An Improved Genome-Scale Metabolic Model of Arthrospira platensis C1 (iAK888) and Its Application in Glycogen Overproduction
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome-Scale Metabolic Network Reconstruction
2.2. Flux Balance Analysis
2.3. Estimation of Glycogen Production Flux
2.4. Model Validation
2.5. Simulation of Glycogen Production Under Nutrient-Limited Conditions
2.6. Experimental Validation
3. Results and Discussion
3.1. Reconstruction of the Updated Genome-Scale Metabolic Network of A. platensis C1
3.2. Characteristics of iAK888 and Comparison
3.3. Validation of iAK888
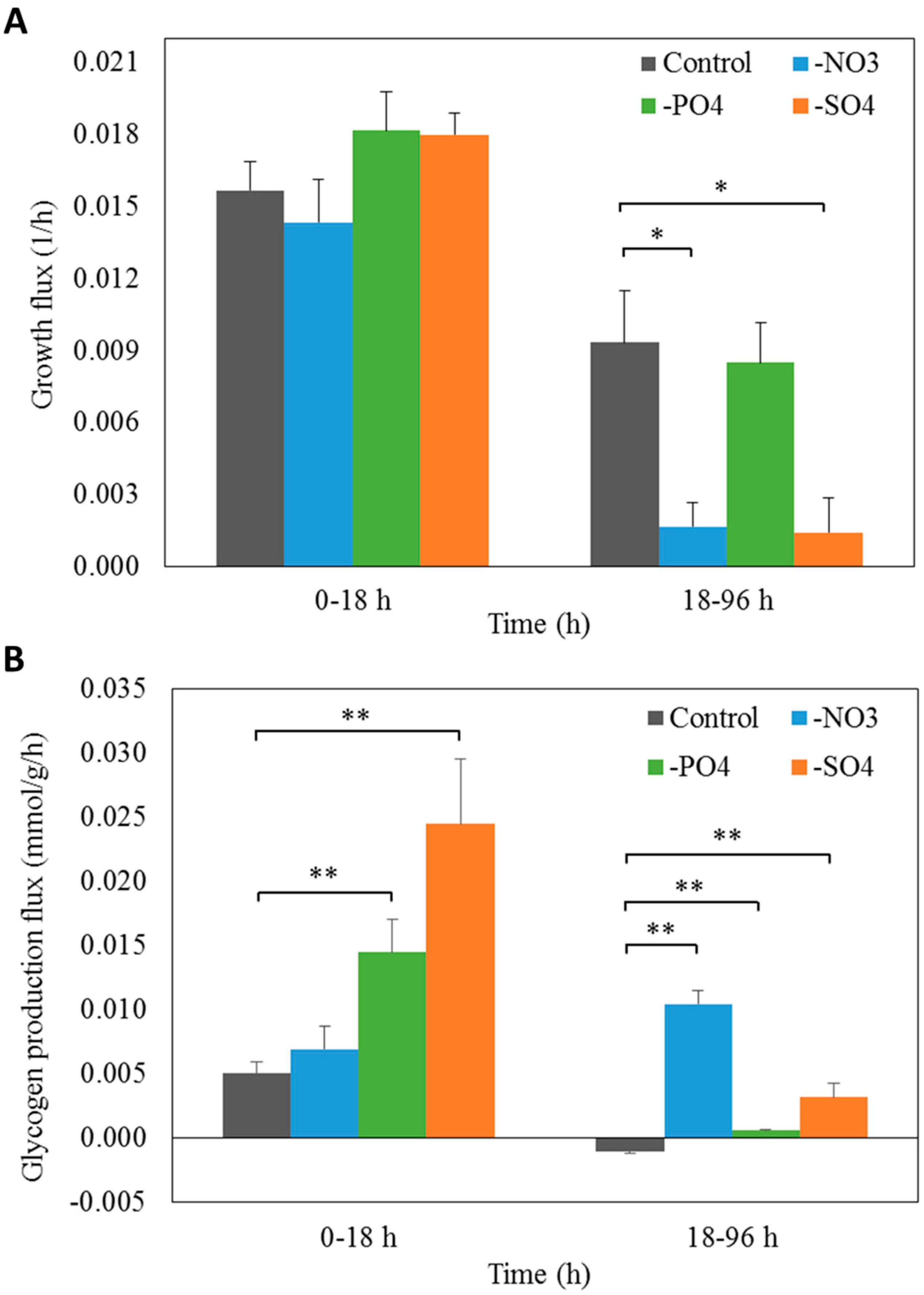
3.4. Prediction of Glycogen Overproduction Using iAK888
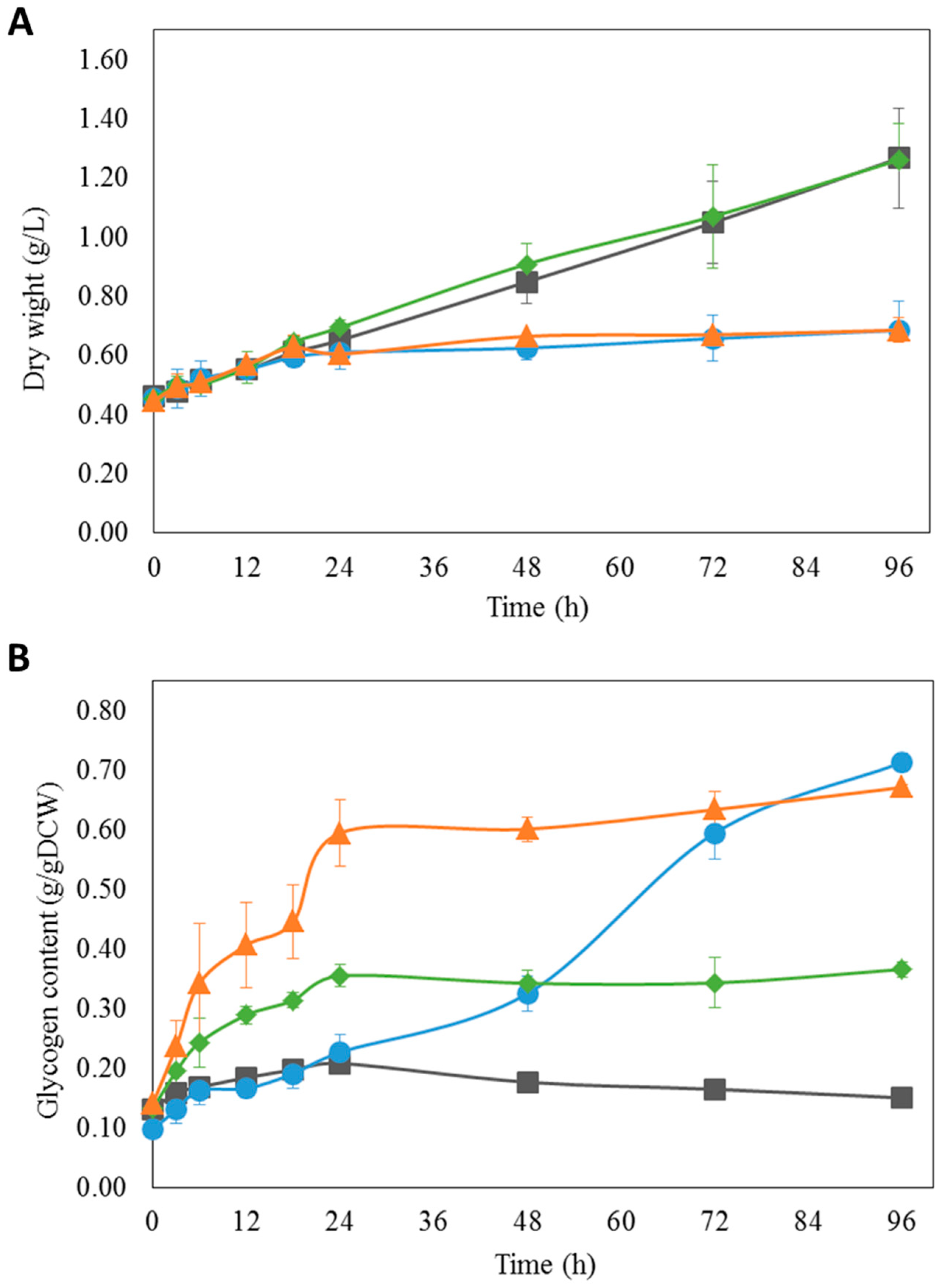
3.5. Experimental Validation of iAK888 Prediction for Glycogen Overproduction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Markou, G.; Angelidaki, I.; Georgakakis, D. Microalgal carbohydrates: An overview of the factors influencing carbohydrates production, and of main bioconversion technologies for production of biofuels. Appl. Microbiol. Biotechnol. 2012, 96, 631–645. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.-H.; Huang, S.-W.; Chen, C.-Y.; Hasunuma, T.; Chang, J.-S. Bioethanol production using carbohydrate-rich microalgae biomass as feedstock. Bioresour. Technol. 2013, 135, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Aikawa, S.; Joseph, A.; Yamad, R.; Izumi, Y.; Yamagishi, T.; Matsuda, F.; Kawai, H.; Chang, J.-S.; Hasunuma, T.; Kondo, A. Direct conversion of Spirulina to ethanol without pretreatment or enzymatic hydrolysis processes. Energy Environ. Sci. 2013, 6, 1844–1849. [Google Scholar] [CrossRef]
- Möllers, K.B.; Cannella, D.; Jørgensen, H.; Frigaard, N.-U. Cyanobacterial biomass as carbohydrate and nutrient feedstock for bioethanol production by yeast fermentation. Biotechnol. Biofuels 2014, 7, 64. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.X.; Zhao, H.; Li, Z.M.; Tan, X.M.; Lu, X. F Photosynthetic production of ethanol from carbon dioxide in genetically engineered cyanobacteria. Energy Environ. Sci. 2012, 5, 9857–9865. [Google Scholar] [CrossRef]
- Wijffels, R.H.; Kruse, O.; Hellingwerf, K.J. Potential of industrial biotechnology with cyanobacteria and eukaryotic microalgae. Curr. Opin. Biotechnol. 2013, 24, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Taton, A.; Unglaub, F.; Wright, N.E.; Zeng, W.Y.; Paz-Yepes, J.; Brahamsha, B.; Palenik, B.; Peterson, T.C.; Haerizadeh, F.; Golden, S.S.; et al. Broad-host-range vector system for synthetic biology and biotechnology in cyanobacteria. Nucleic Acids Res. 2014, 42, e136. [Google Scholar] [CrossRef] [PubMed]
- Aikawa, S.; Izumi, Y.; Matsuda, F.; Hasunuma, T.; Chang, J.S.; Kondo, A. Synergistic Enhancement of Glycogen Production in Arthrospira platensis by Optimization of Light Intensity and Nitrate Supply. Bioresour. Technol. 2012, 108, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Vonshak, A. Spirulina platensis (Arthrospira): Physiology, Cell Biology and Biotechnology. J. Appl. Psychol. 1997, 9, 295–296. [Google Scholar]
- Markou, G.; Chatzipavlidis, I.; Georgakakis, D. Carbohydrates production and bio-flocculation characteristics in cultures of Arthrospira Spirulina platensis: Improvements through phosphorus limitation process. BioEnergy Res. 2012, 5, 915–925. [Google Scholar] [CrossRef]
- Cheevadhanarak, S.; Paithoonrangsarid, K.; Prommeenate, P.; Kaewngam, W.; Musigkain, A.; Tragoonrung, S.; Tabata, S.; Kaneko, T.; Chaijaruwanich, J.; Sangsrakru, D.; et al. Draft Genome Sequence of Arthrospira platensis C1 (PCC 9438). Stand. Genom. Sci. 2012, 6, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Jeamton, W.; Dulsawat, S.; Tanticharoen, M.; Vonshak, A.; Cheevadhanarak, S. Overcoming Intrinsic Restriction Enzyme Barriers Enhances Transformation Efficiency in Arthrospira platensis C1. Plant Cell Physiol. 2017, 58, 822–830. [Google Scholar] [CrossRef] [PubMed]
- Jeamton, W.; Mungpakdee, S.; Sirijuntarut, M.; Prommeenate, P.; Cheevadhanarak, S.; Tanticharoen, M.; Apiradee Hongsthong, A. A combined stress response analysis of Spirulina platensis in terms of global differentially expressed proteins, and mRNA levels and stability of fatty acid biosynthesis genes. FEMS Microbiol. Lett. 2008, 281, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Panyakampol, J.; Cheevadhanarak, S.; Sutheeworapong, S.; Chaijaruwanich, J.; Senachak, J.; Siangdung, W.; Jeamton, W.; Tanticharoen, M.; Paithoonrangsarid, K. Physiological and transcriptional responses to high temperature in Arthrospira (Spirulina) platensis C1. Plant Cell Physiol. 2014, 56, 481–496. [Google Scholar] [CrossRef] [PubMed]
- Hongsthong, A.; Sirijuntarut, M.; Prommeenate, P.; Lertladaluck, K.; Porkaew, K.; Cheevadhanarak, S.; Tanticharoen, M. Proteome analysis at the subcellular level of the cyanobacterium Spirulina platensis in response to low-temperature stress conditions. FEMS Microbiol. Lett. 2008, 288, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Hongsthong, A.; Sirijuntarut, M.; Yutthanasirikul, R.; Senachak, J.; Kurdrid, P.; Cheevadhanarak, S.; Tanticharoen, M. Subcellular proteomic characterization of the high-temperature stress response of the cyanobacterium Spirulina platensis. Proteome Sci. 2009, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Kurdrid, P.; Senachak, J.; Sirijuntarut, M.; Yutthanasirikul, R.; Phuengcharoen, P.; Jeamton, W.; Roytrakul, S.; Cheevadhanarak, S.; Hongsthong, A. Comparative analysis of the Spirulina platensis subcellular proteome in response to low- and high-temperature stresses: Uncovering cross-talk of signaling components. Proteome Sci. 2011, 9, 39. [Google Scholar] [CrossRef] [PubMed]
- Senachak, J.; Cheevadhanarak, S.; Hongsthong, A. SpirPro: A Spirulina proteome database and web-based tools for the analysis of protein-protein interactions at the metabolic level in Spirulina (Arthrospira) platensis C1. BMC Bioinform. 2015, 16, 233. [Google Scholar] [CrossRef] [PubMed]
- Likic, V.A.; McConville, M.J.; Lithgow, T.; Bacic, A. Systems Biology: The Next Frontier for Bioinformatics. Adv. Bioinform. 2010, 2010, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Monk, J.; Nogales, J.; Palsson, B.O. Optimizing genome-scale network reconstructions. Nat. Biotechnol. 2014, 32, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.J.; Kim, H.U.; Lee, S.Y. Current State and Applications of Microbial Genome-scale Metabolic Models. Curr. Opin. Syst. Biol. 2017, 2, 10–18. [Google Scholar] [CrossRef]
- Jeffrey, D.O.; Thiele, I.; Palsson, B.Ø. What Is Flux Balance Analysis? Nat. Biotechnol. 2010, 28, 245–248. [Google Scholar]
- Klanchui, A.; Khannapho, C.; Phodee, A.; Cheevadhanarak, S.; Meechai, A. iAK692: A genome-scale metabolic model of Spirulina platensis C1. BMC Syst. Boil. 2012, 6, 71. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef] [PubMed]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An Automatic Genome Annotation and Pathway Reconstruction Server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef] [PubMed]
- McGinnis, S.; Madden, T.L. Blast: At the Core of a Powerful and Diverse Set of Sequence Analysis Tools. Nucleic Acids Res. 2004, 32, W20–W25. [Google Scholar] [CrossRef] [PubMed]
- Saier, M.H., Jr.; Reddy, V.S.; Tsu, B.V.; Ahmed, M.S.; Li, C.; Moreno-Hagelsieb, G. The Transporter Classification Database (TCDB): Recent Advances. Nucleic Acids Res. 2016, 44, D372–D379. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Billington, R.; Ferrer, L.; Foerster, H.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Mueller, L.A.; et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016, 44, D471–D480. [Google Scholar] [CrossRef] [PubMed]
- Steinhauser, D.; Fernie, A.R.; Araujo, W.L. Unusual cyanobacterial TCA cycles: Not broken just different. Trends Plant Sci. 2012, 17, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Klanchui, A.; Cheevadhanarak, S.; Prommeenate, P.; Meechai, A. Exploring Components of the CO2-Concentrating Mechanism in Alkaliphilic Cyanobacteria through Genome-Based Analysis. Comput. Struct. Biotechnol. 2017, 15, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Satora, P.; Barwińska-Sendra, A.; Duda-Chodak, A.; Wajda, Ł. Strain-dependent production of selected bioactive compounds by Cyanobacteria belonging to the Arthrospira genus. J. Appl. Microbiol. 2015, 119, 736–743. [Google Scholar] [CrossRef] [PubMed]
- Badri, H.; Monsieurs, P.; Coninx, I.; Nauts, R.; Wattiez, R.; Leys, N. Temporal Gene Expression of the Cyanobacterium Arthrospira in Response to Gamma Rays. PLoS ONE 2015, 10, e0135565. [Google Scholar] [CrossRef] [PubMed]
- Knoop, H.; Grundel, M.; Zilliges, Y.; Lehmann, R.; Hoffmann, S.; Lockau, W.; Steuer, R. Flux balance analysis of cyanobacterial metabolism: The metabolic network of Synechocystis sp. PCC 6803. PLoS Comput. Biol. 2013, 9, e1003081. [Google Scholar] [CrossRef] [PubMed]
- Orth, J.D.; Conrad, T.M.; Na, J.; Lerman, J.A.; Nam, H.; Feist, A.M.; Palsson, B.O. A comprehensive genome-scale reconstruction of Escherichia coli metabolism—2011. Mol. Syst Biol. 2011, 7, 535. [Google Scholar] [CrossRef] [PubMed]
- Thiele, I.; Palsson, B.O. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 2010, 5, 93–121. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. The Pfam protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Panyakampol, J. (King Mongkut’s University of Technology Thonburi, Bangkok, Thailand). Unpublished work. 2015.
- Cogne, G.; Gros, J.B.; Dussap, C.G. Identification of a metabolic network structure representative of Arthrospira (spirulina) platensis metabolism. Biotechnol. Bioeng. 2003, 84, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Paithoonrangsarid, K. (King Mongkut’s University of Technology Thonburi, Bangkok, Thailand). Unpublished work. 1997.
- Abd El-Baky, H.H.; El Baz, F.K.; El-Baroty, G.S. Spirulina Species as a Source of Carotenoids and α-tocopherol and its Anticarcinoma Factors. Biotechnology 2003, 2, 222–240. [Google Scholar]
- Shimamatsu, H. Mass production of Spirulina, an edible microalga. Hydrobiologia 2004, 512, 39–44. [Google Scholar] [CrossRef]
- Ali, S.K.; Saleh, A.M. Spirulina—An Overview. Int. J. Pharm. Pharm. Sci. 2012, 4, 9–15. [Google Scholar]
- Ataman, M.; Hatzimanikatis, V. Heading in the right direction: Thermodynamics-based network analysis and pathway engineering. Curr. Opin. Biotechnol. 2015, 36, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Schellenberger, J.; Que, R.; Fleming, R.M.; Thiele, I.; Orth, J.D.; Feist, A.M.; Zielinski, D.C.; Bordbar, A.; Lewis, N.E.; Rahmanian, S.; et al. Quantitative Prediction of Cellular Metabolism with Constraint-based Models: The COBRA Toolbox V2.0. Nat. Protoc. 2011, 6, 1290–1307. [Google Scholar] [CrossRef] [PubMed]
- Phodee, A. (King Mongkut’s University of Technology Thonburi, Bangkok, Thailand). Unpublished work. 2009.
- Panyakampol, J.; Cheevadhanarak, S.; Senachak, J.; Dulsawat, S.; Siangdung, W.; Tanticharoen, M.; Paithoonrangsarid, K. Different Effects of the Combined Stress of Nitrogen Depletion and High Temperature Than an Individual Stress on the Synthesis of Biochemical Compounds in Arthrospira platensis C1 (PCC 9438). J. Appl. Phycol. 2016, 28, 2177–2186. [Google Scholar] [CrossRef]
- Zarrouk, C. Contribution à l’étude d’une cyanophycéeInfluence de divers’ facteurs physiques et chimiques sur la croissance et la photosynthèse de Spirulina maxima. PhD Thesis, Université de Paris, Paris, France, 1996. (In French). [Google Scholar]
- Mahadevan, R.; Schilling, C.H. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab. Eng. 2003, 5, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Smallbone, K.; Simeonidis, E. Flux balance analysis: A geometric perspective. J. Theor. Biol. 2009, 258, 311–315. [Google Scholar] [CrossRef] [PubMed]
- Iwase, T.; Okai, C.; Kamata, Y.; Tajima, A.; Mizunoe, Y. A straightforward assay for measuring glycogen levels and RpoS. J. Microbiol. Meth. 2018, 145, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Karp, P.D. Call for an enzyme genomics initiative. Genome Biol. 2004, 5, 401. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, K.; Aikawa, S.; Kojima, Y.; Toya, Y.; Furusawa, C.; Kondo, A.; Shimizu, H. Construction of a Genome-Scale Metabolic Model of Arthrospira platensis NIES-39 and Metabolic Design for Cyanobacterial Bioproduction. PLoS ONE 2015, 10, e0144430. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Bryant, D.A. The tricarboxylic acid cycle in cyanobacteria. Science 2011, 334, 1551–1553. [Google Scholar] [CrossRef] [PubMed]
- Kramer, D.M.; Evans, J.R. The Importance of Energy Balance in Improving Photosynthetic Productivity. Plant Physiol. 2011, 155, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Nogales, J.; Gudmundsson, S.; Knight, E.M.; Palsson, B.O.; Thiele, I. Detailing the Optimality of Photosynthesis in Cyanobacteria through Systems Biology Analysis. Proc. Natl. Acad. Sci. USA 2012, 109, 2678–2683. [Google Scholar] [CrossRef] [PubMed]
- Cooley, J.W.; Vermaas, W.F. Succinate Dehydrogenase and Other Respiratory Pathways in Thylakoid Membranes of Synechocystis sp. strain PCC 6803: Capacity Comparisons and Physiological Function. J. Bacteriol. 2001, 183, 4251–4258. [Google Scholar] [CrossRef] [PubMed]
- Hasunuma, T.; Kikuyama, F.; Matsuda, M.; Aikawa, S.; Izumi, Y.; Kondo, A. Dynamic metabolic profiling of cyanobacterial glycogen biosynthesis under conditions of nitrate depletion. J. Exp. Bot. 2013, 64, 2943–2954. [Google Scholar] [CrossRef] [PubMed]
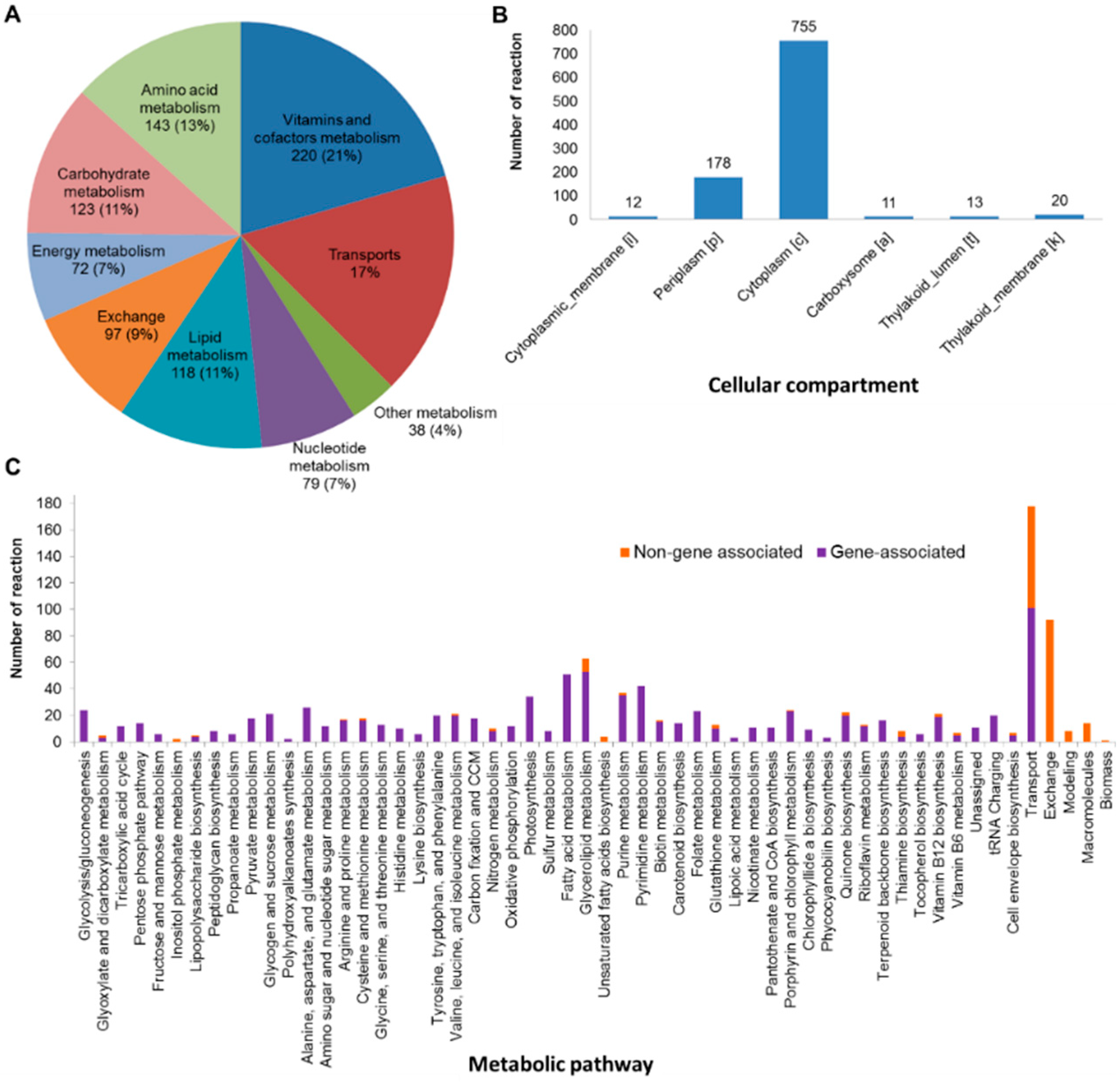
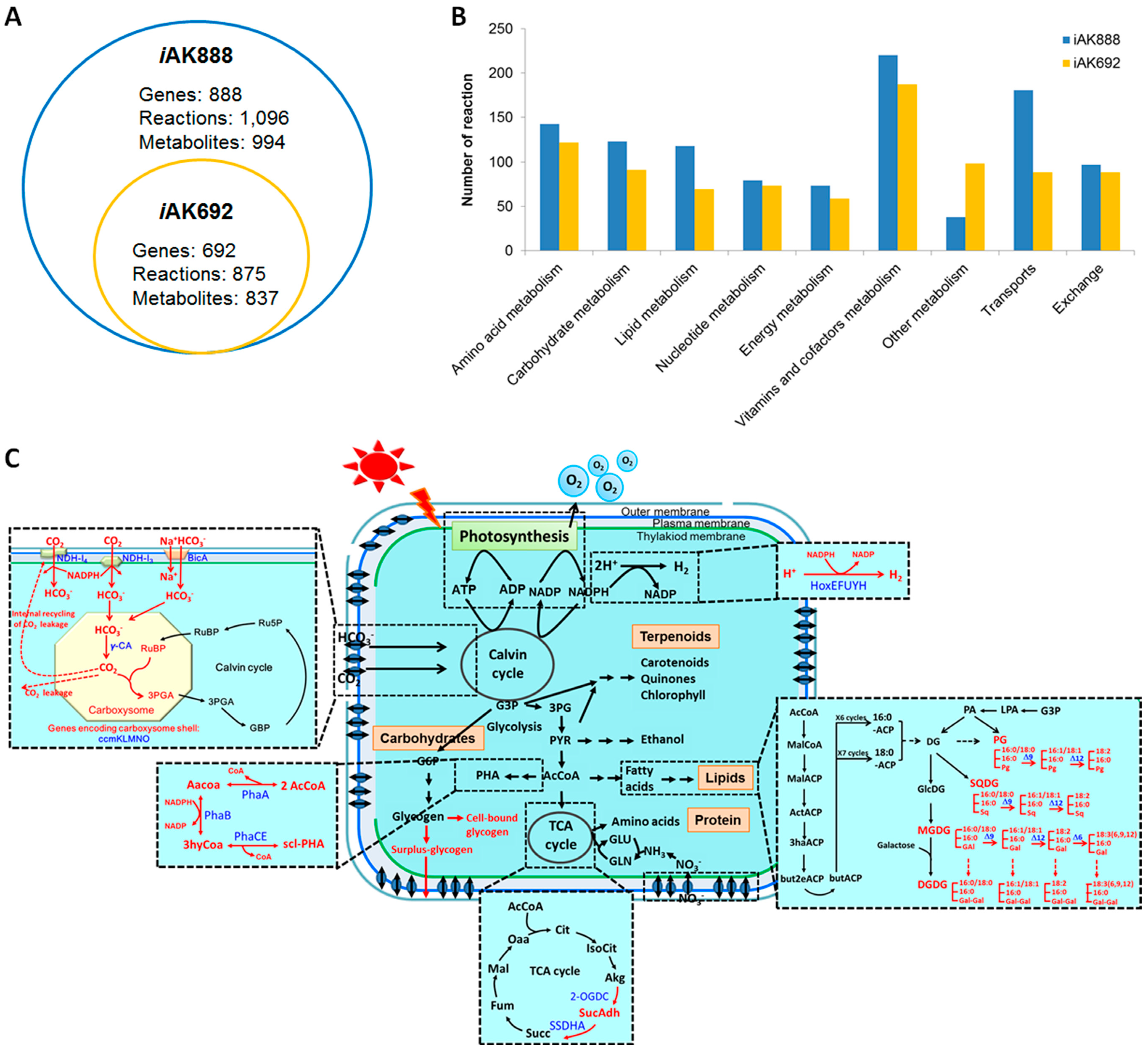
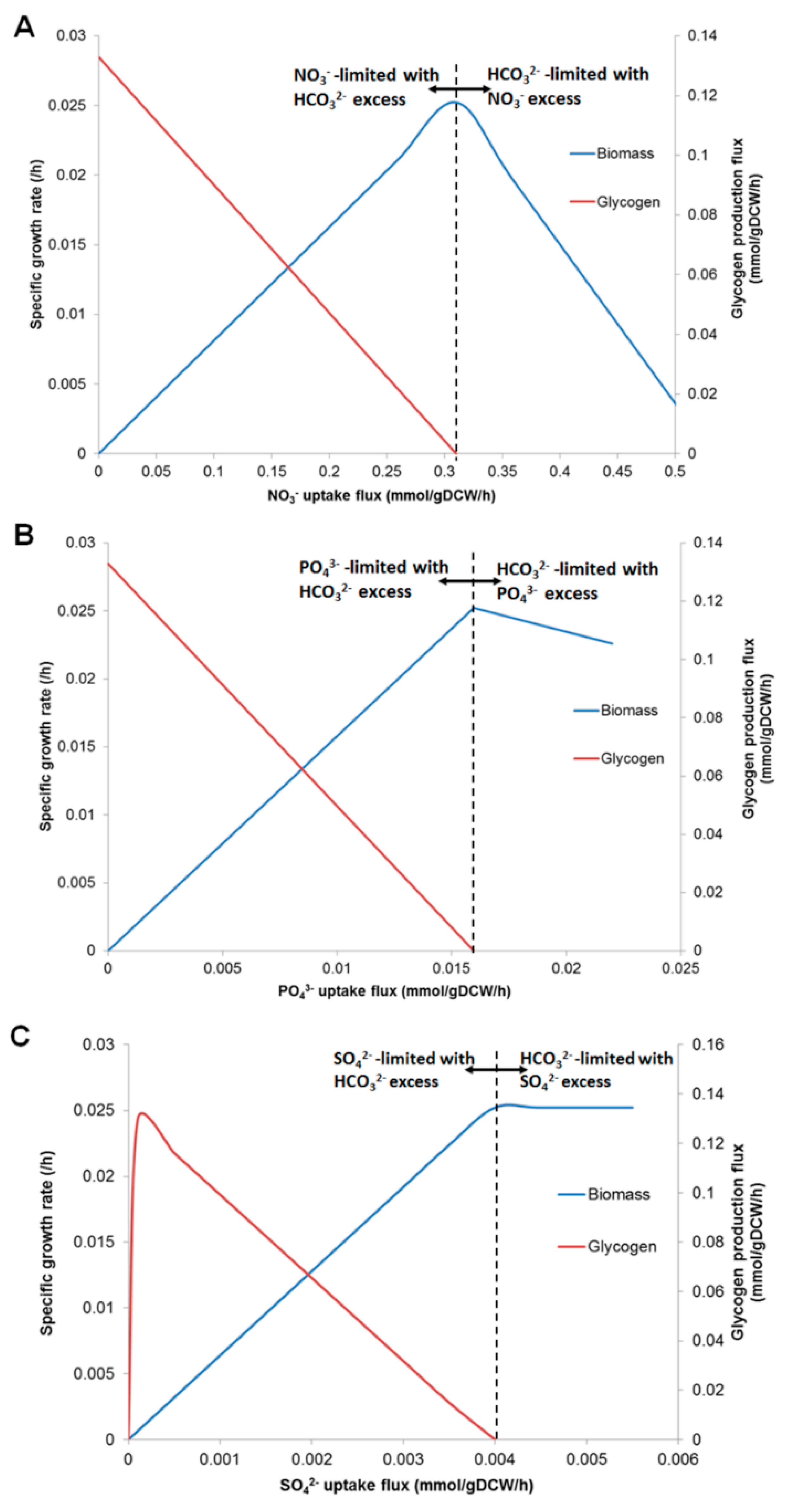
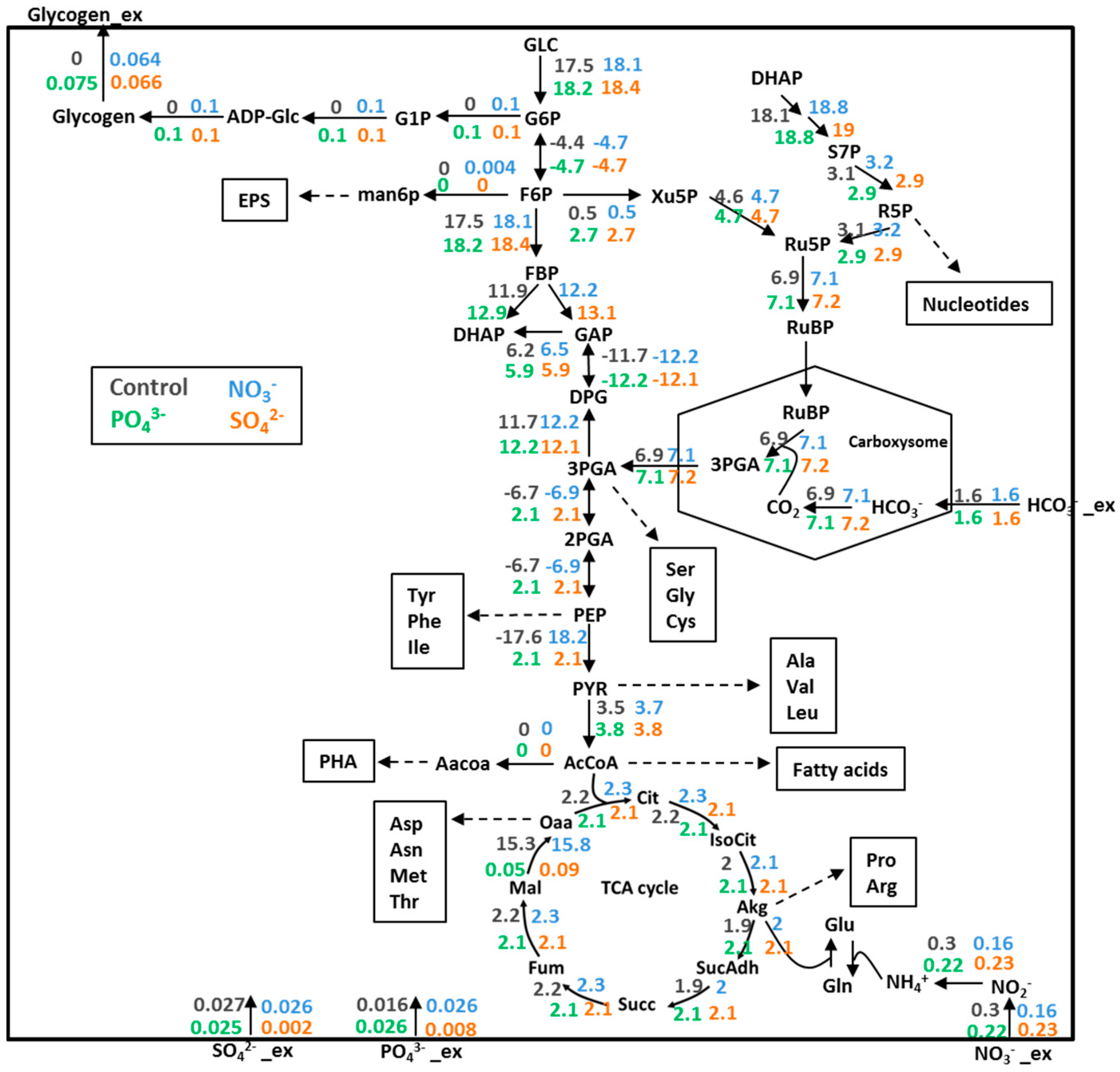
| Component | Content (%w/w) | |
|---|---|---|
| iAK888 | iAK692 [23] | |
| Proteins | 51.44 | 68 |
| Carbohydrates | 31.62 | 16 |
| Lipids | 4.98 | 11 |
| DNA | 0.88 | 0.88 |
| RNA | 3.12 | 3.12 |
| Colorants | 2.84 | 1 |
| Vitamins | 0.11 | - |
| Minerals | 2.79 | - |
| Ash | 2.27 | - |
| Sum | 100 | 100 |
| Arthrospira Species | A. platensis C1 | A. platensis NIE-39 | |
| Genome Statistics | - | - | |
| Genome size (bp) | 6,089,210 | 6,788,435 | |
| Protein coding genes | 6108 | 6630 | |
| Gene with enzymes | 952 | 905 | |
| Transporter genes | 345 | NA | |
| Model Name/Characteristics | iAK888 | iAK692 | - |
| Total genes in model | 888 (15%) | 692 (11%) | 620 (9%) |
| - Metabolic genes | 767 | 692 | 579 |
| - Transporter genes | 121 | 0 | 41 |
| Total biochemical reactions | 1096 | 875 | 746 |
| - Metabolic reactions | 817 | 699 | 652 |
| - Transport reactions | 182 | 88 | 60 |
| - Exchange reactions | 97 | 88 | 34 |
| Metabolites | 994 | 837 | 673 |
| Compartments | 6 | 2 | 2 |
| Reference | This study | [23] | [55] |
| Growth Condition | Constraints of Consumed Metabolites | Maximal Specific Growth Rate (1/h) | % Error | |||
|---|---|---|---|---|---|---|
| Photon Flux (μmol photons m−2 s−1) | HCO3− Uptake Flux (mmol/g DCW/h) | Glucose Uptake Flux (mmol/g DCW/h) | Experiment | In Silico | ||
| Autotroph | 100 | 0.2 | 0 | 0.0255 [23] | 0.0252 | 1.2 |
| Autotroph | 200 | 0.25 | 0 | 0.0331 [48] | 0.0334 | 0.9 |
| Heterotroph | 0 | 0 | 0–0.017 | 0 [23] | 0 | 0 |
| Mixotroph | 100 | 0.2 | 0–0.017 | 0.0262 [23] | 0.0260 | 0.8 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klanchui, A.; Dulsawat, S.; Chaloemngam, K.; Cheevadhanarak, S.; Prommeenate, P.; Meechai, A. An Improved Genome-Scale Metabolic Model of Arthrospira platensis C1 (iAK888) and Its Application in Glycogen Overproduction. Metabolites 2018, 8, 84. https://doi.org/10.3390/metabo8040084
Klanchui A, Dulsawat S, Chaloemngam K, Cheevadhanarak S, Prommeenate P, Meechai A. An Improved Genome-Scale Metabolic Model of Arthrospira platensis C1 (iAK888) and Its Application in Glycogen Overproduction. Metabolites. 2018; 8(4):84. https://doi.org/10.3390/metabo8040084
Chicago/Turabian StyleKlanchui, Amornpan, Sudarat Dulsawat, Kullapat Chaloemngam, Supapon Cheevadhanarak, Peerada Prommeenate, and Asawin Meechai. 2018. "An Improved Genome-Scale Metabolic Model of Arthrospira platensis C1 (iAK888) and Its Application in Glycogen Overproduction" Metabolites 8, no. 4: 84. https://doi.org/10.3390/metabo8040084
APA StyleKlanchui, A., Dulsawat, S., Chaloemngam, K., Cheevadhanarak, S., Prommeenate, P., & Meechai, A. (2018). An Improved Genome-Scale Metabolic Model of Arthrospira platensis C1 (iAK888) and Its Application in Glycogen Overproduction. Metabolites, 8(4), 84. https://doi.org/10.3390/metabo8040084




