SnRK-PP2C-PYL Gene Families in Citrus sinensis: Genomic Characterization and Regulatory Roles in Carotenoid Metabolism
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome-Wide Identification, Characterization, and Phylogenetic Analysis of CsSnRK, CsPP2C, and CsPYL Genes
2.2. Plant Materials and Carotenoid/Chlorophyll Extraction with HPLC Quantification
2.3. Transmission Electron Microscopy (TEM)
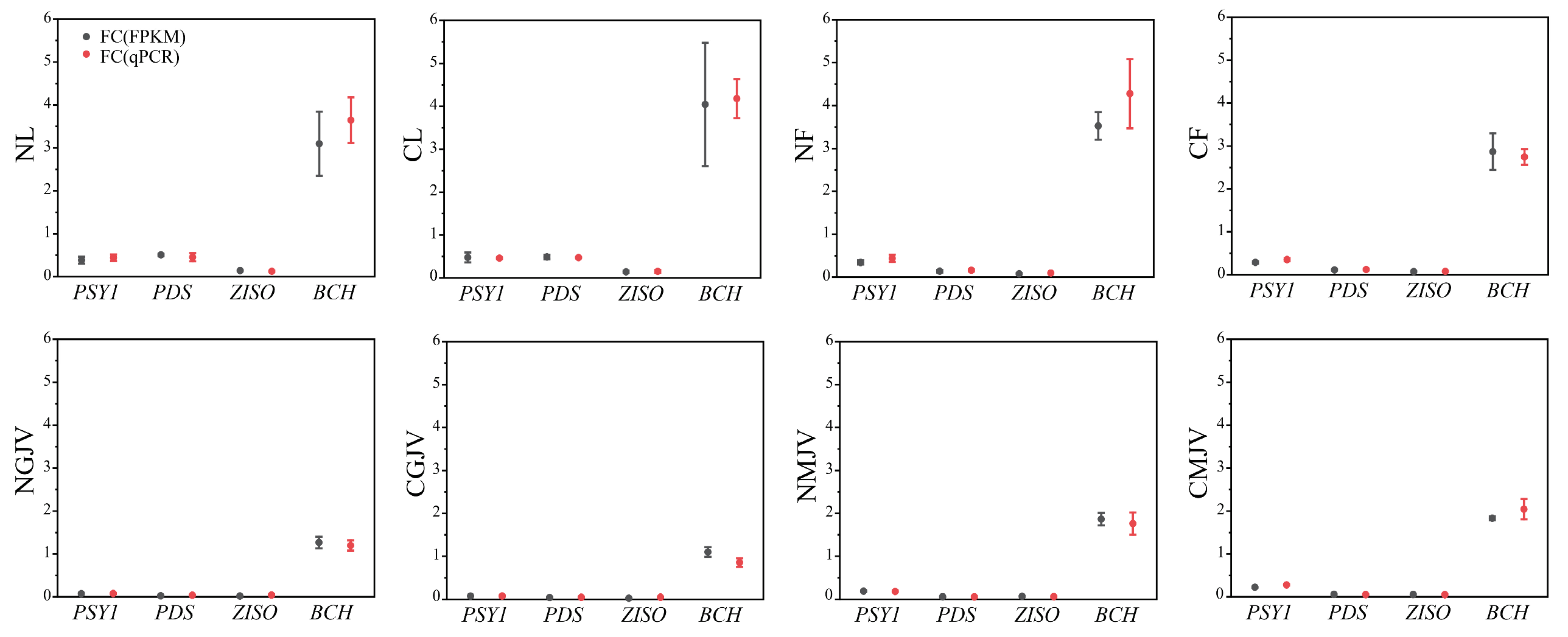
2.4. RNA Extraction, RNA-Seq Analysis, and Quantitative Real-Time PCR (qPCR) Analysis
2.5. Weighted Gene Co-Expression Network Analysis (WGCNA) and Establishment of Co-Expression Networks
3. Results
3.1. Identification and Classification of CsSnRK, CsPYL, and CsPP2C Gene Families in Citrus sinensis
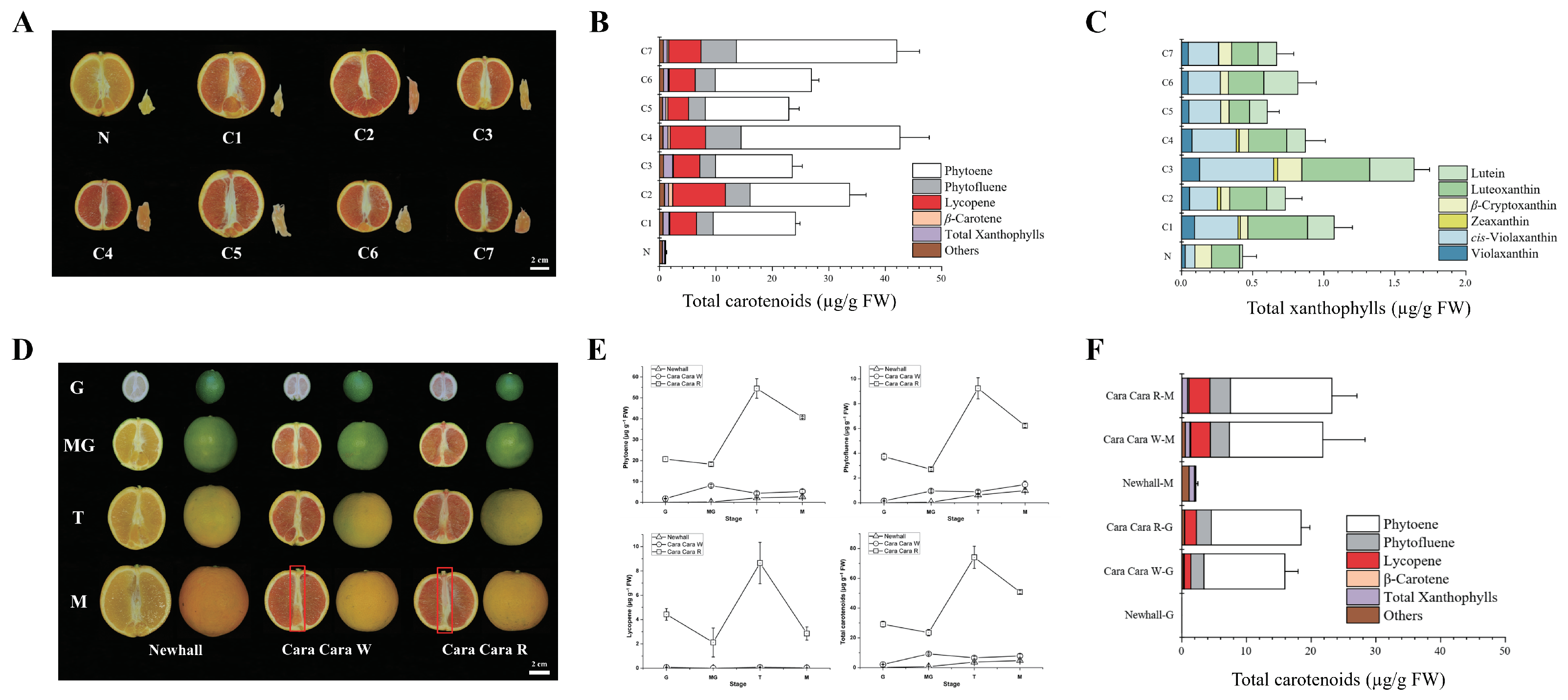
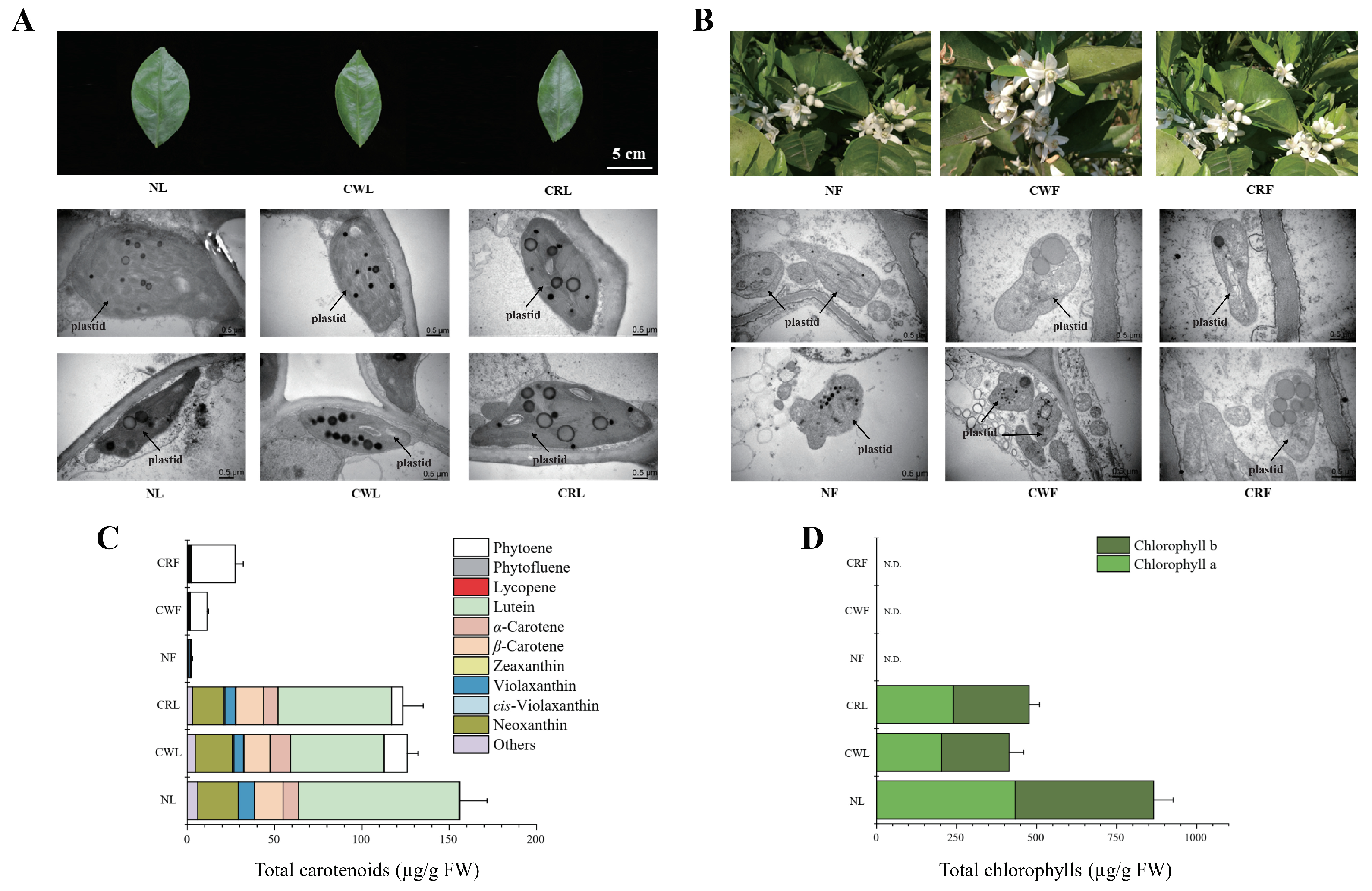
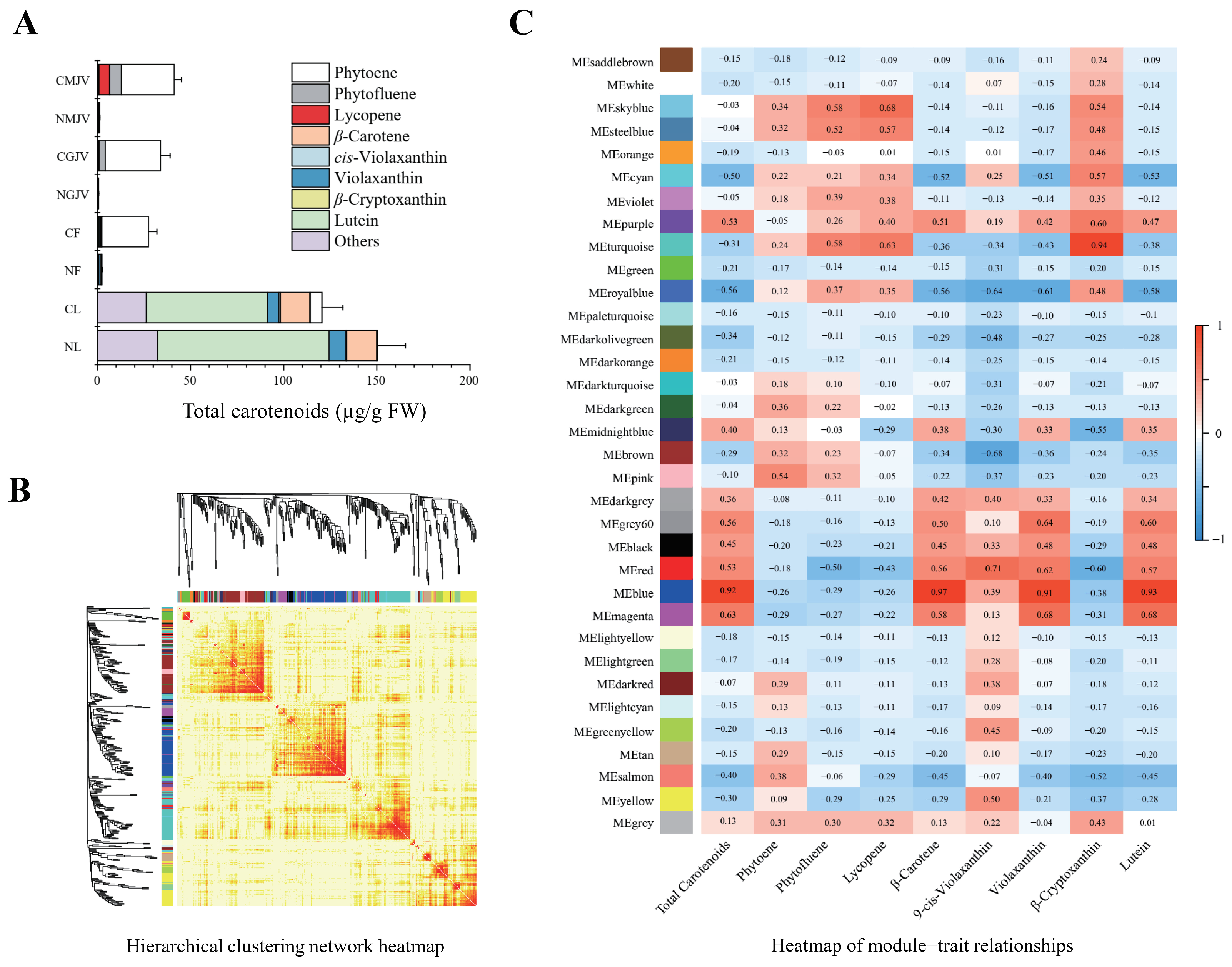
3.2. Comparative Profiling Reveals ‘Cara Cara’ Juice Vesicles Hyperaccumulate Linear Carotenoids (Phytoene, Phytofluene, and Lycopene) over ‘Newhall’, with Parallel Leaf/Flower Phytoene Elevation and Supporting Plastid Ultrastructure
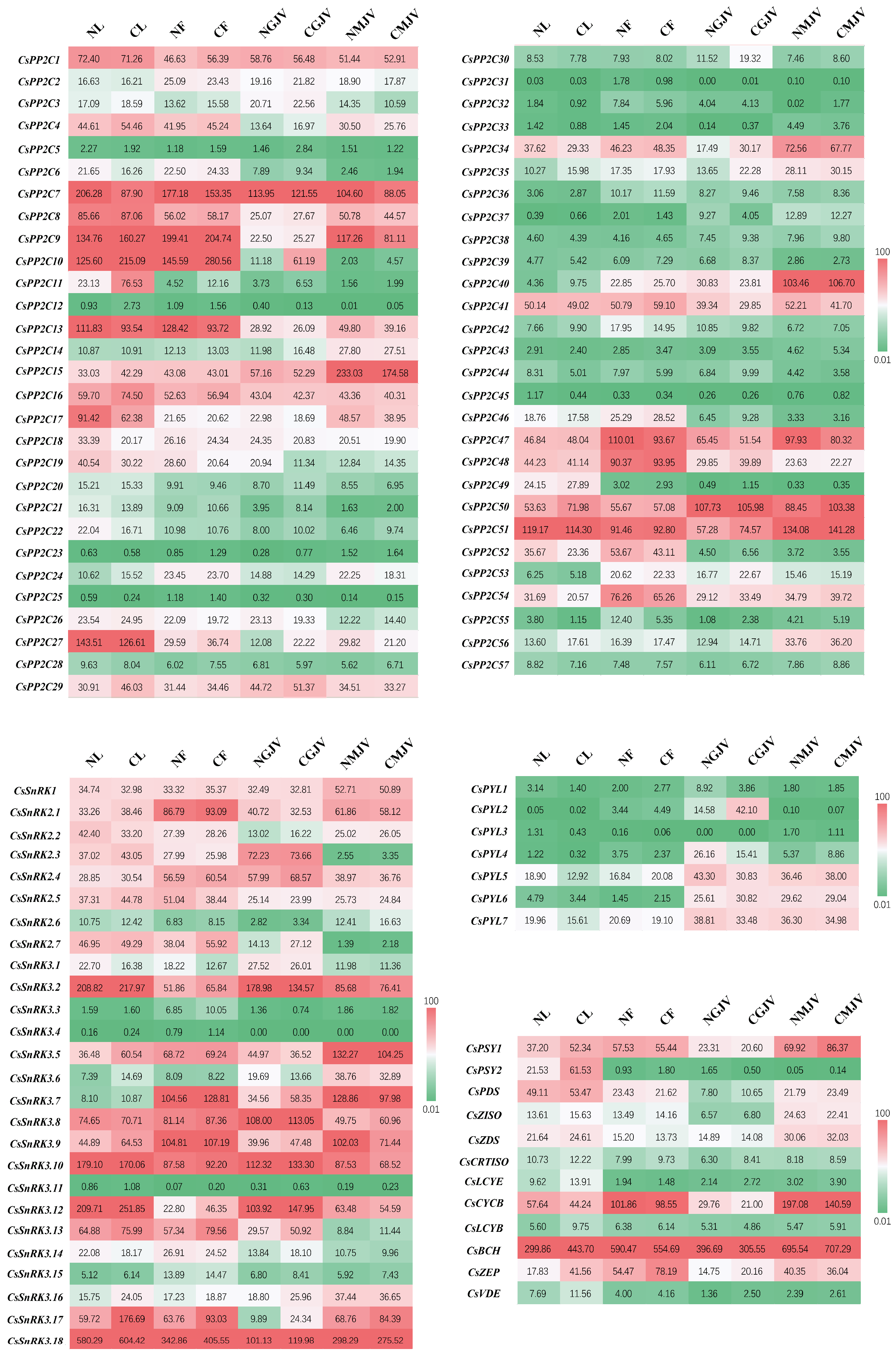
3.3. Spatiotemporal Expression Dynamics of PP2C, SnRK, and PYL Gene Families Across Tissues and Fruit Developmental Stages Between ‘Newhall’ and ‘Cara Cara’
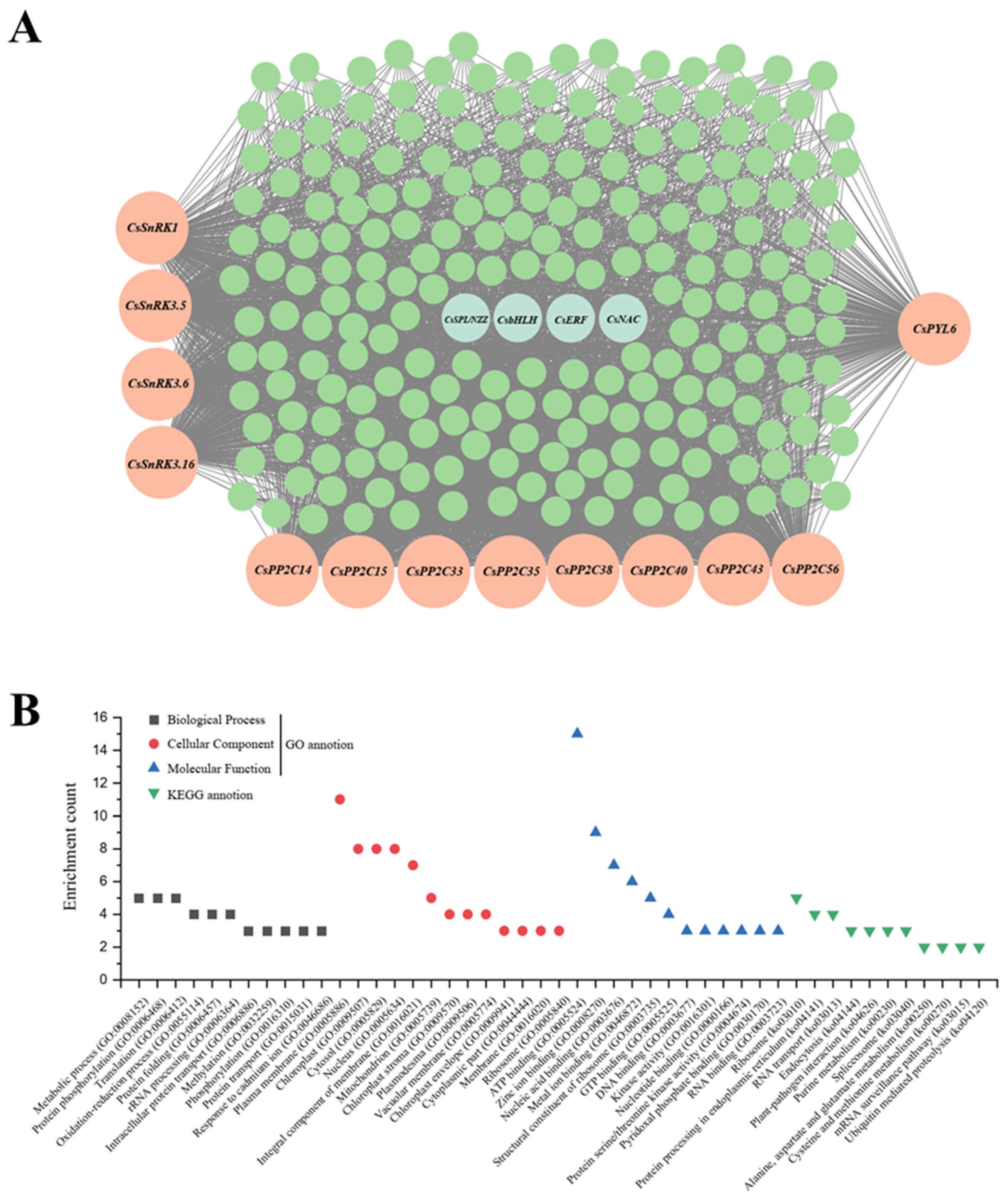
3.4. WGCNA Integrates Transcriptome and Carotenoid Profiles Revealing a ‘Cara Cara’-Specific Module for Linear Carotenoid Accumulation; Network Analysis Identifies Candidate SnRK, PYL, PP2C, and TF Regulators of Post-Transcriptional Control
4. Discussion
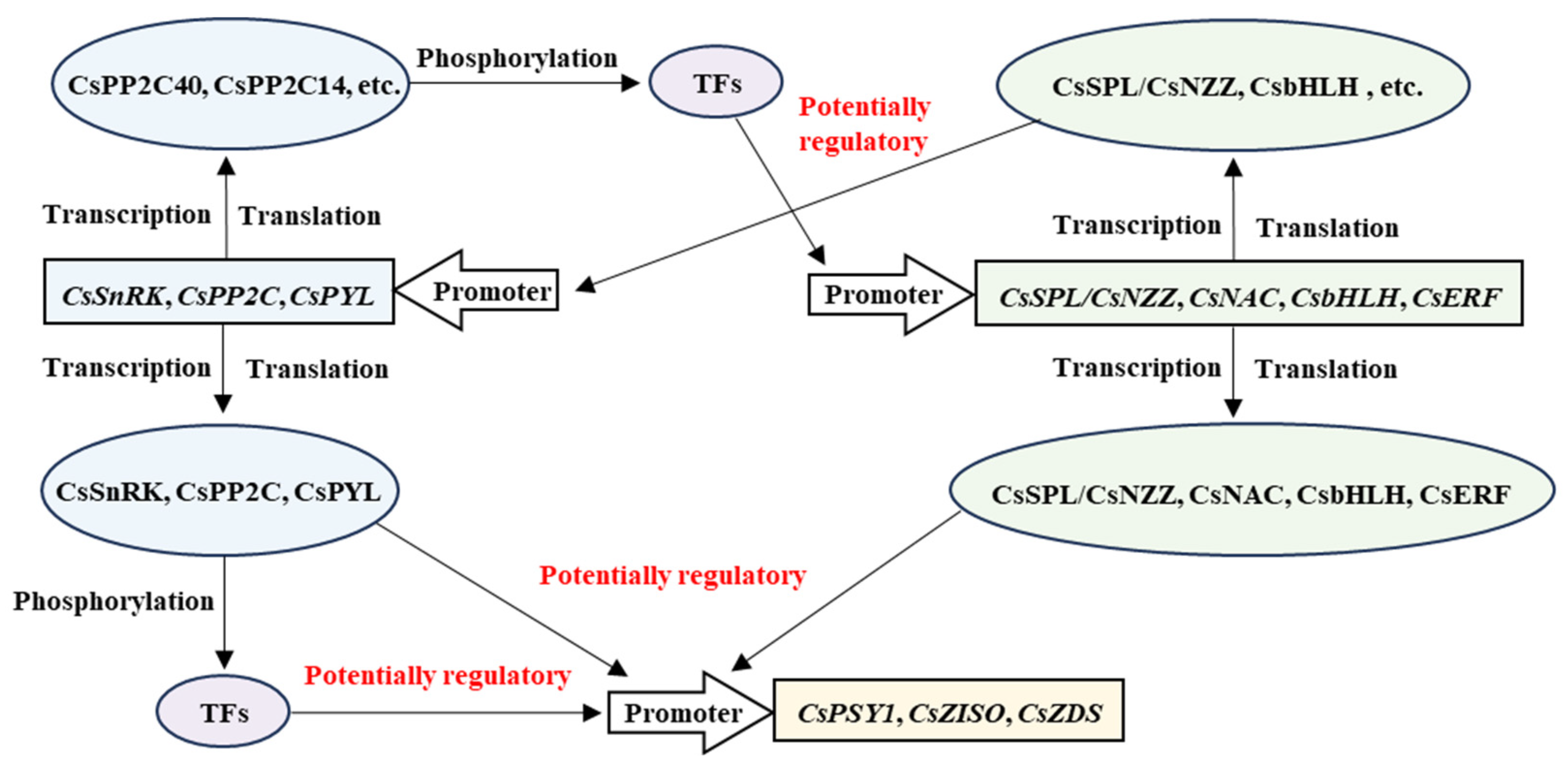
4.1. A Putative Phosphorylation Network Orchestrates Carotenoid Hyperaccumulation
4.2. Bridging ABA Signaling and Carotenoid Metabolism: A Potential Feedback Loop
4.3. Scientific Advance and Comparison with Previous Studies
4.4. Limitations and Future Perspectives
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| SnRK | sucrose non-fermenting related kinases |
| PYR1 | pyrabactin resistance |
| PYL | PYR1-like |
| RCAR | regulatory components of ABA receptors |
| PP2C | protein phosphatases type 2C |
| TFs | transcription factors |
| GGPP | geranylgeranyl pyrophosphate |
| PSY | phytoene synthase |
| PDS | phytoene desaturase |
| Z-ISO | ζ-carotene isomerase |
| ZDS | ζ-carotene desaturase |
| CrtISO | carotene isomerase |
| CYCB | chromoplast-specific lycopene β-cyclase |
| LCYB | lycopene β-cyclase |
| LCYE | lycopene ε-cyclase |
| BCH | β-carotene hydroxylase |
| ZEP | zeaxanthin epoxidase |
| VDE | violaxanthin de-epoxidase |
| NSY | neoxanthin synthase |
| NCED | 9-cis-epoxycarotenoid dioxygenase |
| CCD | carotenoid cleavage dioxygenase |
| NL | ‘Newhall’ leaf |
| CL | ‘Cara Cara’ leaf |
| NF | ‘Newhall’ flower |
| CF | ‘Cara Cara’ flower |
| NGJV | ‘Newhall’ green stage juice vesicle |
| CGJV | ‘Cara Cara’ green stage juice vesicle |
| NMJV | ‘Newhall’ mature stage juice vesicle |
| CMJV | ‘Cara Cara’ mature stage juice vesicle |
| WGCNA | weighted gene co-expression network analysis |
| Co-IP | co-immunoprecipitation |
| BiFC | bimolecular fluorescence complementation |
| EMSA | electrophoretic mobility shift assays |
| qPCR | quantitative real-time PCR |
References
- Luckstead, J.; Devadoss, S. Trends and Issues Facing the US Citrus Industry. Choices 2021, 36, 1–10. [Google Scholar]
- Liu, S.; Xu, Y.; Yang, K.; Huang, Y.; Lu, Z.; Chen, S.; Gao, X.; Xiao, G.; Chen, P.; Zeng, X.; et al. Origin and De Novo Domestication of Sweet Orange. Nat. Genet. 2025, 57, 754–762. [Google Scholar] [CrossRef]
- Seminara, S.; Bennici, S.; Di Guardo, M.; Caruso, M.; Gentile, A.; La Malfa, S.; Distefano, G. Sweet Orange: Evolution, Characterization, Varieties, and Breeding Perspectives. Agriculture 2023, 13, 264. [Google Scholar] [CrossRef]
- Chiu, C.J.; Taylor, A. Nutritional Antioxidants and Age-Related Cataract and Maculopathy. Exp. Eye Res. 2007, 84, 229–245. [Google Scholar]
- Walter, M.H.; Strack, D. Carotenoids and Their Cleavage Products: Biosynthesis and Functions. Nat. Prod. Rep. 2011, 28, 663–692. [Google Scholar] [CrossRef]
- Fiedor, J.; Burda, K. Potential Role of Carotenoids as Antioxidants in Human Health and Disease. Nutrients 2014, 6, 466–488. [Google Scholar] [CrossRef]
- Nisar, N.; Li, L.; Lu, S.; Khin, N.C.; Pogson, B.J. Carotenoid Metabolism in Plants. Mol. Plant 2015, 8, 68–82. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Huq, E.; Rodríguez-Concepción, M. Direct Regulation of Phytoene Synthase Gene Expression and Carotenoid Biosynthesis by Phytochrome-Interacting Factors. Proc. Natl. Acad. Sci. USA 2010, 107, 11626–11631. [Google Scholar] [CrossRef] [PubMed]
- Llorente, B.; D’Andrea, L.; Ruiz-Sola, M.Á.; Botterweg, E.; Pulido, P.; Andilla, J.; Loza-Alvarez, P.; Rodriguez-Concepcion, M. Tomato Fruit Carotenoid Biosynthesis Is Adjusted to Actual Ripening Progression by a Light-Dependent Mechanism. Plant J. 2016, 85, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Vrebalov, J.; Pan, I.L.; Arroyo, A.J.M.; McQuinn, R.; Chung, M.; Poole, M.; Rose, J.; Seymour, G.; Grandillo, S.; Giovannoni, J.; et al. Fleshy Fruit Expansion and Ripening Are Regulated by the Tomato SHATTERPROOF Gene TAGL1. Plant Cell 2009, 21, 3041–3062. [Google Scholar] [CrossRef] [PubMed]
- Martel, C.; Vrebalov, J.; Tafelmeyer, P.; Giovannoni, J.J. The Tomato MADS-Box Transcription Factor RIPENING INHIBITOR Interacts with Promoters Involved in Numerous Ripening Processes in a COLORLESS NONRIPENING-Dependent Manner. Plant Physiol. 2011, 157, 1568–1579. [Google Scholar] [CrossRef] [PubMed]
- Hrabak, E.M.; Chan, C.W.M.; Gribskov, M.; Harper, J.F.; Choi, J.H.; Halford, N.; Kudla, J.; Luan, S.; Nimmo, H.G.; Sussman, M.R.; et al. The Arabidopsis CDPK-SnRK Superfamily of Protein Kinases. Plant Physiol. 2003, 132, 666–680. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, K.; Takezawa, D.; Sakata, Y. Decoding ABA and Osmostress Signalling in Plants from an Evolutionary Point of View. Plant Cell Environ. 2020, 43, 2894–2911. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zhang, P.; Jia, H.; Zhang, S.; Nishawy, E.; Sun, X.; Dai, M. Regulatory Mechanisms and Breeding Strategies for Crop Drought Resistance. New Crops 2024, 1, 100029. [Google Scholar] [CrossRef]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C Phosphatase Activity Function as Abscisic Acid Sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef]
- Melcher, K.; Ng, L.M.; Zhou, X.E.; Soon, F.F.; Xu, Y.; Suino-Powell, K.M.; Park, S.Y.; Weiner, J.J.; Fujii, H.; Chinnusamy, V.; et al. A Gate-Latch-Lock Mechanism for Hormone Signalling by Abscisic Acid Receptors. Nature 2009, 462, 602–608. [Google Scholar] [CrossRef]
- Gonzalez-Guzman, M.; Pizzio, G.A.; Antoni, R.; Vera-Sirera, F.; Merilo, E.; Bassel, G.W.; Fernández, M.A.; Holdsworth, M.J.; Perez-Amador, M.A.; Kollist, H.; et al. Arabidopsis PYR/PYL/RCAR Receptors Play a Major Role in Quantitative Regulation of Stomatal Aperture and Transcriptional Response to Abscisic Acid. Plant Cell 2012, 24, 2483–2496. [Google Scholar] [CrossRef]
- Klingler, J.P.; Batelli, G.; Zhu, J.K. ABA Receptors: The START of a New Paradigm in Phytohormone Signalling. J. Exp. Bot. 2010, 61, 3199–3210. [Google Scholar] [CrossRef]
- Xue, T.; Wang, D.; Zhang, S.; Ehlting, J.; Ni, F.; Jakab, S.; Zheng, C.; Zhong, Y. Genome-Wide and Expression Analysis of Protein Phosphatase 2C in Rice and Arabidopsis. BMC Genom. 2008, 9, 550. [Google Scholar] [CrossRef]
- Lu, P.; Wang, C.; Yin, T.; Zhong, S.L.; Grierson, D.; Chen, K.S.; Xu, C.J. Cytological and Molecular Characterization of Carotenoid Accumulation in Normal and High-Lycopene Mutant Oranges. Sci. Rep. 2017, 7, 761. [Google Scholar] [CrossRef]
- Lu, P.; Wang, S.; Grierson, D.; Xu, C. Transcriptomic Changes Triggered by Carotenoid Biosynthesis Inhibitors and Role of Citrus sinensis Phosphate Transporter 4;2 (CsPHT4;2) in Enhancing Carotenoid Accumulation. Planta 2019, 249, 257–270. [Google Scholar] [CrossRef]
- Romero, P.; Lafuente, M.T.; Rodrigo, M.J. The Citrus ABA Signalosome: Identification and Transcriptional Regulation during Sweet Orange Fruit Ripening and Leaf Dehydration. J. Exp. Bot. 2012, 63, 4931–4945. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER Web Server: Interactive Sequence Similarity Searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.H. Evolview v3: A Webserver for Visualization, Annotation, and Management of Phylogenetic Trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Xu, C.; Fraser, P.D.; Wang, W.; Bramley, P.M. Differences in the Carotenoid Content of Ordinary Citrus and Lycopene-Accumulating Mutants. J. Agric. Food Chem. 2006, 54, 5474–5481. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a Central Hub for Transcription Factors and Regulatory Interactions in Plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Luo, F.; Yu, Z.; Zhou, Q.; Huang, A. Multi-omics-based discovery of plant signaling molecules. Metabolites 2022, 12, 76. [Google Scholar] [CrossRef]
- Monterisi, S.; Zuluaga, M.Y.A.; Senizza, B.; Cardarelli, M.; Rouphael, Y.; Colla, G.; Lucini, L.; Cesco, S.; Pii, Y. The integrated multi-omics analysis unravels distinct roles of Malvaceae-derived protein hydrolysate and its molecular fraction in modulating tomato resilience under limited nitrogen availability. Plant Stress 2025, 15, 100771. [Google Scholar] [CrossRef]
- Pardo-Hernández, M.; García-Pérez, P.; Lucini, L.; Rivero, R.M. Multi-omics exploration of the involvement of ABA in identifying unique molecular markers for single and combined stresses in tomato plants. J. Exp. Bot. 2024, erae372. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Yang, W.; Jing, W.; Shahid, M.O.; Liu, Y.; Qiu, X.; Choisy, P.; Xu, T.; Ma, N.; Gao, J.; et al. Multi-omics analysis reveals key regulatory defense pathways and genes involved in salt tolerance of rose plants. Hortic. Res. 2024, 11, uhae068. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, L.; Mi, X.; Zhao, S.; An, Y.; Xia, X.; Guo, R.; Wei, C. Multi-omics analysis to visualize the dynamic roles of defense genes in the response of tea plants to gray blight. Plant J. 2021, 106, 862–875. [Google Scholar] [CrossRef]
- Kumar, V.; Srivastava, A.K.; AbdElgawad, H. Transcriptional and Post-Transcriptional Regulation of Plant Growth, Development, and Stress Responses. J. Plant Growth Regul. 2025, 44, 1317–1322. [Google Scholar] [CrossRef]
- Mao, X.; Li, Y.; Rehman, S.U.; Zhao, S.; An, Y.; Xia, X.; Guo, R.; Wei, C. The sucrose non-fermenting 1-related protein kinase 2 (SnRK2) genes are multifaceted players in plant growth, development and response to environmental stimuli. Plant Cell Physiol. 2020, 61, 225–242. [Google Scholar] [CrossRef]
- Peixoto, B.; Baena-González, E. Management of plant central metabolism by SnRK1 protein kinases. J. Exp. Bot. 2022, 73, 7068–7082. [Google Scholar] [CrossRef]
- Radani, Y.; Li, R.; Korboe, H.M.; Ma, H.; Yang, L. Transcriptional and post-translational regulation of plant bHLH transcription factors during the response to environmental stresses. Plants 2023, 12, 2113. [Google Scholar] [CrossRef]
- Singh, A.; Roychoudhury, A. Gene regulation at transcriptional and post-transcriptional levels to combat salt stress in plants. Physiol. Plant. 2021, 173, 1556–1572. [Google Scholar] [CrossRef]
- Wang, Q.; Qin, G.; Cao, M.; Chen, R.; He, Y.; Yang, L.; Zeng, Z.; Yu, Y.; Gu, Y.; Xing, W.; et al. A phosphorylation-based switch controls TAA1-mediated auxin biosynthesis in plants. Nat. Commun. 2020, 11, 679. [Google Scholar] [CrossRef] [PubMed]
- Robert, K.Y.; Bieberich, E. Regulation of glycosyltransferases in ganglioside biosynthesis by phosphorylation and dephosphorylation. Mol. Cell. Endocrinol. 2001, 177, 19–24. [Google Scholar] [CrossRef]
- Song, J.; Lin, R.; Tang, M.; Wang, L.; Fan, P.; Xia, X.; Yu, J.; Zhou, Y. SlMPK1- and SlMPK2-mediated SlBBX17 phosphorylation positively regulates CBF-dependent cold tolerance in tomato. New Phytol. 2023, 239, 1887–1902. [Google Scholar] [CrossRef]
- Wang, B.X.; Sun, Z.G.; Liu, Y.; Xu, B.; Li, J.; Chi, M.; Xing, Y.; Yang, B.; Li, J.; Liu, J.; et al. A pervasive phosphorylation cascade modulation of plant transcription factors in response to abiotic stress. Planta 2023, 258, 73. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.J.; Shan, W.; Liu, X.C.; Zhu, L.S.; Wei, W.; Yang, Y.Y.; Guo, Y.F.; Bouzayen, M.; Chen, J.Y.; Lu, W.J.; et al. Phosphorylation of transcription factor bZIP21 by MAP kinase MPK6-3 enhances banana fruit ripening. Plant Physiol. 2022, 188, 1665–1685. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.J.; Zhou, Y.; Zhang, Y.; Su, Y.H.; Xu, T. Protein phosphorylation: A molecular switch in plant signaling. Cell Rep. 2023, 42, 112856. [Google Scholar] [CrossRef] [PubMed]



| F Expressed Genes | GJV Expressed Genes | MJV Expressed Genes | |||
|---|---|---|---|---|---|
| CsSnRK2.1; CsSnRK3.3; CsSnRK3.4; CsSnRK3.7; CsSnRK3.15 | NF/NL≥2 and CF/CL≥2 | CsSnRK2.4; CsSnRK3.7 | NGJV/NL≥2 and CGJV/CL≥2 | CsSnRK3.6; CsSnRK3.7 | NMJV/NL≥2 and CMJV/CL≥2 |
| CsPP2C31; CsPP2C32; CsPP2C36; CsPP2C37; CsPP2C40; CsPP2C48; CsPP2C53; CsPP2C54; CsPP2C55 | CsPP2C36; CsPP2C37; CsPP2C40; CsPP2C53 | CsPP2C14; CsPP2C15; CsPP2C23; CsPP2C31; CsPP2C33; CsPP2C36; CsPP2C37; CsPP2C40; CsPP2C53; CsPP2C56 | |||
| CsPYL2; CsPYL4 | CsPYL1; CsPYL2; CsPYL5; CsPYL6 | CsPYL4; CsPYL6 | |||
| Module | Phytoene | Phytofluene | Lycopene | Sum | Rank |
|---|---|---|---|---|---|
| MEskyblue | 0.34 | 0.58 | 0.68 | 1.60 | 1 |
| MEturquoise | 0.24 | 0.58 | 0.63 | 1.45 | 2 |
| MEsteelblue | 0.32 | 0.52 | 0.57 | 1.41 | 3 |
| MEviolet | 0.18 | 0.39 | 0.38 | 0.95 | 4 |
| MEgrey | 0.31 | 0.30 | 0.32 | 0.93 | 5 |
| MEroyalblue | 0.12 | 0.37 | 0.35 | 0.84 | 6 |
| MEcyan | 0.22 | 0.21 | 0.34 | 0.77 | 7 |
| Module | Count | Percentage |
|---|---|---|
| turquoise | 13 | 13.48% |
| black | 10 | 11.24% |
| red | 10 | 11.24% |
| blue | 9 | 10.11% |
| yellow | 9 | 10.11% |
| brown | 6 | 6.74% |
| cyan | 6 | 6.74% |
| purple | 5 | 5.62% |
| royalblue | 5 | 5.62% |
| salmon | 4 | 4.49% |
| grey60 | 3 | 3.37% |
| midnightblue | 2 | 2.25% |
| darkolivegreen | 1 | 1.12% |
| darkred | 1 | 1.12% |
| greenyellow | 1 | 1.12% |
| lightcyan | 1 | 1.12% |
| lightgreen | 1 | 1.12% |
| lightyellow | 1 | 1.12% |
| magenta | 1 | 1.12% |
| pink | 1 | 1.12% |
| Classification | Gene Name | Gene ID | Connectivity | Rank by Connectivity |
|---|---|---|---|---|
| SnRK | CsSnRK1 | Cs6g21650 | 750.35 | 602 |
| CsSnRK3.5 | Cs4g01450 | 102.98 | 3788 | |
| CsSnRK3.6 | orange1.1t02758 | 522.88 | 1331 | |
| CsSnRK3.16 | Cs2g28990 | 582.86 | 1093 | |
| PP2C | CsPP2C14 | Cs3g20540 | 971.09 | 106 |
| CsPP2C15 | Cs9g18330 | 796.31 | 496 | |
| CsPP2C33 | Cs9g03785 | 455.52 | 1578 | |
| CsPP2C35 | orange1.1t03838 | 337.26 | 2119 | |
| CsPP2C38 | Cs7g22880 | 303.26 | 2288 | |
| CsPP2C40 | Cs9g04470 | 998.44 | 67 | |
| CsPP2C43 | orange1.1t02237 | 230.55 | 2780 | |
| CsPP2C56 | Cs9g06410 | 824.48 | 433 | |
| PYL | CsPYL6 | Cs9g18410 | 348.99 | 2060 |
| Transcription factor | SPOROCYTELESS/NOZZLE (SPL/NZZ) | Cs1g06080 | 705.83 | 711 |
| NAC | Cs1g09660 | 99.36 | 3825 | |
| bHLH | Cs1g02580 | 612.51 | 995 | |
| AP2-EREBP (ERF) | Cs1g03300 | 164.06 | 3236 | |
| Carotenoid biosynthesis structural genes | CsPSY1 | Cs6g15910 | 295.83 | 2332 |
| CsZISO | Cs5g24730 | 343.93 | 2082 | |
| CsZDS | Cs3g11180 | 396.55 | 1847 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, P.; Shi, Z.; Liu, T.; Ji, J.; Li, J.; Li, W.; Sun, C. SnRK-PP2C-PYL Gene Families in Citrus sinensis: Genomic Characterization and Regulatory Roles in Carotenoid Metabolism. Metabolites 2025, 15, 610. https://doi.org/10.3390/metabo15090610
Lu P, Shi Z, Liu T, Ji J, Li J, Li W, Sun C. SnRK-PP2C-PYL Gene Families in Citrus sinensis: Genomic Characterization and Regulatory Roles in Carotenoid Metabolism. Metabolites. 2025; 15(9):610. https://doi.org/10.3390/metabo15090610
Chicago/Turabian StyleLu, Pengjun, Zhenting Shi, Tao Liu, Jianqiu Ji, Jing Li, Wentao Li, and Chongbo Sun. 2025. "SnRK-PP2C-PYL Gene Families in Citrus sinensis: Genomic Characterization and Regulatory Roles in Carotenoid Metabolism" Metabolites 15, no. 9: 610. https://doi.org/10.3390/metabo15090610
APA StyleLu, P., Shi, Z., Liu, T., Ji, J., Li, J., Li, W., & Sun, C. (2025). SnRK-PP2C-PYL Gene Families in Citrus sinensis: Genomic Characterization and Regulatory Roles in Carotenoid Metabolism. Metabolites, 15(9), 610. https://doi.org/10.3390/metabo15090610




