NMR-Based Identification of Metabolites in Polar and Non-Polar Extracts of Avian Liver
Abstract
:1. Introduction
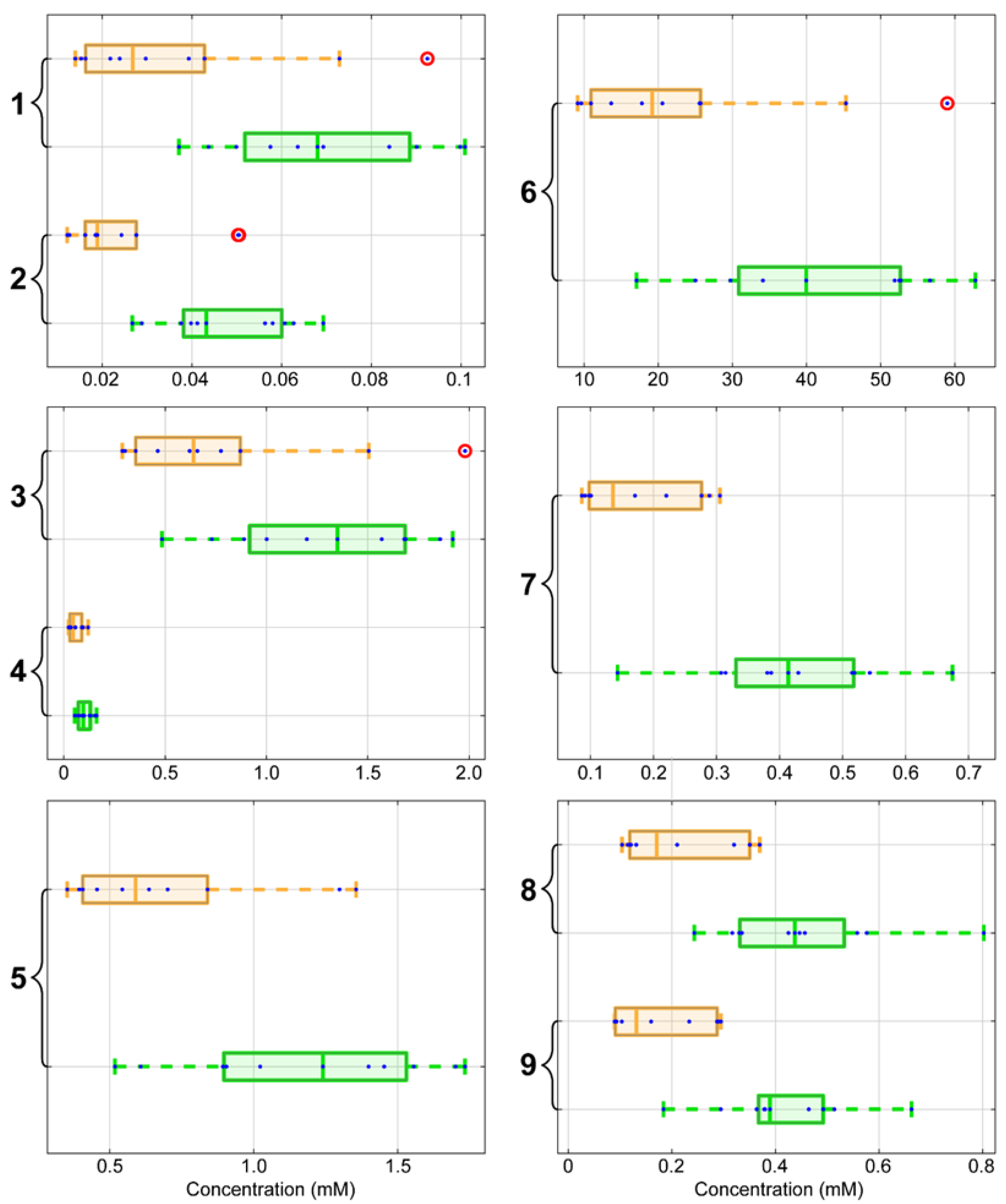
2. Results
3. Discussion
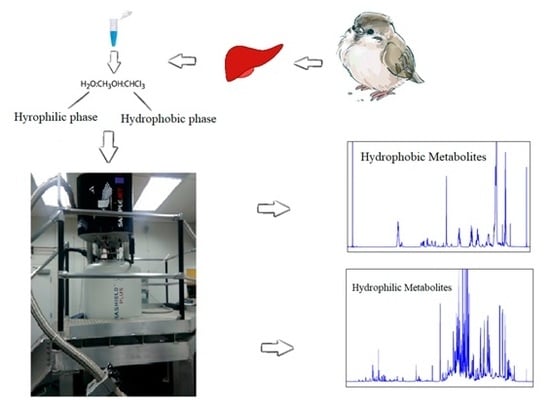
4. Materials and Methods
4.1. Animal Model
4.2. Sample Preparation
4.3. NMR Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bonnefont, C.M.; Guerra, A.; Theron, L.; Molette, C.; Canlet, C.; Fernandez, X. Metabolomic study of fatty livers in ducks: Identification by 1H-NMR of metabolic markers associated with technological quality. Poult. Sci. 2014, 93, 1542–1552. [Google Scholar] [CrossRef] [PubMed]
- Le Roy, C.I.; Mappley, L.J.; La Ragione, R.M.; Woodward, M.J.; Claus, S.P. NMR-based metabolic characterization of chicken tissues and biofluids: A model for avian research. Metabolomics 2016, 12, 157. [Google Scholar] [CrossRef] [PubMed]
- Hoult, D.I.; Busby, S.J.; Gadian, D.G.; Radda, G.K.; Richards, R.E.; Seeley, P.J. Observation of tissue metabolites using 31P nuclear magnetic resonance. Nature 1974, 252, 285–287. [Google Scholar] [CrossRef] [PubMed]
- Markley, J.L.; Brüschweiler, R.; Edison, A.S.; Eghbalnia, H.R.; Powers, R.; Raftery, D.; Wishart, D.S. The future of NMR-based metabolomics. Curr. Opin. Biotechnol. 2016, 43, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. ‘Metabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Southam, A.D.; Hines, A.; Viant, M.R. High-throughput tissue extraction protocol for NMR- and MS-based metabolomics. Anal. Biochem. 2008, 372, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Kruger, N.J.; Troncoso-Ponce, M.A.; Ratcliffe, R.G. 1H NMR metabolite fingerprinting and metabolomic analysis of perchloric acid extracts from plant tissues. Nat. Protoc. 2008, 3, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Beckonert, O.; Keun, H.C.; Ebbels, T.M.D.; Bundy, J.G.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2007, 2, 2692–2703. [Google Scholar] [CrossRef] [PubMed]
- Oostendorp, M.; Engelke, U.F.; Willemsen, M.A.; Wevers, R.A. Diagnosing inborn errors of lipid metabolism with proton nuclear magnetic resonance spectroscopy. Clin. Chem. 2006, 52, 1395–1405. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, E.L.; Akutsu, H.; Doreleijers, J.F.; Harano, Y.; Ioannidis, Y.E.; Lin, J.; Livny, M.; Mading, S.; Maziuk, D.; Miller, Z.; et al. BioMagResBank. Nucleic Acids Res. 2008, 36, D402–D408. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The Human Metabolome Database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef] [PubMed]
- Lei, R.; Wu, C.; Yang, B.; Ma, H.; Shi, C.; Wang, Q.; Wang, Q.; Yuan, Y.; Liao, M. Integrated metabolomic analysis of the nano-sized copper particle-induced hepatotoxicity and nephrotoxicity in rats: A rapid in vivo screening method for nanotoxicity. Toxicol. Appl. Pharmacol. 2008, 232, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Wu, H.; Tjeerdema, R.S.; Viant, M.R. Evaluation of metabolite extraction strategies from tissue samples using NMR metabolomics. Metabolomics 2007, 3, 55–67. [Google Scholar] [CrossRef]
- Moazzami, A.A.; Andersson, R.; Kamal-Eldin, A. Changes in the metabolic profile of rat liver after α-tocopherol deficiency as revealed by metabolomics analysis. NMR Biomed. 2011, 24, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Samino, S.; Revuelta-Cervantes, J.; Vinaixa, M.; Rodriguez, M.A.; Valverde, A.M.; Correig, X. A 1H NMR metabolic profiling to the assessment of protein tyrosine phosphatase 1B role in liver regeneration after partial hepatectomy. Biochimie 2013, 95, 808–816. [Google Scholar] [CrossRef] [PubMed]
- Wagner, L.; Trattner, S.; Pickova, J.; Gomez-Requeni, P.; Moazzami, A.A. 1H NMR-based metabolomics studies on the effect of sesamin in Atlantic salmon (Salmo salar). Food Chem. 2014, 147, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Waters, N.J.; Holmes, E.; Waterfield, C.J.; Farrant, R.D.; Nicholson, J.K. NMR and pattern recognition studies on liver extracts and intact livers from rats treated with alpha-naphthylisothiocyanate. Biochem. Pharmacol. 2002, 64, 67–77. [Google Scholar] [CrossRef]
- Rott, K.H.; Caviedes-Vidal, E.; Karasov, W.H. Intestinal digestive enzyme modulation in house sparrow nestlings occurs within 24 h of a change in diet composition. J. Exp. Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
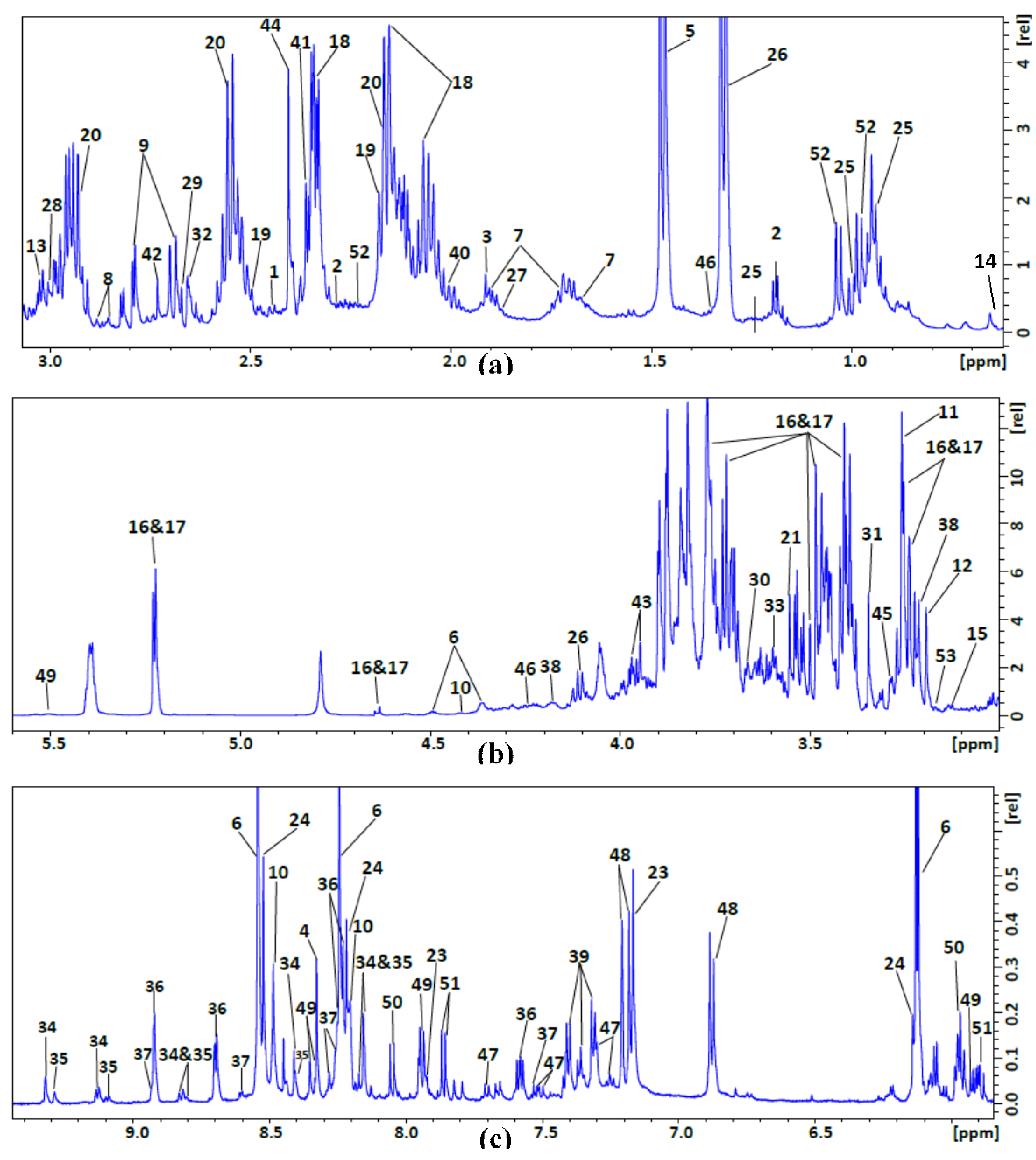
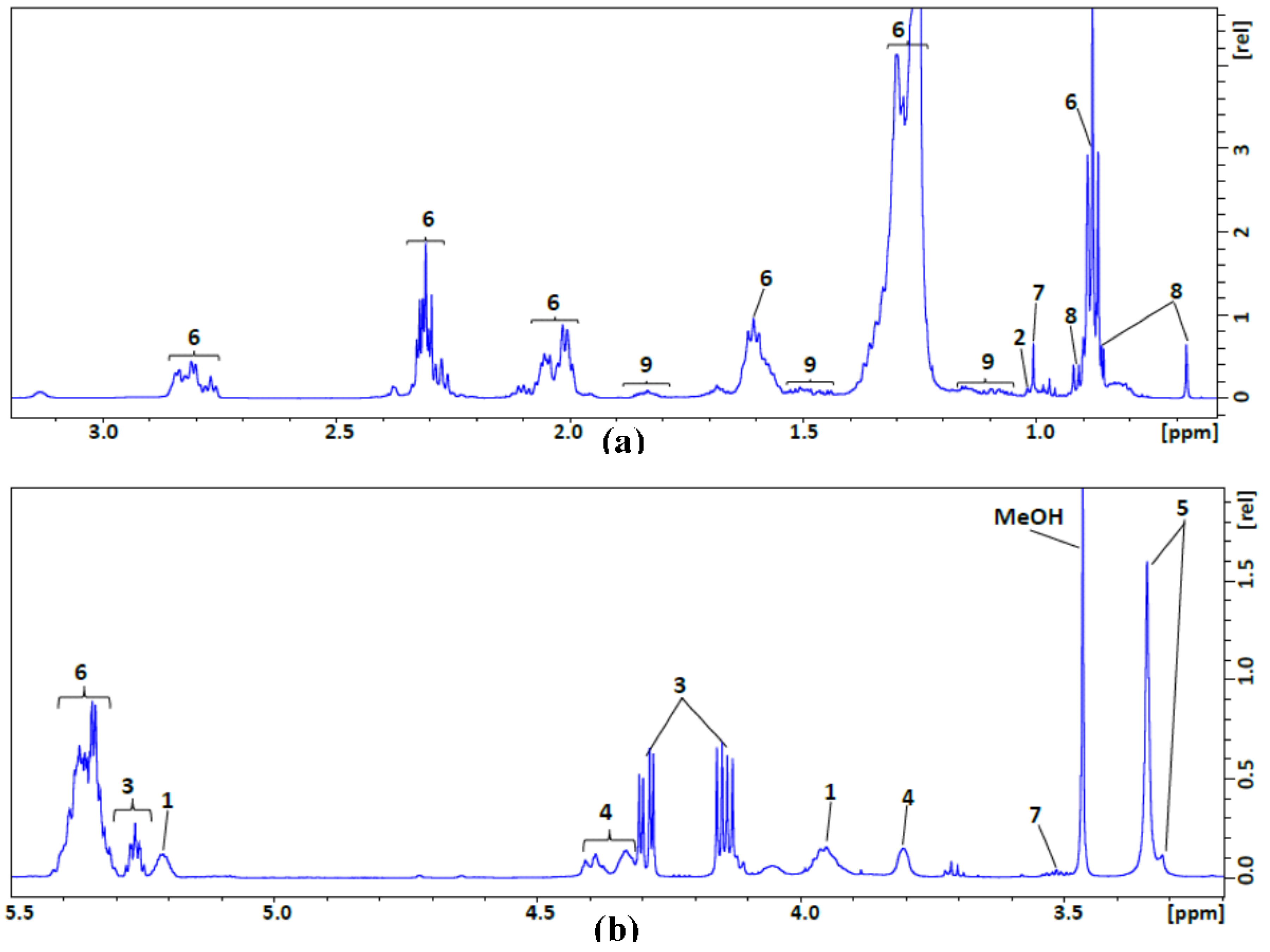
| No. | Compound Name | 1H NMR Chemical Shift (ppm), Multiplicity |
|---|---|---|
| 1 | 2-Oxoglutarate | 2.4 t, 3 t |
| 2 | 3-Hydroxybutyrate | 1.2 d, 2.3 m, 2.4 m, 4.1 m |
| 3 | Acetate | 1.9 s |
| 4 | Adenosine | 8.3 s, 8.2 s, 6.1 d, 4.8 t, 4.4 m, 4.3 m, 3.9 dd, 3.8 dd |
| 5 | Alanine | 1.47 d, 3.78 q |
| 6 | AMP | 4.01 m, 4.36 m, 4.50 q, 4.79 t, 6.12 d, 8.25 s, 8.58 s |
| 7 | Arginine | 1.66 m, 1.91 m, 3.27 t, 3.77 t |
| 8 | Asparagine | 2.85 dd, 2.94 dd, 4.00 dd |
| 9 | Aspartate | 2.66 dd, 2.80 dd, 3.91 dd |
| 10 | ATP | 8.49 s, 8.24 s, 6.12 d, 4.73 t, 4.56 t, 4.27 m, 4.23 m |
| 11 | Betaine | 3.25 s, 3.88 s |
| 12 | Choline | 3.19 s, 3.50 dd, 4.05 t |
| 13 | Creatine | 3.02 s, 3.91 s |
| 14 | DSS | 0.00 s, 0.62 t, 1.75 m, 2.90 m |
| 15 | Ethanolamine | 3.13 t, 3.83 t |
| 16 | α-Glucose | 3.42 m, 3.54 m, 3.72 m, 3.73 m, 3.77 m, 3.87 m, 5.23 d |
| 17 | β-Glucose | 3.25 m, 3.49 m, 3.49 m, 3.50 m, 3.88 m, 3.91 m, 4.66 d |
| 18 | Glutamate | 2.04 m, 2.12 m, 2.32 m, 2.32 m, 3.76 dd |
| 19 | Glutamine | 2.15 m, 2.18 m, 2.42 m, 2.46 m, 3.76 t |
| 20 | Glutathione | 2.17 m, 2.50 m, 2.56 m, 2.94 dd, 2.98 dd, 3.83 m, 4.56 q |
| 21 | Glycine | 3.54 s |
| 22 | GTP | 8.13 s, 5.93 d, 4.74 t, 4.54 t, 4.35 m, 4.22 m, 4.25 m |
| 23 | Histidine | 3.16 dd, 3.23 dd, 3.98 dd 7.11 s, 7.86 s |
| 24 | IMP | 8.53 s, 8.21 s, 6.13 d, 4.49 t, 4.36 m, 4.03 m |
| 25 | Isoleucine | 0.93 t, 1.02 d, CH2 1.26 m, 1.46 dd, 1.97 m, 3.66 d |
| 26 | Lactate | 1.31 d, 4.10 q |
| 27 | Leucine | 0.94 d, 0.95 d, 1.71 m, 3.73 m |
| 28 | Lysine | 1.43 m, 1.49 m, 1.71 m 1.87 m, 1.91 m, 3.01 t, 3.7 t |
| 29 | Malate | 2.36 dd, 2.66 dd, 4.30 dd |
| 30 | Mannitol | 3.66 m, 3.75 m, 3.80 d, 3.86 dd |
| 31 | Methanol | 3.33 s |
| 32 | Methionine | 2.11 s ,2.12 s, 2.19 m, 2.63 t, 3.85 t |
| 33 | Myo-inositol | 3.26 t, 3.51 dd, 3.60 t, 4.04 t |
| 34 | NAD+ | 9.3 s, 9.13 d, 8.84 d, 8.42 s, 8.20 t, 8.16 s, 8.08 d,6.02 d, 4.75 t, 4.53 t, 4.49 m, 4.48 t, 4.36 m, 4.2 m |
| 35 | NADP+ | 9.28 s, 9.09 d, 8.81 d, 8.40 s, 8.18 m, 8.14 s, 6.09 d, 6.04 d, 4.97 m, 4.60 m, 4.49 m, 4.45 t, 4.39 q , 4.36 m, 4.31 m, 4.27 m, 4.2 m |
| 36 | Niacinamide | 8.92 d, 8.70 d, 8.25 d, 7.58 q |
| 37 | Nicotinate | 8.92 d, 8.59 d, 8.24 d, 7.51 q |
| 38 | O-Phosphocholine | 3.21 s, 3.58 m, 4.16 m |
| 39 | Phenylalanine | 3.11 dd, 3.28 dd, 3.99 dd, 7.31 m, 7.36 m, 7.41 m |
| 40 | Proline | 1.97 m, 2.03 m, 2.33 m, 3.33 m, 3.41 m, 4.12 dd |
| 41 | Pyruvate | 2.35 s |
| 42 | Sarcosine | 2.72 s, 3.60 s |
| 43 | Serine | 3.83 dd, 3.93 dd, 3.97 dd |
| 44 | Succinate | 2.39 s |
| 45 | Taurine | 3.26 t, 3.43 t |
| 46 | Threonine | 1.32 d, 3.57 d, 4.25 m |
| 47 | Tryptophan | 3.30 dd, 3.48 dd, 4.05 dd, 7.19 t, 7.27 t, 7.30 s, 7.52 d, 7.72 d |
| 48 | Tyrosine | 3.04 dd, 3.19 dd, 3.93 dd, 6.88 m, 7.18 m |
| 49 | UDP-N-Acetylglucosamine | 2.06 s, 3.55 t, 3.80 m, 3.87 m, 3.91 d, 3.98 t, 4.17 m, 4.23 m, 4.27 m, 4.35 m, 5.5 dd, 5.96 d, 5.98 d, 7.94 d, 8.34 d |
| 50 | UMP | 3.96 d, 4.03 d, 4.25 s, 4.34 t, 4.41 t, 5.97 d, 5.98 d, 8.08 d |
| 51 | Uridine | 3.80 dd, 3.90 dd, 4.12 dt, 4.21 dd, 4.34 dd, 5.89 d, 5.90 m, 7.86 d |
| 52 | Valine | 0.98 d, 1.03 d, 2.26 m, 3.60 d |
| 53 | β-Alanine | 2.53 t, 3.17 t |
| No. | Compound Name | Assignment, 1H NMR Chemical Shift (ppm), Multiplicity |
|---|---|---|
| 1 | Glycerophospholipid backbone | C-3 H2 (3.96, s); C-2 H (5.17–5.24, m) |
| 2 | Esterified cholesterol | C-19 H3 (1.02, s) |
| 3 | Glycerol backbone | C-1 H2/C-3 H2 (4.15/4.29, m); C-2 H (5.26, p) |
| 4 | Phosphatidyl choline | N-CH2 (3.81, s-broad); PO-CH2 (4.32–4.43, m) |
| 5 | Sphingomyelin and choline | N(CH3)3 (3.32/3.35, s/s) |
| 6 | Fatty acyl chain | CH3(CH2)n (0.88, t); (CH2)n (1.24–1.37, m); –CH2CH2CO (1.55–1.65, m); –CH2CH= (1.98–2.09, m); –CH2CO (2.24–2.35, m); =CHCH2CH= (2.77–2.87, m); –HC=CH– (5.29–5.43,m) |
| 7 | Free cholesterol | C-19 H3(1.01, s); C-3 H(3.48–3.57, m) |
| 8 | Total cholesterol | C-18 H3( 0.68, s); C-26 H3/C-27 H3 (0.86/0.87, d/d); C-21 H3 (0.91, d) |
| 9 | Multiple cholesterol protons | 1.05–1.19; 1.42–1.55; 1.79–1.88 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fathi, F.; Brun, A.; Rott, K.H.; Falco Cobra, P.; Tonelli, M.; Eghbalnia, H.R.; Caviedes-Vidal, E.; Karasov, W.H.; Markley, J.L. NMR-Based Identification of Metabolites in Polar and Non-Polar Extracts of Avian Liver. Metabolites 2017, 7, 61. https://doi.org/10.3390/metabo7040061
Fathi F, Brun A, Rott KH, Falco Cobra P, Tonelli M, Eghbalnia HR, Caviedes-Vidal E, Karasov WH, Markley JL. NMR-Based Identification of Metabolites in Polar and Non-Polar Extracts of Avian Liver. Metabolites. 2017; 7(4):61. https://doi.org/10.3390/metabo7040061
Chicago/Turabian StyleFathi, Fariba, Antonio Brun, Katherine H. Rott, Paulo Falco Cobra, Marco Tonelli, Hamid R. Eghbalnia, Enrique Caviedes-Vidal, William H. Karasov, and John L. Markley. 2017. "NMR-Based Identification of Metabolites in Polar and Non-Polar Extracts of Avian Liver" Metabolites 7, no. 4: 61. https://doi.org/10.3390/metabo7040061
APA StyleFathi, F., Brun, A., Rott, K. H., Falco Cobra, P., Tonelli, M., Eghbalnia, H. R., Caviedes-Vidal, E., Karasov, W. H., & Markley, J. L. (2017). NMR-Based Identification of Metabolites in Polar and Non-Polar Extracts of Avian Liver. Metabolites, 7(4), 61. https://doi.org/10.3390/metabo7040061