Univariate Statistical Analysis as a Guide to 1H-NMR Spectra Signal Assignment by Visual Inspection
Abstract
1. Introduction
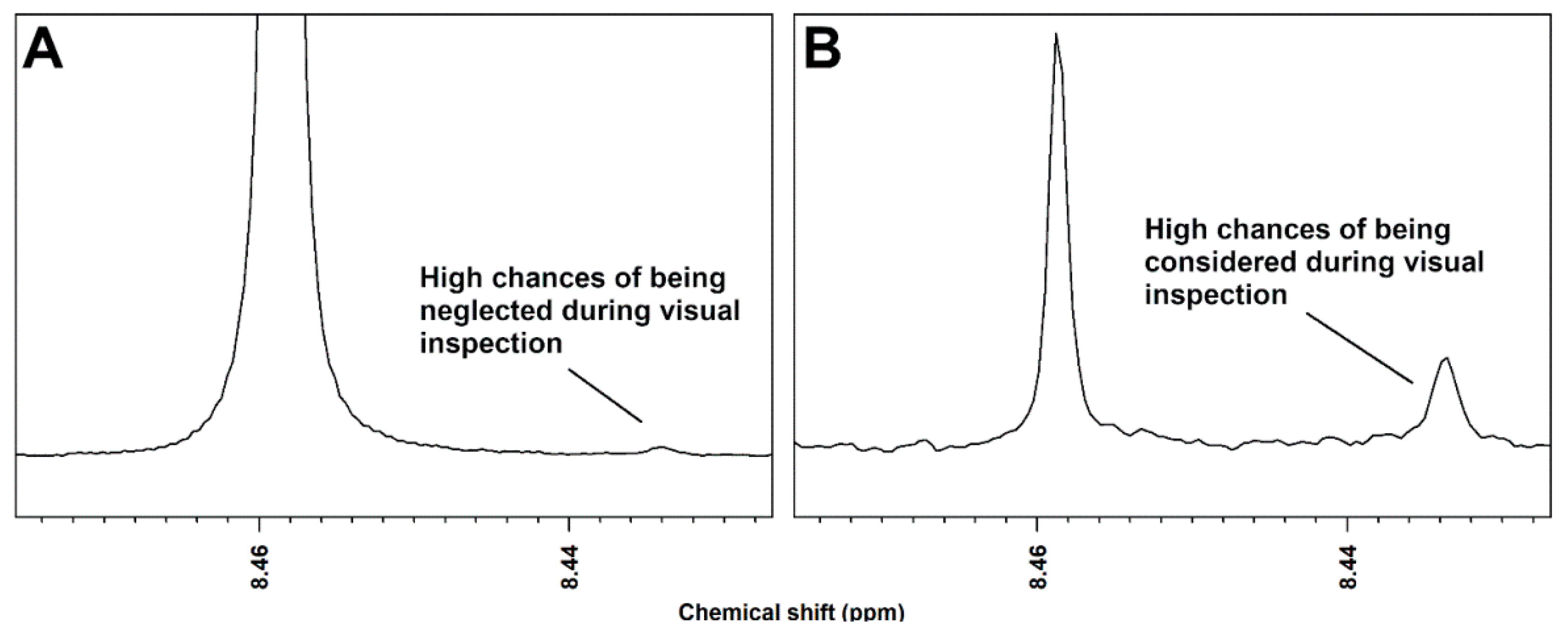
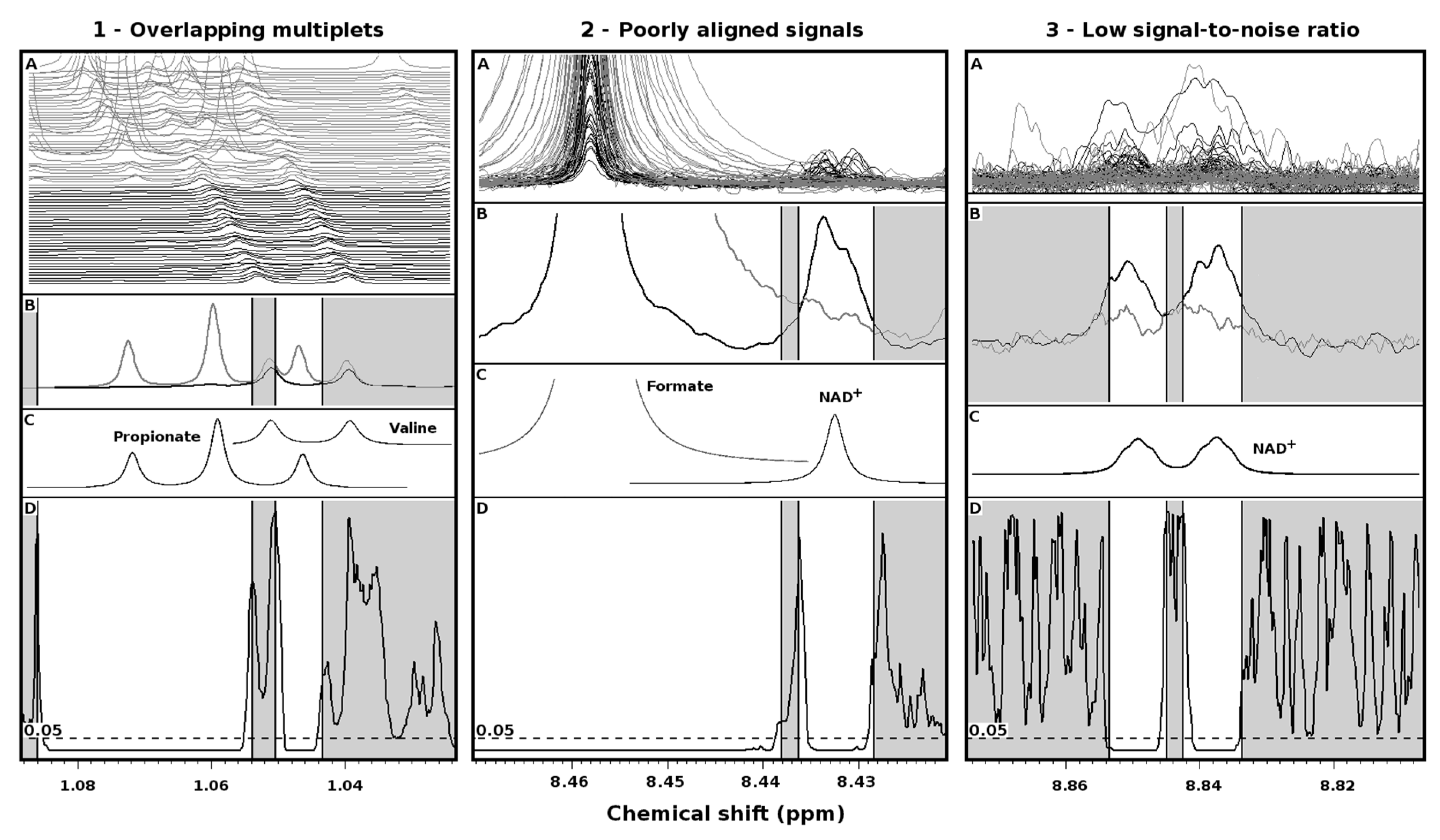
2. Results
3. Discussion
4. Materials and Methods
4.1. Samples, Spectra and Statistics
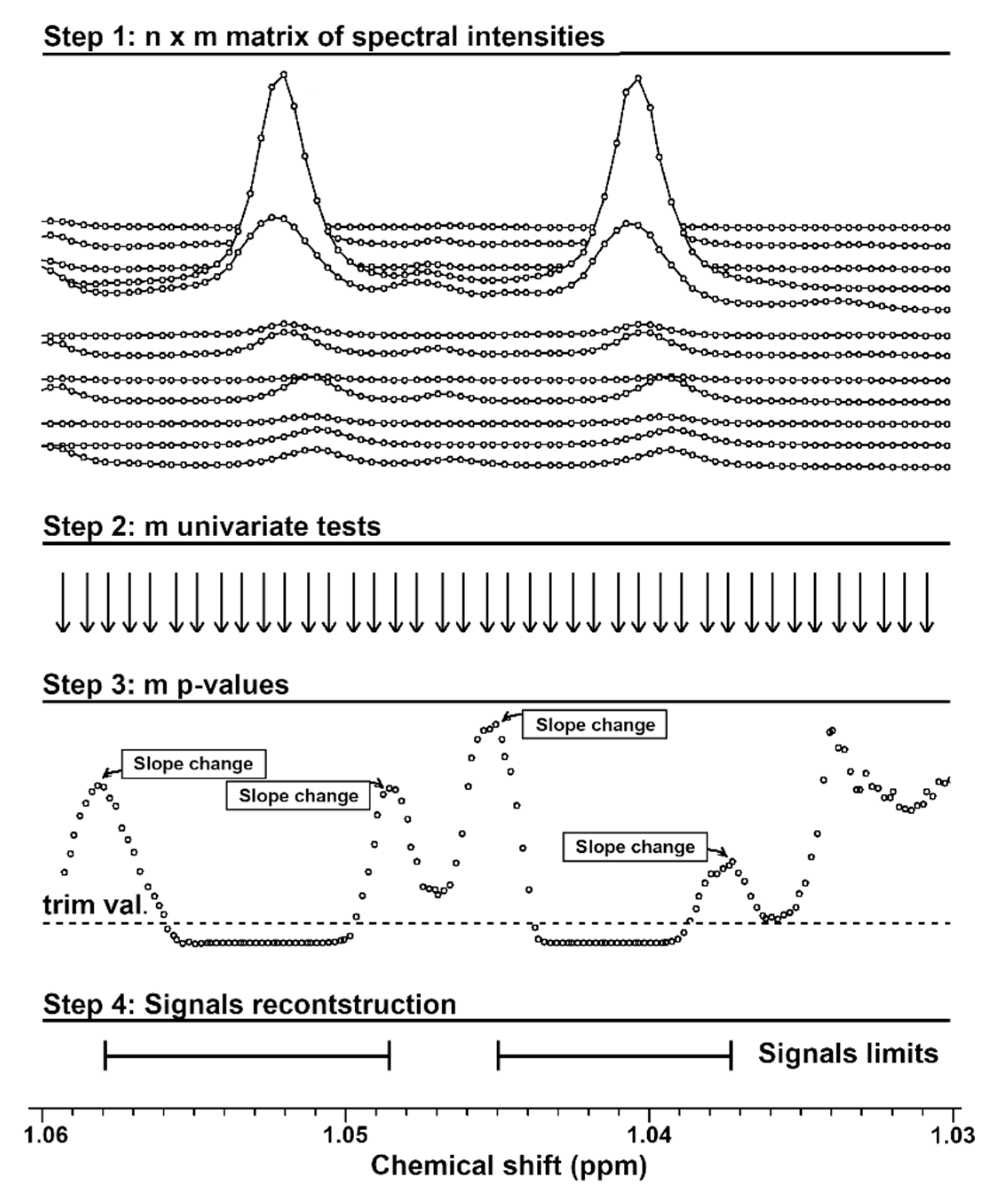
4.2. Rationale of the Procedure for Signals Reconstruction
4.3. A Hands-on Example
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Klassen, A.; Faccio, A.T.; Canuto, G.A.B.; da Cruz, P.L.R.; Ribeiro, H.C.; Tavares, M.F.M.; Sussulini, A. Metabolomics: Definitions and significance in systems biology. In Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2017; Volume 965, pp. 3–17. ISBN 1471-2415-1471-2415. [Google Scholar]
- Picone, G.; Laghi, L.; Gardini, F.; Lanciotti, R.; Siroli, L.; Capozzi, F. Evaluation of the effect of carvacrol on the Escherichia coli 555 metabolome by using 1H-NMR spectroscopy. Food Chem. 2013, 141, 4367–4374. [Google Scholar] [CrossRef] [PubMed]
- Bordoni, A.; Laghi, L.; Babini, E.; Di Nunzio, M.; Picone, G.; Ciampa, A.; Valli, V.; Danesi, F.; Capozzi, F. The foodomics approach for the evaluation of protein bioaccessibility in processed meat upon in vitro digestion. Electrophoresis 2014, 35, 1607–1614. [Google Scholar] [CrossRef] [PubMed]
- Marcolini, E.; Babini, E.; Bordoni, A.; Di Nunzio, M.; Laghi, L.; Maczó, A.; Picone, G.; Szerdahelyi, E.; Valli, V.; Capozzi, F. Bioaccessibility of the Bioactive Peptide Carnosine during in Vitro Digestion of Cured Beef Meat. J. Agric. Food Chem. 2015, 63, 4973–4978. [Google Scholar] [CrossRef] [PubMed]
- Larive, C.K.; Barding, G.A.; Dinges, M.M. NMR spectroscopy for metabolomics and metabolic profiling. Anal. Chem. 2015, 87, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.W.M. Metabolite profiling by one- and two-dimensional NMR analysis of complex mixtures. Prog. Nucl. Magn. Reson. Spectrosc. 1996, 28, 161–219. [Google Scholar] [CrossRef]
- Good, B.M.; Su, A.I. Games with a scientific purpose. Genome Biol. 2011, 12, 135. [Google Scholar] [CrossRef] [PubMed]
- Laghi, L.; Picone, G.; Cruciani, F.; Brigidi, P.; Calanni, F.; Donders, G.; Capozzi, F.; Vitali, B. Rifaximin modulates the vaginal microbiome and metabolome in women affected by bacterial vaginosis. Antimicrob. Agents Chemother. 2014, 58, 3411–3420. [Google Scholar] [CrossRef] [PubMed]
- Vitali, B.; Cruciani, F.; Picone, G.; Parolin, C.; Donders, G.; Laghi, L. Vaginal microbiome and metabolome highlight specific signatures of bacterial vaginosis. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 2367–2376. [Google Scholar] [CrossRef] [PubMed]
- Nardini, P.; Nãhui Palomino, R.A.; Parolin, C.; Laghi, L.; Foschi, C.; Cevenini, R.; Vitali, B.; Marangoni, A. Lactobacillus crispatus inhibits the infectivity of Chlamydia trachomatis elementary bodies, in vitro study. Sci. Rep. 2016, 6, 29024. [Google Scholar] [CrossRef] [PubMed]
- Foschi, C.; Laghi, L.; Parolin, C.; Giordani, B.; Compri, M.; Cevenini, R.; Marangoni, A.; Vitali, B. Novel approaches for the taxonomic and metabolic characterization of lactobacilli: Integration of 16S rRNA gene sequencing with MALDI-TOF MS and 1H-NMR. PLoS One 2017, 12, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Cruciani, F.; Brigidi, P.; Calanni, F.; Lauro, V.; Tacchi, R.; Donders, G.; Peters, K.; Guaschino, S.; Vitali, B. Efficacy of rifaximin vaginal tablets in treatment of bacterial vaginosis: A molecular characterization of the vaginal microbiota. Antimicrob. Agents Chemother. 2012, 56, 4062–4070. [Google Scholar] [CrossRef] [PubMed]
- Spratlin, J.L.; Serkova, N.J.; Eckhardt, S.G. Clinical applications of metabolomics in oncology: A review. Clin. Cancer Res. 2009, 15, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, D.M.; Deltimple, N.; van Velzen, E.; van Dorsten, F.A.; Bingham, M.; Vaughan, E.E.; van Duynhoven, J. 1H NMR metabolite profiling of feces as a tool to assess the impact of nutrition on the human microbiome. NMR Biomed. 2008, 21, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Tankou, S.K.; Regev, K.; Healy, B.C.; Tjon, E.; Laghi, L.; Cox, L.M.; Kivisäkk, P.; Pierre, I.V.; Hrishikesh, L.; Gandhi, R.; et al. A probiotic modulates the microbiome and immunity in multiple sclerosis. Ann. Neurol. 2018, 83, 1147–1161. [Google Scholar] [CrossRef] [PubMed]
- Emwas, A.H.; Saccenti, E.; Gao, X.; McKay, R.T.; dos Santos, V.A.P.M.; Roy, R.; Wishart, D.S. Recommended strategies for spectral processing and post-processing of 1D 1H-NMR data of biofluids with a particular focus on urine. Metabolomics 2018, 14, 31. [Google Scholar] [CrossRef] [PubMed]
- Nobeli, I.; Ponstingl, H.; Krissinel, E.B.; Thornton, J.M. A structure-based anatomy of the E. coli metabolome. J. Mol. Biol. 2003, 334, 697–719. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, C.; Vitali, B.; Donders, G.; Parolin, C.; Li, Y.; Laghi, L. Univariate Statistical Analysis as a Guide to 1H-NMR Spectra Signal Assignment by Visual Inspection. Metabolites 2019, 9, 15. https://doi.org/10.3390/metabo9010015
Zhu C, Vitali B, Donders G, Parolin C, Li Y, Laghi L. Univariate Statistical Analysis as a Guide to 1H-NMR Spectra Signal Assignment by Visual Inspection. Metabolites. 2019; 9(1):15. https://doi.org/10.3390/metabo9010015
Chicago/Turabian StyleZhu, Chenglin, Beatrice Vitali, Gilbert Donders, Carola Parolin, Yan Li, and Luca Laghi. 2019. "Univariate Statistical Analysis as a Guide to 1H-NMR Spectra Signal Assignment by Visual Inspection" Metabolites 9, no. 1: 15. https://doi.org/10.3390/metabo9010015
APA StyleZhu, C., Vitali, B., Donders, G., Parolin, C., Li, Y., & Laghi, L. (2019). Univariate Statistical Analysis as a Guide to 1H-NMR Spectra Signal Assignment by Visual Inspection. Metabolites, 9(1), 15. https://doi.org/10.3390/metabo9010015







