1D “Spikelet” Projections from Heteronuclear 2D NMR Data—Permitting 1D Chemometrics While Preserving 2D Dispersion
Abstract
1. Introduction
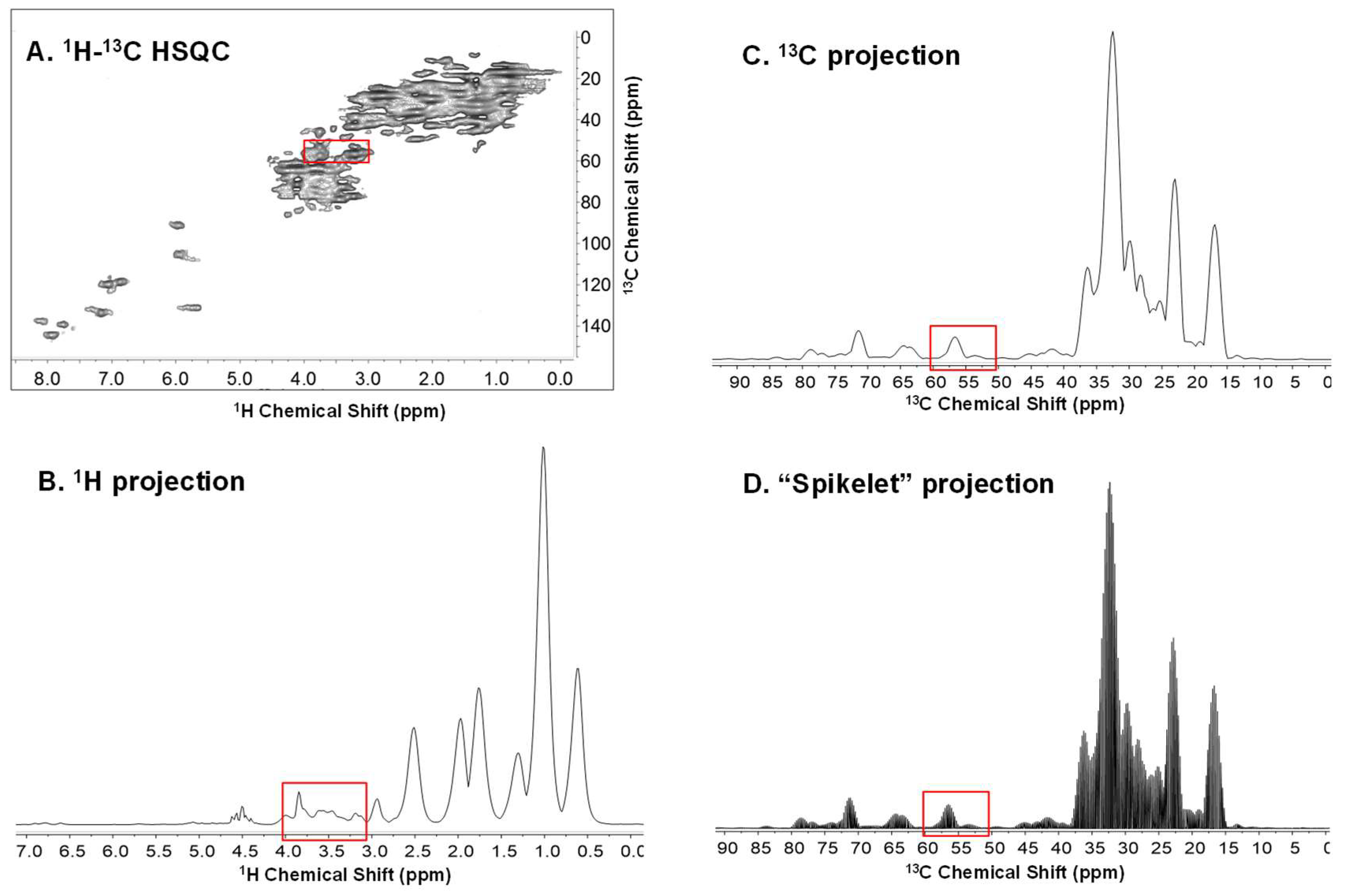
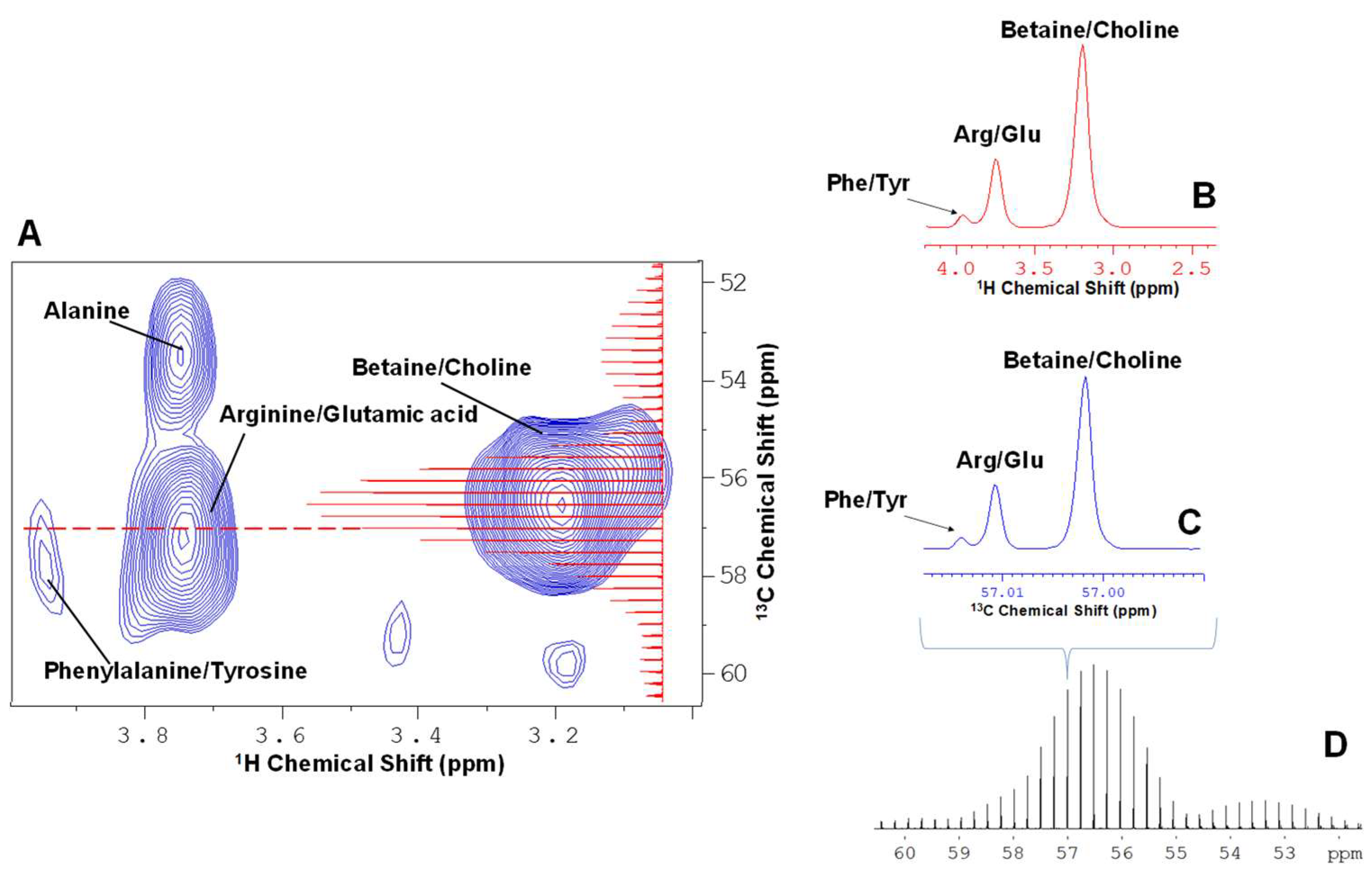
2. Results and Discussion
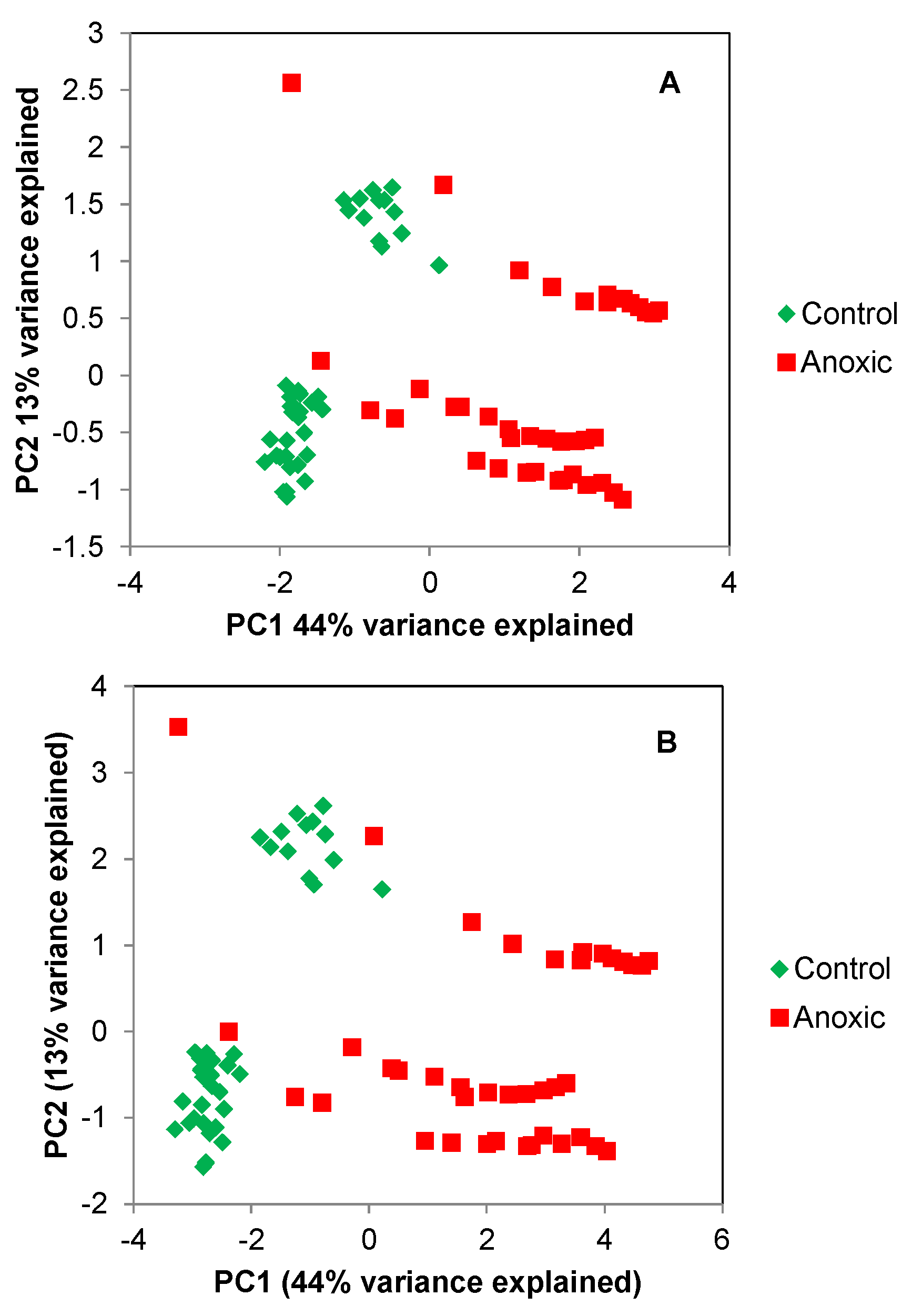
2.1. Principle Component Analysis (PCA) of the Spikelet Projections
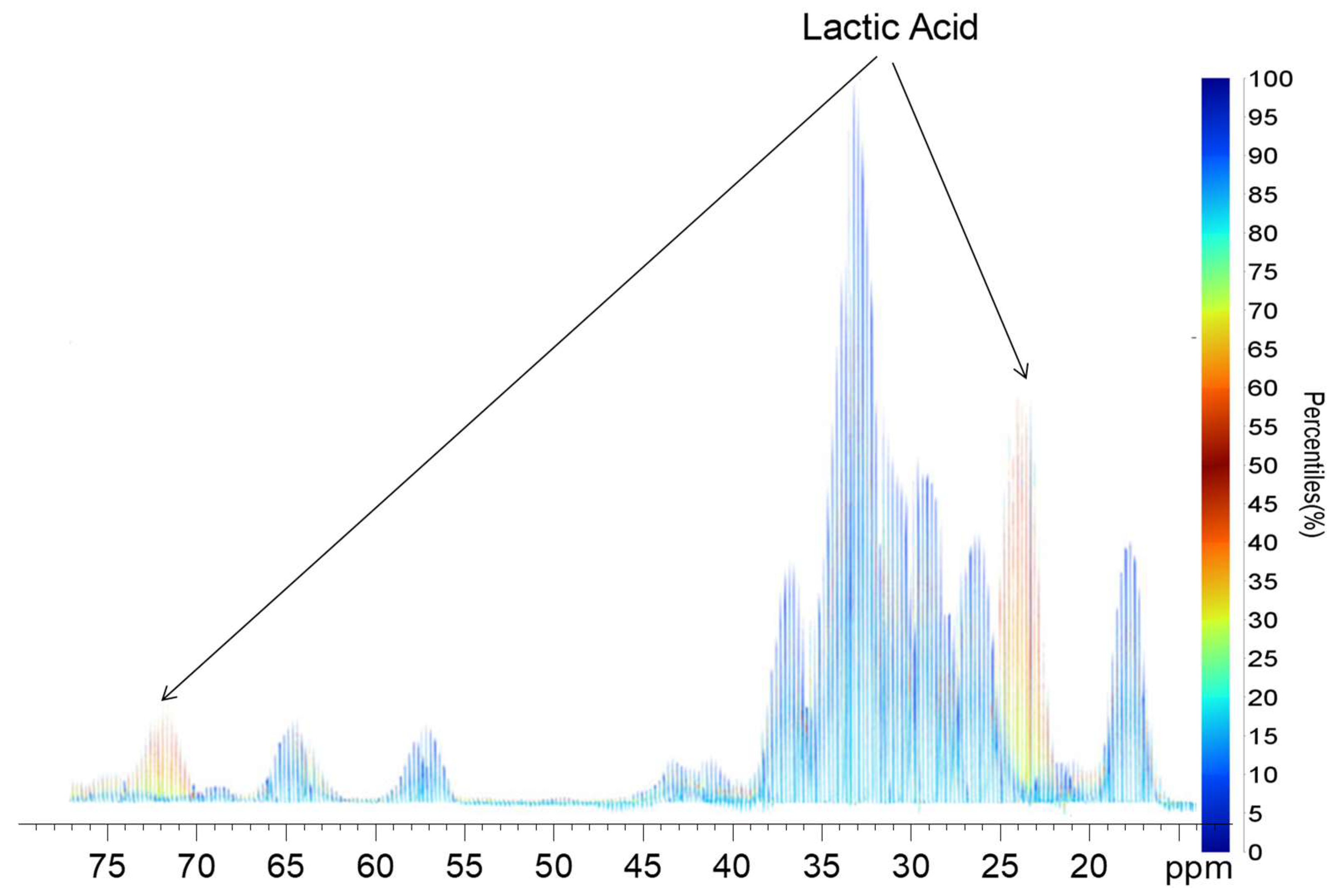
2.2. Quantile Plot from the Spikelet Projections
3. Materials and Methods
3.1. Daphnia and Algae Culturing
3.2. NMR Spectroscopy
3.3. In Vivo NMR Spectroscopy
3.4. Data and Statistical Analysis
4. Conclusions
Funding
Conflicts of Interest
Appendix A
References
- Hertkorn, N.; Ruecker, C.; Meringer, M.; Gugisch, R.; Frommberger, M.; Perdue, E.M.; Witt, M.; Schmitt-Kopplin, P. High-precision frequency measurements: Indispensable tools at the core of the molecular-level analysis of complex systems. Anal. Bioanal. Chem. 2007, 389, 1311–1327. [Google Scholar] [CrossRef] [PubMed]
- Lewis, I.A.; Schommer, S.C.; Hodis, B.; Robb, K.A.; Tonelli, M.; Westler, W.M.; Sussman, M.R.; Markley, J.L. Method for determining molar concentrations of metabolites in complex solutions from two-dimensional1H-13C NMR spectra. Anal. Chem. 2007, 79, 9385–9390. [Google Scholar] [CrossRef] [PubMed]
- Bollard, M.E.; Garrod, S.; Holmes, E.; Lindon, J.C.; Humpfer, E.; Spraul, M.; Nicholson, J.K. High-resolution 1H and 1H-13C magic angle spinning NMR spectroscopy of rat liver. Magn. Reson. Med. 2000. [Google Scholar] [CrossRef]
- An, Y.J.; Xu, W.J.; Jin, X.; Wen, H.; Kim, H.; Lee, J.; Park, S. Metabotyping of the C. elegans sir-2.1 mutant using in vivo labeling and13C-heteronuclear multidimensional NMR metabolomics. ACS Chem. Biol. 2012, 7, 2012–2018. [Google Scholar] [CrossRef] [PubMed]
- Dumas, M.E.; Canlet, C.; Andŕ, F.; Vercauteren, J.; Paris, A. Metabonomic assessment of physiological disruptions using1H-13C HMBC-NMR spectroscopy combined with pattern recognition procedures performed on filtered variables. Anal. Chem. 2002. [Google Scholar] [CrossRef]
- Yuk, J.; Simpson, M.J.; Simpson, A.J. 1-D and 2-D NMR-based metabolomics of earthworms exposed to endosulfan and endosulfan sulfate in soil. Environ. Pollut. 2013, 175, 35–44. [Google Scholar] [CrossRef]
- Yuk, J.; Simpson, M.J.; Simpson, A.J. 1-D and 2-D NMR metabolomics of earthworm responses to sub-lethal trifluralin and endosulfan exposure. Environ. Chem. 2011. [Google Scholar] [CrossRef]
- Yuk, J.; McKelvie, J.R.; Simpson, M.J.; Spraul, M.; Simpson, A.J. Comparison of 1-D and 2-D NMR techniques for screening earthworm responses to sub-lethal endosulfan exposure. Environ. Chem. 2010, 7, 524–536. [Google Scholar] [CrossRef]
- Creek, D.J.; Chokkathukalam, A.; Jankevics, A.; Burgess, K.E.V.; Breitling, R.; Barrett, M.P. Stable isotope-assisted metabolomics for network-wide metabolic pathway elucidation. Anal. Chem. 2012, 84, 8442–8447. [Google Scholar] [CrossRef]
- Clendinen, C.S.; Stupp, G.S.; Ajredini, R.; Lee-McMullen, B.; Beecher, C.; Edison, A.S. An overview of methods using 13C for improved compound identification in metabolomics and natural products. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef]
- Clendinen, C.S.; Stupp, G.S.; Wang, B.; Garrett, T.J.; Edison, A.S. 13C Metabolomics: NMR and IROA for Unknown Identification. Curr. Metab. 2016, 4, 116–120. [Google Scholar] [CrossRef]
- Chokkathukalam, A.; Kim, D.H.; Barrett, M.P.; Breitling, R.; Creek, D.J. Stable isotope-labeling studies in metabolomics: New insights into structure and dynamics of metabolic networks. Bioanalysis 2014, 6, 511–524. [Google Scholar] [CrossRef]
- Tabatabaei Anaraki, M.; Dutta Majumdar, R.; Wagner, N.; Soong, R.; Kovacevic, V.; Reiner, E.J.; Bhavsar, S.P.; Ortiz Almirall, X.; Lane, D.; Simpson, M.J.; et al. Development and Application of a Low-Volume Flow System for Solution-State in Vivo NMR. Anal. Chem. 2018, 90, 7912–7921. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, R.D.; Akhter, M.; Fortier-McGill, B.; Soong, R.; Liaghati-Mobarhan, Y.; Simpson, A.J.; Spraul, M.; Schmidt, S.; Heumann, H. In vivo solution-state NMR-based environmental metabolomics. eMagRes 2017, 6, 133–148. [Google Scholar] [CrossRef]
- Yang, C.; Harrison, C.; Jin, E.S.; Chuang, D.T.; Sherry, A.D.; Malloy, C.R.; Merritt, M.E.; DeBerardinis, R.J. Simultaneous steady-state and dynamic13C NMR can differentiate alternative routes of pyruvate metabolism in living cancer cells. J. Biol. Chem. 2014, 289, 6212–6224. [Google Scholar] [CrossRef] [PubMed]
- Féraud, B.; Govaerts, B.; Verleysen, M.; de Tullio, P. Statistical treatment of 2D NMR COSY spectra in metabolomics: Data preparation, clustering-based evaluation of the Metabolomic Informative Content and comparison with1H-NMR. Metabolomics 2015, 11, 1756–1768. [Google Scholar] [CrossRef]
- Ludwig, C.; Viant, M.R. Two-dimensional J-resolved NMR spectroscopy: Review of a key methodology in the metabolomics toolbox. Phytochem. Anal. 2010, 21, 22–32. [Google Scholar] [CrossRef]
- Arbogast, L.W.; Delaglio, F.; Schiel, J.E.; Marino, J.P. Multivariate Analysis of Two-Dimensional 1 H, 13 C Methyl NMR Spectra of Monoclonal Antibody Therapeutics to Facilitate Assessment of Higher Order Structure. Anal. Chem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Worley, B.; Powers, R. Generalized adaptive intelligent binning of multiway data. Chemom. Intell. Lab. Syst. 2015, 146, 42–46. [Google Scholar] [CrossRef]
- Tabatabaei Anaraki, M.; Simpson, M.J.; Simpson, A.J. Reducing impacts of organism variability in metabolomics via time trajectory in vivo NMR. Magn. Reson. Chem. 2018, 56, 1117–1123. [Google Scholar] [CrossRef]
- Soong, R.; Nagato, E.; Sutrisno, A.; Fortier-Mcgill, B.; Akhter, M.; Schmidt, S.; Heumann, H.; Simpson, A.J. In vivo NMR spectroscopy: Toward real time monitoring of environmental stress. Magn. Reson. Chem. 2015, 53, 774–779. [Google Scholar] [CrossRef] [PubMed]
- Barjat, H.; Morris, G.A.; Swanson, A.G. A Three-Dimensional DOSY-HMQC Experiment for the High-Resolution Analysis of Complex Mixtures. J. Magn. Reson. 1998. [Google Scholar] [CrossRef]
- Peng, J.W.; Wagner, G. Investigation of protein motions via relaxation measurements. Methods Enzymol. 1994. [Google Scholar] [CrossRef]
- Schober, D.; Jacob, D.; Wilson, M.; Cruz, J.A.; Marcu, A.; Grant, J.R.; Moing, A.; Deborde, C.; De Figueiredo, L.F.; Haug, K.; et al. NmrML: A Community Supported Open Data Standard for the Description, Storage, and Exchange of NMR Data. Anal. Chem. 2018, 90, 649–656. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tabatabaei Anaraki, M.; Bermel, W.; Dutta Majumdar, R.; Soong, R.; Simpson, M.; Monnette, M.; Simpson, A.J. 1D “Spikelet” Projections from Heteronuclear 2D NMR Data—Permitting 1D Chemometrics While Preserving 2D Dispersion. Metabolites 2019, 9, 16. https://doi.org/10.3390/metabo9010016
Tabatabaei Anaraki M, Bermel W, Dutta Majumdar R, Soong R, Simpson M, Monnette M, Simpson AJ. 1D “Spikelet” Projections from Heteronuclear 2D NMR Data—Permitting 1D Chemometrics While Preserving 2D Dispersion. Metabolites. 2019; 9(1):16. https://doi.org/10.3390/metabo9010016
Chicago/Turabian StyleTabatabaei Anaraki, Maryam, Wolfgang Bermel, Rudraksha Dutta Majumdar, Ronald Soong, Myrna Simpson, Martine Monnette, and André J. Simpson. 2019. "1D “Spikelet” Projections from Heteronuclear 2D NMR Data—Permitting 1D Chemometrics While Preserving 2D Dispersion" Metabolites 9, no. 1: 16. https://doi.org/10.3390/metabo9010016
APA StyleTabatabaei Anaraki, M., Bermel, W., Dutta Majumdar, R., Soong, R., Simpson, M., Monnette, M., & Simpson, A. J. (2019). 1D “Spikelet” Projections from Heteronuclear 2D NMR Data—Permitting 1D Chemometrics While Preserving 2D Dispersion. Metabolites, 9(1), 16. https://doi.org/10.3390/metabo9010016




