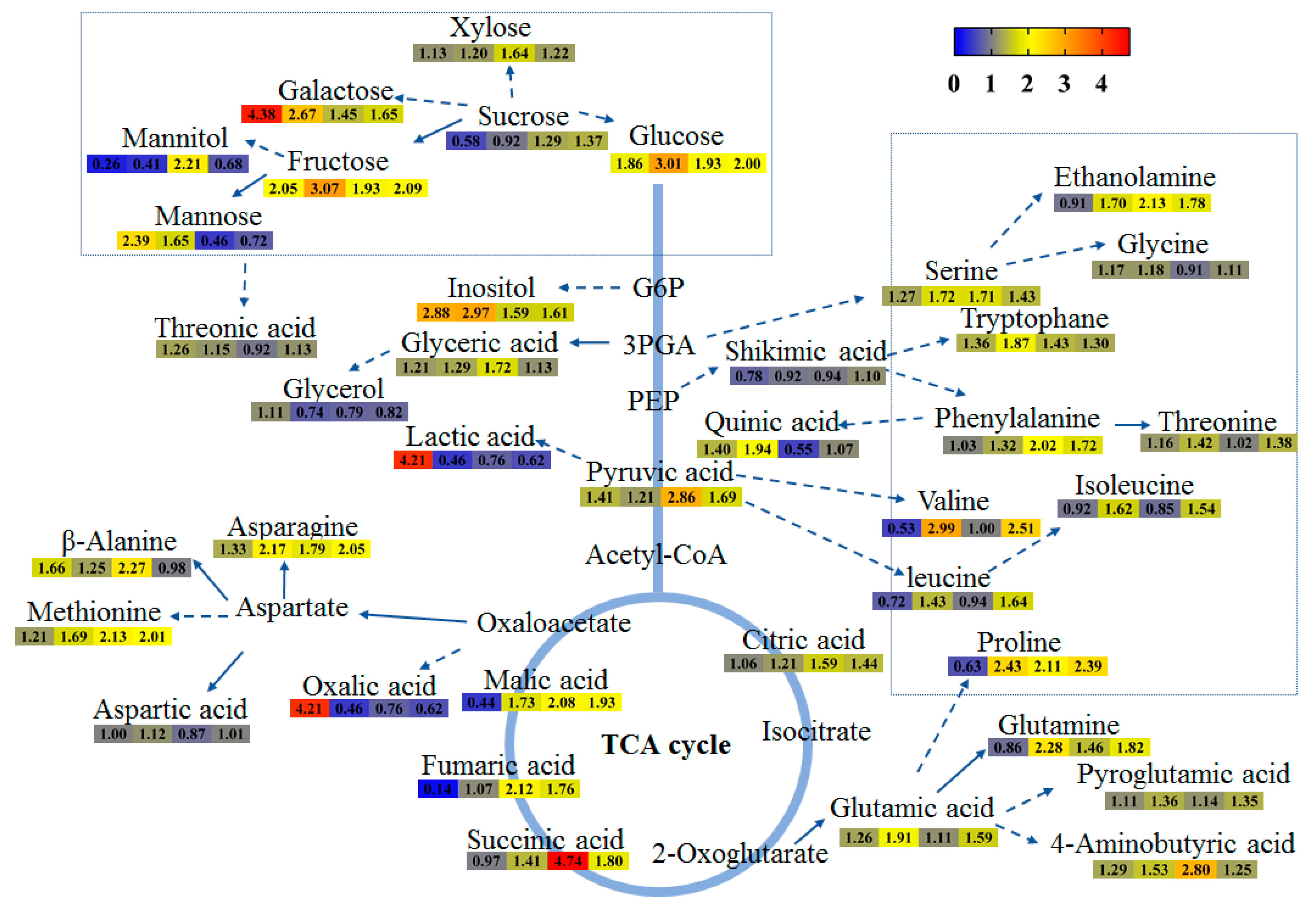
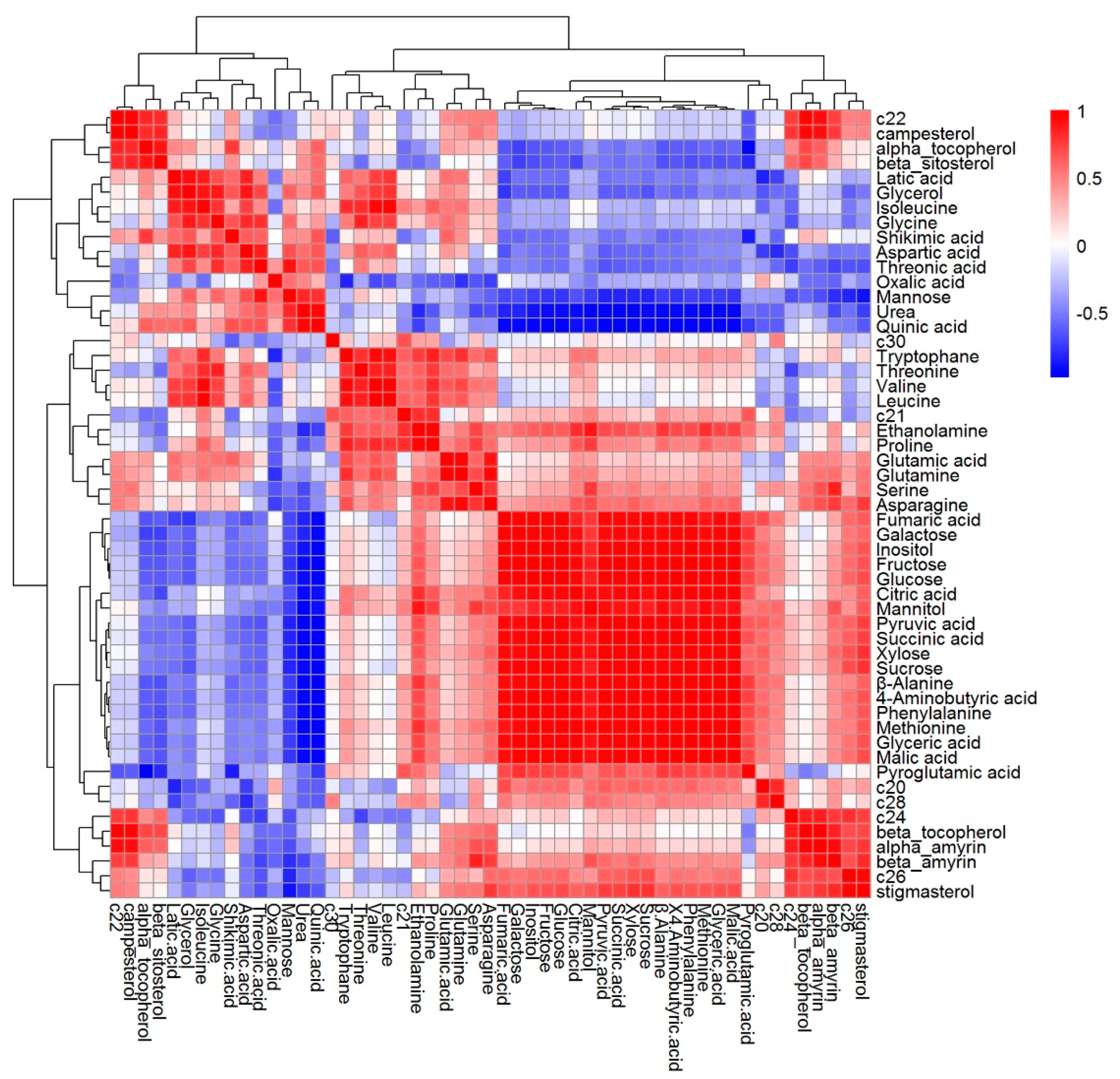
2.4. The Content of Metabolites in Tartary Buckwheat in Response to Salt Stress
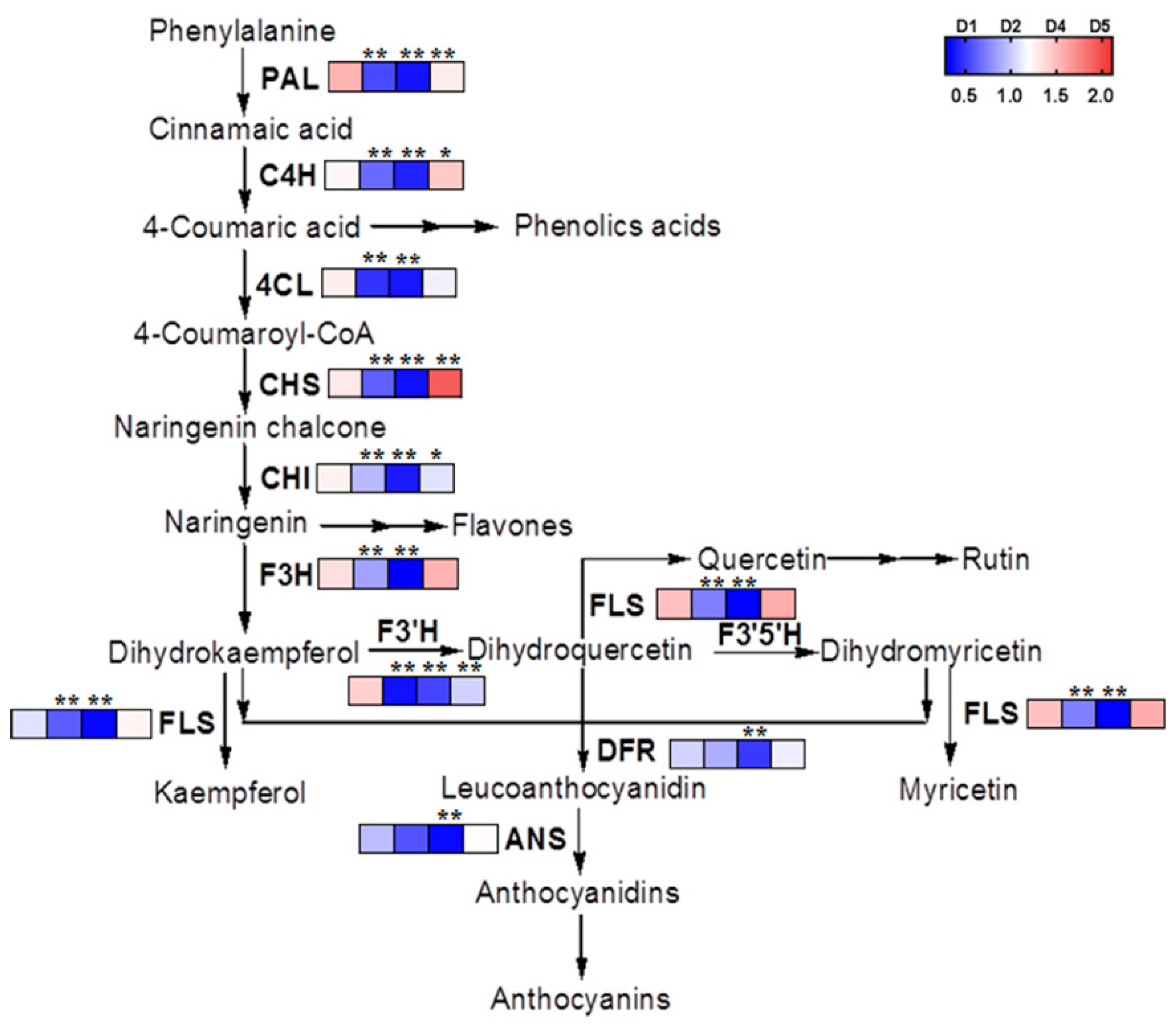
2.5. The Transcriptional and Metabolic Regulation of Phenylpropanoid Biosynthetic Pathway of Buckwheat in Response to Salt Stress
2.6. The Transcriptional and Metabolic Regulation of Carotenoid Biosynthesis of Buckwheat in Response to Salt Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Culture Conditions and Salinity Treatment
4.2. RNA Isolation, Library Preparation and Transcriptome Sequencing
4.3. Illumina Sequencing and De Novo ASSEMBLY
4.4. Availability of Supporting Data
4.5. Total RNA Extraction and cDNA Synthesis
4.6. Quantitative Real-Time PCR (qRT-PCR) Analysis
4.7. Extraction and Quantitative HPLC Analysis for Flavonoids
4.8. Liphophilic and Hydrophilic Metabolites Analysis Using GC-TOF-MS
4.9. Carotenoid Extraction and Analysis
4.10. Data Processing and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| GO | Gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KOG | Eukaryotic Ortholog Groups |
| Nr | NCBI non-redundant protein sequences |
| Nt | NCBI non-redundant nucleotide sequences |
| DEGs | Differential expressed genes |
| HPLC | High-performance liquid chromatography |
| GC-TOF-MS | Gas Chromatography Time-Of-Flight Mass Spectrometry |
References
- Tester, M.; Davenport, R. Na+ tolerance and Na+ transport in higher plants. Ann. Bot. 2003, 91, 503–527. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.P. Characterisation of the DNA-binding profile of barley HvCBF1 using an enzymatic method for rapid, quantitative and high-throughput analysis of the DNA-binding activity. Nucleic Acids Res. 2002, 30, e77. [Google Scholar] [CrossRef] [PubMed]
- Shokri-Gharelo, R.; Noparvar, P.M. Molecular response of canola to salt stress: Insights on tolerance mechanisms. PeerJ 2018, 6, e4822. [Google Scholar] [CrossRef] [PubMed]
- Golldack, D.; Li, C.; Mohan, H.; Probst, N. Tolerance to drought and salt stress in plants: Unraveling the signaling networks. Front. Plant Sci. 2014, 5, 151. [Google Scholar] [CrossRef] [PubMed]
- Stoeva, N.; Kaymakanova, M. Effect of salt stress on the growth and photosynthesis rate of bean plants (Phaseolus vulgaris L.). J. Cent. Eur. Agric. 2008, 9, 385–391. [Google Scholar]
- Zhang, H.; Han, B.; Wang, T.; Chen, S.; Li, H.; Zhang, Y.; Dai, S. Mechanisms of plant salt response: Insights from proteomics. J. Proteome Res. 2012, 11, 49–67. [Google Scholar] [CrossRef] [PubMed]
- Payyavula, R.S.; Navarre, D.A.; Kuhl, J.C.; Pantoja, A.; Pillai, S.S. Differential effects of environment on potato phenylpropanoid and carotenoid expression. BMC Plant Biol. 2012, 12, 39. [Google Scholar] [CrossRef]
- Petridis, A.; Therios, I.; Samouris, G.; Tananaki, C. Salinity-induced changes in phenolic compounds in leaves and roots of four olive cultivars (Olea europaea L.) and their relationship to antioxidant activity. Environ. Exp. Bot. 2012, 79, 37–43. [Google Scholar] [CrossRef]
- Sreenivasulu, N.; Grimm, B.; Wobus, U.; Weschke, W. Differential response of antioxidant compounds to salinity stress in salt-tolerant and salt-sensitive seedlings of foxtail millet (Setaria italica). Physiol. Plant 2000, 109, 435–442. [Google Scholar] [CrossRef]
- Borghesi, E.; Gonzà lez-Miret, M.L.; Escudero-Gilete, M.L.; Malorgio, F.; Heredia, F.J.; Meléndez-Martínez, A.J. Effects of salinity stress on carotenoids, anthocyanins and color of diverse tomato genotypes. J. Agric. Food Chem. 2011, 59, 11676–11682. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, S.B.; Aung, B.; Amyot, L.; Lalin, I.; Lachâal, M.; Karray-Bouraoui, N.; Hannoufa, A. Salt stress (NaCl) affects plant growth and branch pathways of carotenoid and flavonoid biosyntheses in Solanum nigrum. Acta Physiol. Plant. 2016, 38, 1–13. [Google Scholar] [CrossRef]
- Tuteja, N. Mechanisms of high salinity tolerance in plants. Methods Enzymol. 2007, 428, 419–438. [Google Scholar] [PubMed]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Tian, X.; Wang, F.; Zhang, L.; Xin, M.; Hu, Z.; Yao, Y.; Ni, Z.; Sun, Q.; Peng, H. Characterization of wheat MYB genes responsive to high temperatures. BMC Plant Biol. 2017, 17, 208. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Lu, J.; Xu, J.; Duan, B.; He, X.; Liu, J. Genome-wide identification of WRKY genes in the desert Poplar Populus euphratica and adaptive evolution of the genes in response to salt stress. Evol. Bioinform. 2015, 11, 47–55. [Google Scholar]
- He, Y.; Li, W.; Lv, J.; Jia, Y.; Wang, M.; Xia, G. Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salinity stress tolerance in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 1511–1522. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.H.; Yoo, K.S.; Hyoung, S.; Nguyen, H.T.; Kim, Y.Y.; Kim, H.J.; Ok, S.H.; Yoo, S.D.; Shin, J.S. An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Lett. 2013, 587, 1773–1778. [Google Scholar] [CrossRef]
- Zhong, H.; Guo, Q.Q.; Chen, L.; Ren, F.; Wang, Q.Q.; Zheng, Y.; Li, X.B. Two Brassica napus genes encoding NAC transcription factors are involved in response to high-salinity stress. Plant Cell Rep. 2012, 31, 1991–2003. [Google Scholar] [CrossRef]
- Nakashima, K.; Tran, L.S.; Van Nguyen, D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef]
- Xu, Z.; Zhaxi, G.; Wang, C.; Xue, F.; Zhang, H.; Ji, W. Wheat NAC transcription factor TaNAC29 is involved in response to salt stress. Plant Physiol. Biochem. 2015, 96, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Chen, G.; Zhang, J.; Zhang, Y.; Xie, Q.; Zhao, Z.; Pan, Y.; Hu, Z. The abiotic stress-responsive NAC-type transcription factor SlNAC4 regulates salt and drought tolerance and stress-related genes in tomato (Solanum lycopersicum). Plant Cell Rep. 2014, 33, 1851–1863. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Gao, J.; Feng, Y.; Zhang, Z.; Liu, Y.; Fang, W.; Chen, S.; Chen, F.; Jiang, J. The chrysanthemum leaf and root transcript profiling in response to salinity stress. Gene 2018, 674, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Golldack, D.; Luking, I.; Yang, O. Plant tolerance to drought and salinity: Stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 2011, 30, 1383–1391. [Google Scholar] [CrossRef] [PubMed]
- Park, N.I.; Li, X.; Suzuki, T.; Kim, S.J.; Woo, S.H.; Park, C.H.; Park, S.U. Differential expression of anthocyanin biosynthetic genes and anthocyanin accumulation in tartary buckwheat cultivars ‘Hokkai t8’ and ‘Hokkai t10’. J. Agric. Food Chem. 2011, 59, 2356–2361. [Google Scholar] [CrossRef]
- Mendler-Drienyovszki, N.; Cal, A.J.; Dobranszki, J. Progress and prospects for interspecific hybridization in buckwheat and the genus Fagopyrum. Biotechnol. Adv. 2013, 31, 1768–1775. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Maeda, T.; Sarker, M.Z.; Takigawa, S.; Matsuura-Endo, C.; Yamauchi, H.; Mukasa, Y.; Saito, K.; Hashimoto, N.; Noda, T.; et al. Identification of anthocyanins in the sprouts of buckwheat. J. Agric. Food Chem. 2007, 55, 6314–6318. [Google Scholar] [CrossRef]
- Zheng, C.; Hu, C.; Ma, X.; Peng, C.; Zhang, H.; Qin, L. Cytotoxic phenylpropanoid glycosides from Fagopyrum tataricum (L.) Gaertn. Food Chem. 2012, 132, 433–438. [Google Scholar] [CrossRef]
- Matsui, T.; Kudo, A.; Tokuda, S.; Matsumoto, K.; Hosoyama, H. Identification of a new natural vasorelaxatant compound, (+)-osbeckic acid, from rutin-free tartary buckwheat extract. J. Agric. Food Chem. 2010, 58, 10876–10879. [Google Scholar] [CrossRef]
- Yang, N.; Ren, G. Determination of D-chiro-Inositol in tartary buckwheat using high-performance liquid chromatography with an evaporative light-scattering detector. J. Agric. Food Chem. 2008, 56, 757–760. [Google Scholar] [CrossRef]
- Ren, Q.; Wu, C.; Ren, Y.; Zhang, J. Characterization and identification of the chemical constituents from tartary buckwheat (Fagopyrum tataricum Gaertn) by high performance liquid chromatography/photodiode array detector/linear ion trap FTICR hybrid mass spectrometry. Food Chem. 2013, 136, 1377–1389. [Google Scholar] [CrossRef] [PubMed]
- Wijngaard, H.H.; Arendt, E.K. Buckwheat. Cereal Chem. 2006, 83, 391–401. [Google Scholar] [CrossRef]
- Middleton, E., Jr.; Kandaswami, C.; Theoharides, T.C. The effects of plant flavonoids on mammalian cells: Implications for inflammation, heart disease and cancer. Pharmacol. Rev. 2000, 52, 673–751. [Google Scholar] [PubMed]
- Panwar, A.; Gupta, N.; Chauhan, R.S. Biosynthesis and accumulation of flavonoids in Fagopyrum spp. Eur. J. Plant Sci. Biotechnol. 2012, 6, 17–26. [Google Scholar]
- Suzuki, T.; Morishita, T.; Mukasa, Y.; Takigawa, S.; Yokota, S.; Ishiguro, K.; Noda, T. Breeding of ‘Manten-Kirari,’ a non-bitter and trace-rutinosidase variety of Tartary buckwheat (Fagopyrum tataricum Gaertn.). Breed. Sci. 2014, 64, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, M.; Ohkawara, T.; Sato, Y.; Satoh, H.; Suzuki, T.; Ishiguro, K.; Noda, T.; Morishita, T.; Nishihira, J. Effectiveness of rutin-rich Tartary buckwheat (Fagopyrum tataricum Gaertn.) ‘Manten-Kirari’ in body weight reduction related to its antioxidant properties: A randomised, double-blind, placebo-controlled study. J. Funct. Foods 2016, 26, 460–469. [Google Scholar] [CrossRef]
- Watanabe, M. Catechins as antioxidants from buckwheat (Fagopyrum esculentum Moench) groats. J. Agric. Food Chem. 1998, 46, 839–845. [Google Scholar] [CrossRef]
- Holasova, M.; Fiedlerova, V.H.; Orsak, M.; Lachman, J. Vavreinova Buckwheat—The source of antioxidant activity in functional foods. Food Res. Int. 2002, 35, 207–211. [Google Scholar] [CrossRef]
- Krkošková, B.; Mrázová, Z. Prophylactic components of buckwheat. Food Res. Int. 2005, 38, 561–568. [Google Scholar] [CrossRef]
- Park, J.W.; Kang, D.B.; Kim, C.W.; Koh, S.H.; Yum, H.Y.; Kim, K.E.; Hong, C.S.; Lee, K.Y. Identification and characterization of the major allergens of buckwheat. Allergy 2015, 55, 1035–1041. [Google Scholar] [CrossRef]
- Karki, R.; Park, C.H.; Kim, D.W. Extract of buckwheat sprouts scavenges oxidation and inhibits pro-inflammatory mediators in lipopolysaccharide-stimulated macrophages (RAW264.7). J. Integr. Med. 2013, 11, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.M.; Wei, P. Anti-fatigue properties of tartary buckwheat extracts in mice. Int. J. Mol. Sci. 2011, 12, 4770–4780. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Qiao, Z.; Wang, H.; Zhu, L.; Zhang, L.; Lu, Y.; Cui, Y.; Zhang, Z.; Wang, Z. Tartary buckwheat flavonoid activates caspase 3 and induces HL-60 cell apoptosis. Methods Find. Exp. Clin. Pharmacol. 2001, 23, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Qiao, Z.; Wang, H.; Zhu, L.; Zhang, L.; Lu, Y.; Zhang, Z.; Wang, Z. Molecular basis of fas and cytochrome c pathways of apoptosis induced by tartary buckwheat flavonoid in HL-60 cells. Methods Find. Exp. Clin. Pharmacol. 2003, 25, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhu, K.; Hui, Z.; Yao, H. Anti-tumor activity of a novel protein obtained from tartary buckwheat. Int. J. Mol. Sci. 2010, 11, 5201–5211. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-J.; Park, K.-J.; Lim, J.-H. Metabolomic analysis of phenolic compounds in buckwheat (Fagopyrum esculentum M.) sprouts treated with methyl Jasmonate. J. Agric. Food Chem. 2011, 59, 5707–5713. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.J.; Lim, J.H.; Park, K.J.; Kang, S.; Kim, D.S.; Park, S. Methyl jasmolate treated buckwheat sprout powder enhances glucose metabolism by potentiating hepatic insulin signaling in estrogen-deficient rats. Nutrition 2016, 32, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.-M.; Arasu, M.V.; Kim, Y.-B.; Park, S.U.; Kim, S.-J. Phenylalanine and LED lights enhance phenolic compound production in Tartary buckwheat sprouts. Food Chem. 2015, 177, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.H.; Zhang, B.; Xu, Z.Q. Salt tolerance conferred by overexpression of Arabidopsis vacuolar Na(+)/H (+) antiporter gene AtNHX1 in common buckwheat (Fagopyrum esculentum). Transgenic Res. 2008, 17, 121–132. [Google Scholar] [CrossRef]
- Qin, P.; Wang, Q.; Shan, F.; Hou, Z.; Ren, G. Nutritional composition and flavonoids content of flour from different buckwheat cultivars. Int. J. Food Sci. Technol. 2010, 45, 951–958. [Google Scholar] [CrossRef]
- Wan, Y.; Xiang, D.B.; Zeng, X.L.; Wang, X.; Yuan, Z.H.; Tan, M.L.; Zou, L.; Zhao, G. Salt stress influence on yield of tartary buckwheat sprouts and flavonoids content. Sci. Technol. Food Ind. 2016, 37, 328–332. [Google Scholar]
- Zhu, Y.; Guo, Y. Optimization of culture conditions for accumulating gamma-aminobutyric acid (GABA) in germinated tartary buckwheat under salt stress by response surface methodology. Food Sci. 2015, 36, 96–100. [Google Scholar]
- Li, X.; Kim, Y.B.; Kim, Y.; Zhao, S.; Kim, H.H.; Chung, E.; Lee, J.H.; Park, S.U. Differential stress-response expression of two flavonol synthase genes and accumulation of flavonols in tartary buckwheat. J. Plant Physiol. 2013, 170, 1630–1636. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.H.; Wang, Y.Q.; Song, J.N.; Yang, H.B. Transcriptomic identification of salt-related genes and de novo assembly in common buckwheat (F. esculentum). Plant Physiol. Biochem. 2018, 127, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Lim, S.H.; Ha, S.H.; Yeo, Y.; Park, W.T.; Kwon, D.Y.; Park, S.U.; Kim, J.K. Metabolite profiling approach reveals the interface of primary and secondary metabolism in colored cauliflowers (Brassica oleracea, L. ssp. botrytis). J. Agric. Food Chem. 2013, 61, 6999–7007. [Google Scholar] [CrossRef] [PubMed]
- Tuan, P.A.; Thwe, A.A.; Kim, J.K.; Kim, Y.B.; Lee, S.; Park, S.U. Molecular characterisation and the light-dark regulation of carotenoid biosynthesis in sprouts of tartary buckwheat (Fagopyrum tataricum Gaertn.). Food Chem. 2013, 141, 3803–3812. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Rivero, R.M.; Shulaev, V.; Blumwald, E.; Mittler, R. Abiotic and biotic stress combinations. New Phytol. 2014, 203, 32–43. [Google Scholar] [CrossRef]
- Jeon, J.; Kim, J.K.; Wu, Q.; Park, S.U. Effects of cold stress on transcripts and metabolites in tartary buckwheat (Fagopyrum tataricum). Environ. Exp. Bot. 2018, 155, 488–496. [Google Scholar] [CrossRef]
- Li, S.-J.; Bai, Y.-C.; Li, C.-L.; Yao, H.-P.; Chen, H.; Zhao, H.-X.; Wu, Q.J.A.P.P. Anthocyanins accumulate in tartary buckwheat (Fagopyrum tataricum) sprout in response to cold stress. Acta Physiol. Plant. 2015, 37, 159. [Google Scholar] [CrossRef]
- Guo, R.; Yang, Z.; Li, F.; Yan, C.; Zhong, X.; Liu, Q.; Xia, X.; Li, H.; Zhao, L. Comparative metabolic responses and adaptive strategies of wheat (Triticum aestivum) to salt and alkali stress. BMC Plant Biol. 2015, 15, 170. [Google Scholar] [CrossRef] [PubMed]
- Hernández, J.A.; Jiménez, A.; Mullineaux, P.; Sevilia, F. Tolerance of pea (Pisum sativum L.) to long-term salt stress is associated with induction of antioxidant defences. Plant Cell Environ. 2000, 23, 853–862. [Google Scholar] [CrossRef]
- Pascale, S.D.; Maggio, A.; Angelino, G.; Graziani, G. Effect of salt stress on water relations and antioxidant activity in tomato. Acta Hortic. 2003, 613, 39–46. [Google Scholar] [CrossRef]
- Huang, C.; He, W.; Guo, J.; Chang, X.; Su, P.; Zhang, L. Increased sensitivity to salt stress in an ascorbate-deficient Arabidopsis mutant. J. Exp. Bot. 2005, 56, 3041–3049. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; He, L.; Cai, L. Transcriptome Sequencing: RNA-Seq. Methods Mol. Biol. 2018, 1754, 15–27. [Google Scholar]
- Upadhyay, A.; Gaonkar, T.; Upadhyay, A.K.; Jogaiah, S.; Shinde, M.P.; Kadoo, N.Y.; Gupta, V.S. Global transcriptome analysis of grapevine (Vitis vinifera L.) leaves under salt stress reveals differential response at early and late stages of stress in table grape cv. Thompson Seedless. Plant Physiol. Biochem. 2018, 129, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Bai, X.; Zhao, W.; Xiang, D.; Wan, Y.; Yan, J.; Zou, L.; Zhao, G. De novo assembly and analysis of tartary buckwheat (Fagopyrum tataricum Garetn.) transcriptome discloses key regulators involved in salt-stress response. Genes 2017, 8, 255. [Google Scholar] [CrossRef] [PubMed]
- Linic, I.; Samec, D.; Gruz, J.; Vujcic Bok, V.; Strnad, M.; Salopek-Sondi, B. Involvement of phenolic acids in short-term adaptation to salinity stress is species-specific among Brassicaceae. Plants 2019, 8, 155. [Google Scholar] [CrossRef]
- Enfissi, E.M.A.; Barneche, F.; Ahmed, I.; Lichtlé, C.; Gerrish, C.; McQuinn, R.P.; Giovannoni, J.J.; Lopez-Juez, E.; Bowler, C.; Bramley, P.M.; et al. Integrative transcript and metabolite analysis of nutritionally enhanced DE-ETIOLATED1 downregulated tomato fruit. Plant Cell 2010, 22, 1190–1215. [Google Scholar] [CrossRef] [PubMed]
- Ayenew, B.; Degu, A.; Manela, N.; Perl, A.; Shamir, M.O.; Fait, A. Metabolite profiling and transcript analysis reveal specificities in the response of a berry derived cell culture to abiotic stresses. Front. Plant Sci. 2015, 6, 728. [Google Scholar] [CrossRef]
- Park, C.H.; Morgan, A.M.A.; Park, B.B.; Lee, S.Y.; Lee, S.; Kim, J.K.; Park, S.U. Metabolic analysis of four cultivars of Liriope platyphylla. Metabolites 2019, 9, 59. [Google Scholar] [CrossRef]
- Park, C.H.; Yeo, H.J.; Park, Y.E.; Baek, S.A.; Kim, J.K.; Park, S.U. Transcriptome analysis and metabolic profiling of Lycoris Radiata. Biology 2019, 8, 63. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.H.; Park, K.J.; Kim, B.K.; Jeong, J.W.; Kim, H.J. Effect of salinity stress on phenolic compounds and carotenoids in buckwheat (Fagopyrum esculentum M.) sprout. Food Chem. 2012, 135, 1065–1070. [Google Scholar] [CrossRef] [PubMed]
- Sudhir, P.; Murthy, S.D.S. Effects of salt stress on basic processes of photosynthesis. Photosynthetica 2004, 42, 481–486. [Google Scholar] [CrossRef]
- Eulgem, T.; Somssich, I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007, 10, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Dong, D.; Yang, Q.; Zhu, D. Salt-responsive transcriptome profiling of suaeda glaucavia RNA sequencing. PLoS ONE 2016, 11, e0150504. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Guo, X.; Guo, X.; Zhao, D.; Zhao, W.; Chen, J.; Li, T. PacMYBA, a sweet cherry R2R3-MYB transcription factor, is a positive regulator of salt stress tolerance and pathogen resistance. Plant Physiol. Biochem. 2017, 112, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Xu, Y.; Ma, Q.; Xu, W.; Wang, T.; Xue, Y.; Chong, K. Overexpression of an R1R2R3 MYB Gene, OsMYB3R-2, Increases tolerance to freezing, drought and salt stress in transgenic arabidopsis. Plant Physiol. 2007, 143, 1739–1751. [Google Scholar] [CrossRef]
- Gao, F.; Zhao, H.X.; Yao, H.P.; Li, C.L.; Chen, H.; Wang, A.H.; Park, S.U.; Wu, Q. Identification, isolation and expression analysis of eight stress-related R2R3-MYB genes in tartary buckwheat (Fagopyrum tataricum). Plant Cell Rep. 2016, 35, 1385–1396. [Google Scholar] [CrossRef]
- Gao, F.; Yao, H.; Zhao, H.; Zhou, J.; Luo, X.; Huang, Y.; Li, C.; Chen, H.; Wu, Q. Tartary buckwheat FtMYB10 encodes an R2R3-MYB transcription factor that acts as a novel negative regulator of salt and drought response in transgenic Arabidopsis. Plant Physiol. Biochem. 2016, 109, 387–396. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Zhang, L.; Li, X.; Ma, B.; Gao, Q.; Du, H.; Han, Y.; Li, Y.; Cao, Y.; Qi, M.; Zhu, Y.; et al. The tartary buckwheat genome provides insights into rutin biosynthesis and abiotic stress tolerance. Mol. Plant 2017, 10, 1224–1237. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Kim, J.K.; Park, S.Y.; Zhao, S.; Kim, Y.B.; Lee, S.; Park, S.U. Comparative analysis of flavonoids and polar metabolite profiling of Tanno-original and Tanno-high rutin buckwheat. J. Agric. Food Chem. 2014, 62, 2701–2708. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.J.; Choi, J.; Kim, K.W.; Ahn, S.K.; Ha, S.-H.; Choi, Y.; Park, N.I.; Kim, J.K. Metabolite profiling of peppers of various colors reveals relationships between tocopherol, carotenoid and phytosterol content. J. Food Sci. 2017, 82, 2885–2893. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.A.; Jung, Y.H.; Lim, S.H.; Park, S.U.; Kim, J.K. Metabolic profiling in chinese cabbage (Brassica rapa, L. subsp. pekinensis) cultivars reveals that glucosinolate content is correlated with carotenoid content. J. Agric. Food Chem. 2016, 64, 4426–4434. [Google Scholar] [CrossRef] [PubMed]
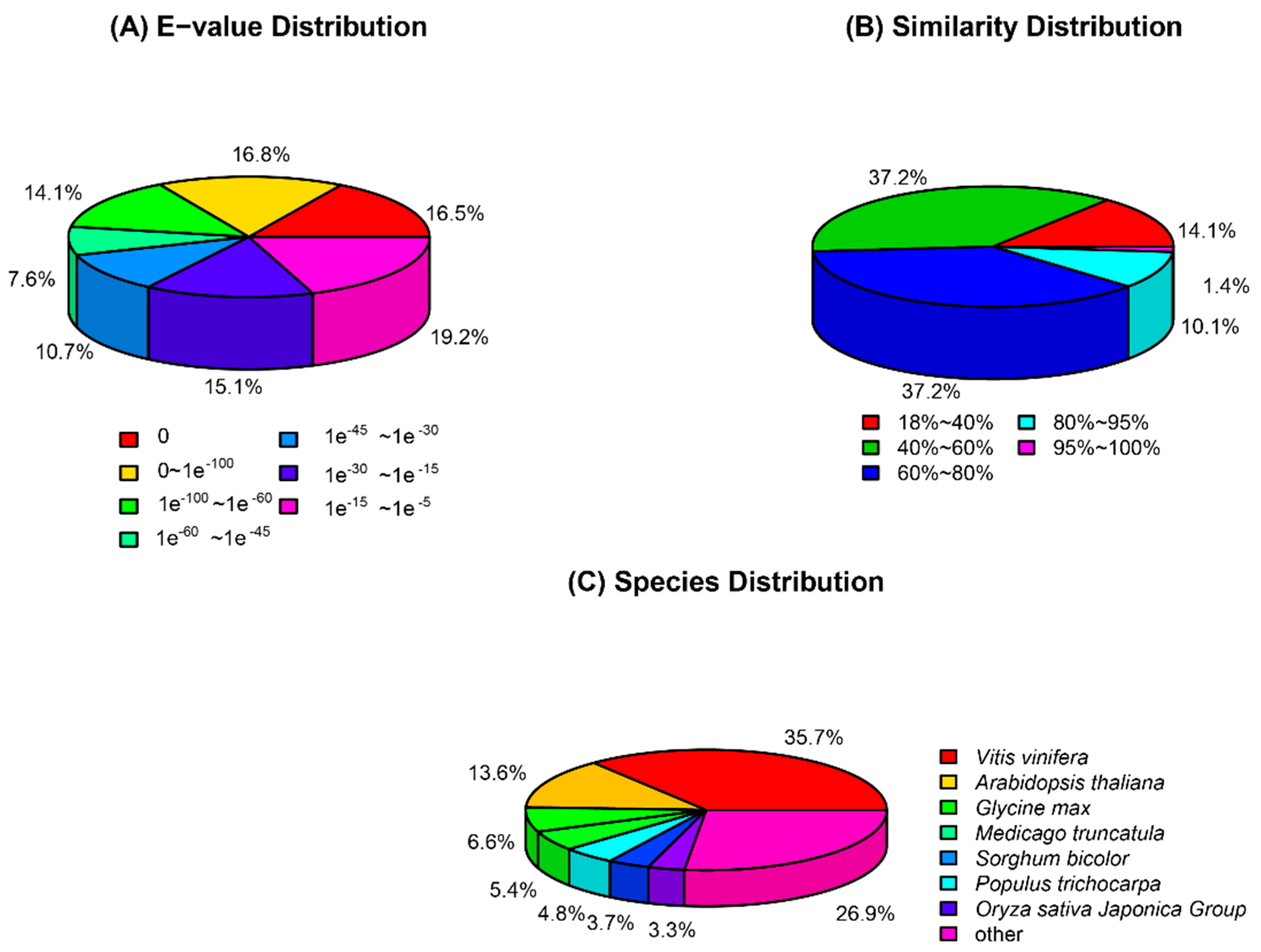
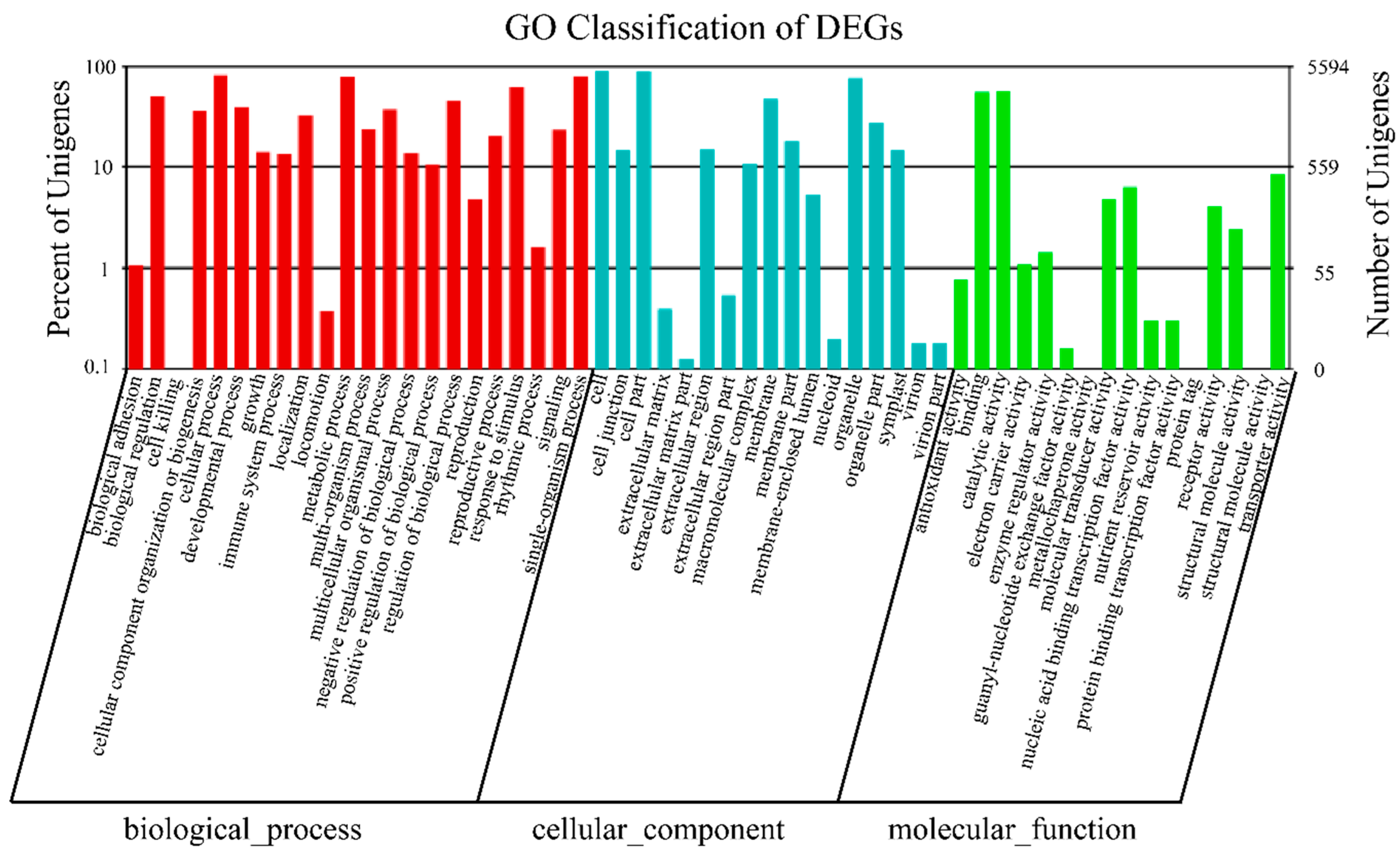
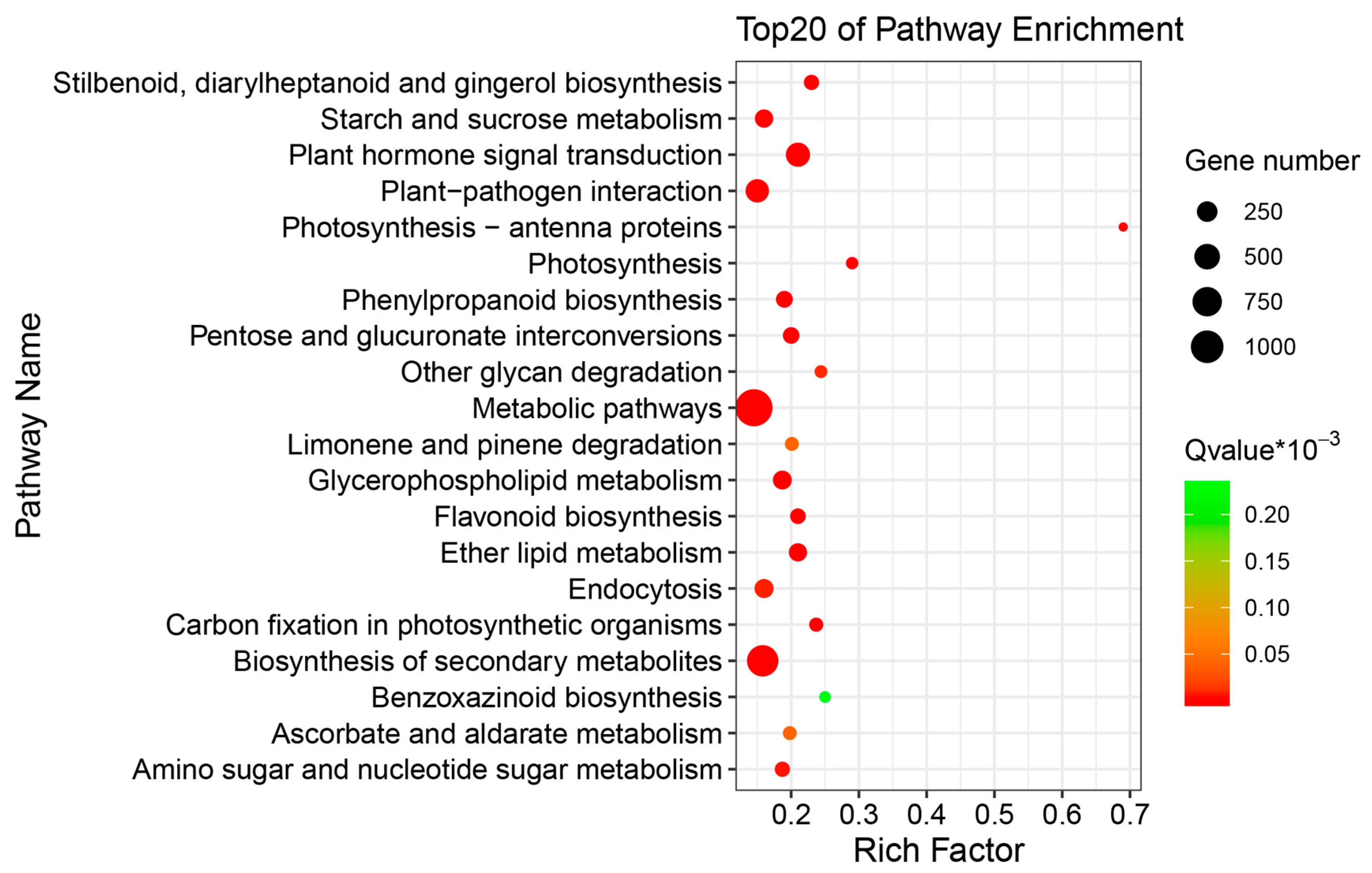
| Sample | Total Number | Total Length (nt) | Mean Length (nt) | N50 | Total Consensus Sequences | Distinct Clusters | Distinct Singletons | |
|---|---|---|---|---|---|---|---|---|
| Contigs | C1 | 97,017 | 48,783,474 | 503 | 1133 | - | - | - |
| C2 | 103,961 | 51,398,910 | 494 | 1104 | - | - | - | |
| C3 | 98,588 | 49,113,021 | 498 | 1130 | - | - | - | |
| T1 | 117,182 | 52,926,262 | 452 | 1007 | - | - | - | |
| T2 | 104,429 | 49,990,958 | 479 | 1081 | - | - | - | |
| T3 | 118,868 | 53,985,959 | 454 | 1001 | - | - | - | |
| Unigenes | C1 | 58,789 | 60,570,998 | 1030 | 1680 | 58,789 | 21,105 | 37,684 |
| C2 | 62,559 | 64,289,688 | 1028 | 1686 | 62,559 | 22,445 | 40,114 | |
| C3 | 58,644 | 60,081,202 | 1025 | 1668 | 58,644 | 20,637 | 38,007 | |
| T1 | 64,470 | 62,513,110 | 970 | 1639 | 64,470 | 21,291 | 43,179 | |
| T2 | 60,146 | 60,524,742 | 1006 | 1675 | 60,146 | 20,498 | 39,648 | |
| T3 | 65,749 | 63,160,040 | 961 | 1631 | 65,749 | 20,948 | 44,801 | |
| All | 94,950 | 107,581,109 | 1133 | 1900 | 94,950 | 38,242 | 56,708 |
| Samples | Catechin | Chlorogenic Acid | Caffeic Acid | p-Coumaric Acid | Benzoic Acid | Rutin | Quercetin | Kaempferol | |
|---|---|---|---|---|---|---|---|---|---|
| Con. | 1D | 12.04 ± 2.36a | 625.66 ± 28.28cd | 46.32 ± 0.03ab | 12.73 ± 2.72abc | 176.59 ± 8.75c | 12115.99 ± 132.05f | 54.84 ± 10.24c | 34.28 ± 4.42bc |
| 2D | 11.56 ± 0.72a | 596.23 ± 3.06c | 37.29 ± 6.07ab | 14.2 ± 2.06bc | 162.52 ± 8.19b | 10406.33 ± 41.03c | 36.55 ± 14.71bc | 32.74 ± 6.92ab | |
| 4D | 19.21 ± 1.99bc | 776.19 ± 26.62d | 49.46 ± 0.02b | 10.32 ± 0.72a | 144.29 ± 3.23a | 9506.46 ± 212.82a | 10.52 ± 4.88a | 39.45 ± 1.21bc | |
| 5D | 14.61 ± 0.07ab | 535.18 ± 18.28ab | 38.83 ± 1.13ab | 10.24 ± 0.27a | 148.43 ± 4.77a | 9906.37 ± 78.64b | 52.76 ± 6.32c | 26.81 ± 1.09a | |
| Tr. | 1D | 13.62 ± 5.58ab | 543 ± 31.06b | 48.32 ± 5.86b | 15.85 ± 2.44c | 200.01 ± 3.07d | 13599.51 ± 112.49g | 22.76 ± 4.03ab | 38.02 ± 4.34bc |
| 2D | 21.66 ± 2.00c | 516.64 ± 10.10ab | 44.26 ± 0.43ab | 10.61 ± 0.21ab | 184.06 ± 2.38c | 11743.61 ± 110.46e | 46.73 ± 19.37c | 32.67 ± 1.83ab | |
| 4D | 23.04 ± 3.32c | 639.35 ± 6.33c | 39.99±14.99ab | 11.6 ± 0.48ab | 146.85 ± 1.43a | 9975.95 ± 93.02b | 111.04 ± 11.74d | 41.14 ± 5.04c | |
| 5D | 13.47 ± 5.87ab | 503.38 ± 1.84a | 33.46 ± 9.32a | 13.07 ± 3.55abc | 167.23 ± 5.67b | 11370.11 ± 0.01d | 47.5 ± 15.36c | 37.12 ± 1.94bc | |
| Sample | Lutein | Zeaxanthin | 13Z-β-Carotene | α-Carotene | E-β-Carotene | 9Z-β-Carotene | |
|---|---|---|---|---|---|---|---|
| Con. | 1D | 244.38 ± 7.96a | 1.01 ± 0.25a | 38.58 ± 4.24a | 5.46 ± 0.46a | 193.78 ± 9.61b | 26.63 ± 1.88ab |
| 2D | 236.83 ± 7.12a | 1.16 ± 0.19a | 36.34 ± 1.91a | 4.32 ± 0.59a | 172.21 ± 12.27ab | 22.57 ± 0.73a | |
| 4D | 299.29 ± 20.97b | 2.68 ± 0.41d | 47.27 ± 2.02b | 4.88 ± 0.09a | 235.54 ± 25.51c | 29.11 ± 3.31b | |
| 5D | 225.37 ± 17.38a | 1.80 ± 0.15bc | 33.19 ± 1.53a | 4.61 ± 0.72a | 156.30 ± 14.33a | 21.86 ± 2.42a | |
| Tr. | 1D | 242.01 ± 14.83a | 1.42 ± 0.25abc | 38.46 ± 0.67a | 4.86 ± 0.29a | 179.58 ± 19.13ab | 25.89 ± 2.49ab |
| 2D | 253.28 ± 36.49a | 1.27 ± 0.13ab | 37.51 ± 5.69a | 5.11 ± 1.57a | 177.76 ± 25.53ab | 26.02 ± 6.26ab | |
| 4D | 249.16 ± 31.94a | 3.24 ± 0.55e | 37.24 ± 1.20a | 5.54 ± 0.19a | 164.47 ± 23.28ab | 29.88 ± 3.85b | |
| 5D | 241.95 ± 9.48a | 1.89 ± 0.22c | 34.61 ± 0.52a | 5.02 ± 0.06a | 157.60 ± 11.81a | 26.12 ± 2.43ab | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, W.; Kim, J.K.; Jia, C.; Yin, F.; Kim, H.J.; Akram, W.; Hu, X.; Li, X. Comparative Transcriptome and Metabolic Profiling Analysis of Buckwheat (Fagopyrum Tataricum (L.) Gaertn.) under Salinity Stress. Metabolites 2019, 9, 225. https://doi.org/10.3390/metabo9100225
Ma W, Kim JK, Jia C, Yin F, Kim HJ, Akram W, Hu X, Li X. Comparative Transcriptome and Metabolic Profiling Analysis of Buckwheat (Fagopyrum Tataricum (L.) Gaertn.) under Salinity Stress. Metabolites. 2019; 9(10):225. https://doi.org/10.3390/metabo9100225
Chicago/Turabian StyleMa, Weibo, Jae Kwang Kim, Caihua Jia, Feifan Yin, Hyo Jin Kim, Waheed Akram, Xuebo Hu, and Xiaohua Li. 2019. "Comparative Transcriptome and Metabolic Profiling Analysis of Buckwheat (Fagopyrum Tataricum (L.) Gaertn.) under Salinity Stress" Metabolites 9, no. 10: 225. https://doi.org/10.3390/metabo9100225
APA StyleMa, W., Kim, J. K., Jia, C., Yin, F., Kim, H. J., Akram, W., Hu, X., & Li, X. (2019). Comparative Transcriptome and Metabolic Profiling Analysis of Buckwheat (Fagopyrum Tataricum (L.) Gaertn.) under Salinity Stress. Metabolites, 9(10), 225. https://doi.org/10.3390/metabo9100225