Untargeted Metabolomics Reveals Acylcarnitines as Major Metabolic Targets of Resveratrol in Breast Cancer Cells
Abstract
1. Introduction
2. Methods
2.1. Materials
2.2. Cell Culture
2.3. Treatment of MCF-7 and MDA-MB-231 Cells with Resveratrol
2.4. Metabolite Extraction
2.5. UHPLC-HRMS Data Acquisition and Multivariate Statistics
2.6. Pathway Analysis
2.7. Metabolite Identification
2.8. Univariate Statistics
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arnold, M.; Morgan, E.; Rumgay, H.; Mafra, A.; Singh, D.; Laversanne, M.; Vignat, J.; Gralow, J.R.; Cardoso, F.; Siesling, S.; et al. Current and future burden of breast cancer: Global statistics for 2020 and 2040. Breast 2022, 66, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Petrucelli, N.; Daly, M.B.; Pal, T. BRCA1- and BRCA2-Associated Hereditary Breast and Ovarian Cancer Summary Genetic counseling Suggestive Findings. Gene Rev. 1998, 1–37. [Google Scholar]
- Redig, A.J.; McAllister, S.S. Breast cancer as a systemic disease: A view of metastasis. J. Intern. Med. 2013, 274, 113–126. [Google Scholar] [CrossRef]
- Zeichner, S.B.; Terawaki, H.; Gogineni, K. A review of systemic treatment in metastatic triple-negative breast cancer. Breast Cancer Basic Clin. Res. 2016, 10, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Dent, R.; Trudeau, M.; Pritchard, K.I.; Hanna, W.M.; Kahn, H.K.; Sawka, C.A.; Lickley, L.A.; Rawlinson, E.; Sun, P.; Narod, S.A. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin. Cancer Res. 2007, 13, 4429–4434. [Google Scholar] [CrossRef]
- Zhou, Y.; Zheng, J.; Li, Y.; Xu, D.P.; Li, S.; Chen, Y.M.; Li, H. Bin Natural polyphenols for prevention and treatment of cancer. Nutrients 2016, 8, 515. [Google Scholar] [CrossRef]
- Avtanski, D.; Poretsky, L. Phyto-polyphenols as potential inhibitors of breast cancer metastasis. Mol. Med. 2018, 24, 29. [Google Scholar] [CrossRef]
- Dayem, A.A.; Choi, H.Y.; Yang, G.M.; Kim, K.; Saha, S.K.; Cho, S.G. The anti-cancer effect of polyphenols against breast cancer and cancer stem cells: Molecular mechanisms. Nutrients 2016, 8, 581. [Google Scholar] [CrossRef]
- Keating, E.; Martel, F. Antimetabolic Effects of Polyphenols in Breast Cancer Cells: Focus on Glucose Uptake and Metabolism. Front. Nutr. 2018, 5, 25. [Google Scholar] [CrossRef]
- Rushing, B.R.; Wiggs, A.; Molina, S.; Schroder, M.; Sumner, S. Metabolomics Analysis Reveals Novel Targets of Chemosensitizing Polyphenols and Omega-3 Polyunsaturated Fatty Acids in Triple Negative Breast Cancer Cells. Int. J. Mol. Med. 2023, 24, 4406. [Google Scholar]
- Niedzwiecki, A.; Roomi, M.W.; Kalinovsky, T.; Rath, M. Anticancer efficacy of polyphenols and their combinations. Nutrients 2016, 8, 552. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Virmani, T.; Sharma, A.; Pathak, K. Codelivery of Phytochemicals with Conventional Anticancer Drugs in Form of Nanocarriers. Pharmaceutics 2023, 15, 889. [Google Scholar] [CrossRef]
- Singh, S.; Sharma, B.; Kanwar, S.S.; Kumar, A. Lead phytochemicals for anticancer drug development. Front. Plant Sci. 2016, 7, 1667. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, J.; Abbasi, B.A.; Mahmood, T.; Kanwal, S.; Ali, B.; Shah, S.A.; Khalil, A.T. Plant-derived anticancer agents: A green anticancer approach. Asian Pac. J. Trop. Biomed. 2017, 7, 1129–1150. [Google Scholar] [CrossRef]
- Naeem, A.; Hu, P.; Yang, M.; Zhang, J.; Liu, Y.; Zhu, W.; Zheng, Q. Natural Products as Anticancer Agents: Current Status and Future Perspectives. Molecules 2022, 27, 8367. [Google Scholar] [CrossRef]
- Zhang, M.; Chen, X.; Radacsi, N. New tricks of old drugs: Repurposing non-chemo drugs and dietary phytochemicals as adjuvants in anti-tumor therapies. J. Control. Release 2021, 329, 96–120. [Google Scholar] [CrossRef]
- Berman, A.Y.; Motechin, R.A.; Wiesenfeld, M.Y.; Holz, M.K. The therapeutic potential of resveratrol: A review of clinical trials. npj Precis. Oncol. 2017, 1, 35. [Google Scholar] [CrossRef]
- Shukla, Y.; Singh, R. Resveratrol and cellular mechanisms of cancer prevention. Ann. N. Y. Acad. Sci. 2011, 1215, 1–8. [Google Scholar] [CrossRef]
- Sun, Y.; Zhou, Q.M.; Lu, Y.Y.; Zhang, H.; Chen, Q.L.; Zhao, M.; Su, S.B. Resveratrol inhibits the migration and metastasis of MDA-MB-231 human breast cancer by reversing TGF-β1-induced epithelial-mesenchymal transition. Molecules 2019, 24, 1131. [Google Scholar] [CrossRef] [PubMed]
- Horgan, X.J.; Tatum, H.; Brannan, E.; Paull, D.H.; Rhodes, L.V. Resveratrol analogues surprisingly effective against triple-negative breast cancer, independent of ERα. Oncol. Rep. 2019, 41, 3517–3526. [Google Scholar] [CrossRef]
- Vladu, A.F.; Ficai, D.; Ene, A.G.; Ficai, A. Combination Therapy Using Polyphenols: An Efficient Way to Improve Antitumoral Activity and Reduce Resistance. Int. J. Mol. Sci. 2022, 23, 10244. [Google Scholar] [CrossRef] [PubMed]
- Brockmueller, A.; Sameri, S.; Liskova, A.; Zhai, K.; Varghese, E.; Samuel, S.M.; Büsselberg, D.; Kubatka, P.; Shakibaei, M. Resveratrol’s anti-cancer effects through the modulation of tumor glucose metabolism. Cancers 2021, 13, 188. [Google Scholar] [CrossRef]
- Blanquer-Rosselló, M.d.M.; Hernández-López, R.; Roca, P.; Oliver, J.; Valle, A. Resveratrol induces mitochondrial respiration and apoptosis in SW620 colon cancer cells. Biochim. Biophys. Acta—Gen. Subj. 2017, 1861, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Gimeno-Mallench, L.; Mas-Bargues, C.; Inglés, C.; Olaso, G.; Borras, C.; Gambini, J.; Vina, J. Resveratrol shifts energy metabolism to increase lipid oxidation in healthy old mice. Biomed. Pharmacother. 2019, 118, 109130. [Google Scholar] [CrossRef] [PubMed]
- Jäger, W.; Gruber, A.; Giessrigl, B.; Krupitza, G.; Szekeres, T.; Sonntag, D. Metabolomic analysis of resveratrol-induced effects in the human breast cancer cell lines MCF-7 and MDA-MB-231. OMICS J. Integr. Biol. 2011, 15, 9–14. [Google Scholar] [CrossRef]
- Rushing, B.R.; Fogle, H.M.; Sharma, J.; You, M.; Mccormac, J.P.; Molina, S.; Sumner, S.; Krupenko, N.I.; Krupenko, S.A. Exploratory Metabolomics Underscores the Folate Enzyme ALDH1L1 as a Regulator of Glycine and Methylation Reactions. Molecules 2022, 27, 8394. [Google Scholar] [CrossRef]
- Rushing, B.R.; Schroder, M.; Sumner, S.C.J. Comparison of Lysis and Detachment Sample Preparation Methods for Cultured Triple-Negative Breast Cancer Cells Using UHPLC–HRMS-Based Metabolomics. Metabolites 2022, 12, 168. [Google Scholar] [CrossRef]
- Rushing, B.R.; Tilley, S.; Molina, S.; Schroder, M.; Sumner, S. Commonalities in Metabolic Reprogramming Between Tobacco Use and Oral Cancer. Int. J. Environ. Res. Public Health 2022, 19, 10261. [Google Scholar] [CrossRef]
- Joseph, S.; Zhang, X.; Droby, G.N.; Wu, D.; Bae-Jump, V.; Lyons, S.; Mordant, A.; Mills, A.; Herring, L.; Rushing, B.; et al. MAPK14/p38α shapes the molecular landscape of endometrial cancer and promotes tumorigenic characteristics. Cell Rep. 2025, 44, 115104. [Google Scholar] [CrossRef]
- Rushing, B.R.; Molina, S.; Sumner, S. Metabolomics Analysis Reveals Altered Metabolic Pathways and Response to Doxorubicin in Drug-Resistant Triple-Negative Breast Cancer Cells. Metabolites 2023, 13, 865. [Google Scholar] [CrossRef]
- Rushing, B.R.; McRitchie, S.; Arbeeva, L.; Nelson, A.E.; Azcarate-Peril, M.A.; Li, Y.Y.; Qian, Y.; Pathmasiri, W.; Sumner, S.C.J.; Loeser, R.F. Fecal metabolomics reveals products of dysregulated proteolysis and altered microbial metabolism in obesity-related osteoarthritis. Osteoarthr. Cartil. 2022, 30, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Välikangas, T.; Suomi, T.; Elo, L.L. A systematic evaluation of normalization methods in quantitative label-free proteomics. Brief. Bioinform. 2018, 19, 1–11. [Google Scholar] [CrossRef]
- Jodynis-liebert, J.; Kujawska, M. Biphasic dose-response induced by phytochemicals: Experimental evidence. J. Clin. Med. 2020, 9, 718. [Google Scholar] [CrossRef]
- Calabrese, E.J.; Mattson, M.P.; Calabrese, V. Dose response biology: The case of resveratrol. Hum. Exp. Toxicol. 2010, 29, 1034–1037. [Google Scholar] [CrossRef] [PubMed]
- Shaito, A.; Posadino, A.M.; Younes, N.; Hasan, H.; Halabi, S.; Alhababi, D.; Al-Mohannadi, A.; Abdel-Rahman, W.M.; Eid, A.H.; Nasrallah, G.K.; et al. Potential adverse effects of resveratrol: A literature review. Int. J. Mol. Sci. 2020, 21, 2084. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Dudley, J.I.; Das, D.K. Dose-dependency of resveratrol in providing health benefits. Dose-Response 2010, 8, 478–500. [Google Scholar] [CrossRef]
- Salehi, B.; Mishra, A.P.; Nigam, M.; Sener, B.; Kilic, M.; Sharifi-Rad, M.; Fokou, P.V.T.; Martins, N.; Sharifi-Rad, J. Resveratrol: A double-edged sword in health benefits. Biomedicines 2018, 6, 91. [Google Scholar] [CrossRef]
- Jiang, L.; Yu, H.; Wang, C.; He, F.; Shi, Z.; Tu, H.; Ning, N.; Duan, S.; Zhao, Y. The Anti-Cancer Effects of Mitochondrial-Targeted Triphenylphosphonium–Resveratrol Conjugate on Breast Cancer Cells. Pharmaceuticals 2022, 15, 1271. [Google Scholar] [CrossRef]
- Gomez, L.S.; Zancan, P.; Marcondes, M.C.; Ramos-Santos, L.; Meyer-Fernandes, J.R.; Sola-Penna, M.; Da Silva, D. Resveratrol decreases breast cancer cell viability and glucose metabolism by inhibiting 6-phosphofructo-1-kinase. Biochimie 2013, 95, 1336–1343. [Google Scholar] [CrossRef]
- Dambrova, M.; Makrecka-Kuka, M.; Kuka, J.; Vilskersts, R.; Nordberg, D.; Attwood, M.M.; Smesny, S.; Sen, Z.D.; Guo, A.C.; Oler, E.; et al. Acylcarnitines: Nomenclature, Biomarkers, Therapeutic Potential, Drug Targets, and Clinical Trials. Pharmacol. Rev. 2022, 74, 506–551. [Google Scholar] [CrossRef]
- Melone, M.A.B.; Valentino, A.; Margarucci, S.; Galderisi, U.; Giordano, A.; Peluso, G. The carnitine system and cancer metabolic plasticity. Cell Death Dis. 2018, 9, 228. [Google Scholar] [CrossRef] [PubMed]
- Reuter, S.E.; Evans, A.M. Carnitine and acylcarnitines: Pharmacokinetic, pharmacological and clinical aspects. Clin. Pharmacokinet. 2012, 51, 553–572. [Google Scholar] [CrossRef] [PubMed]
- Schooneman, M.G.; Vaz, F.M.; Houten, S.M.; Soeters, M.R. Acylcarnitines: Reflecting or inflicting insulin resistance? Diabetes 2013, 62, 1–8. [Google Scholar] [CrossRef]
- Longo, N.; Frigeni, M.; Pasquali, M. Carnitine transport and fatty acid oxidation. Biochim Biophys Acta 2016, 1863, 2422–2435. [Google Scholar] [CrossRef]
- McCoin, C.S.; Knotts, T.A.; Adams, S.H. Acylcarnitines-old actors auditioning for new roles in metabolic physiology. Nat. Rev. Endocrinol. 2015, 11, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Knottnerus, S.J.G.; Bleeker, J.C.; Wüst, R.C.; Ferdinandusse, S.; IJlst, L.; Wijburg, F.A.; Wanders, R.J.A.; Visser, G.; Houtkooper, R.H. Disorders of mitochondrial long-chain fatty acid oxidation and the carnitine shuttle. Rev. Endocr. Metab. Disord. 2018, 19, 93–106. [Google Scholar]
- Aires, V.; Delmas, D.; Le Bachelier, C.; Latruffe, N.; Schlemmer, D.; Benoist, J.F.; Djouadi, F.; Bastin, J. Stilbenes and resveratrol metabolites improve mitochondrial fatty acid oxidation defects in human fibroblasts. Orphanet J. Rare Dis. 2014, 9, 79. [Google Scholar] [CrossRef] [PubMed]
- Price, N.L.; Gomes, A.P.; Ling, A.J.Y.; Duarte, F.V.; Martin-Montalvo, A.; North, B.J.; Agarwal, B.; Ye, L.; Ramadori, G.; Teodoro, J.S.; et al. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 2012, 15, 675–690. [Google Scholar] [CrossRef]
- Khan, A.; Aljarbou, A.N.; Aldebasi, Y.H.; Faisal, S.M.; Khan, M.A. Resveratrol suppresses the proliferation of breast cancer cells by inhibiting fatty acid synthase signaling pathway. Cancer Epidemiol. 2014, 38, 765–772. [Google Scholar] [CrossRef]
- Wajner, M.; Amaral, A.U. Mitochondrial dysfunction in fatty acid oxidation disorders: Insights from human and animal studies. Biosci. Rep. 2016, 36, e00281. [Google Scholar] [CrossRef]
- Muoio, D.M.; Neufer, P.D. Lipid-induced mitochondrial stress and insulin action in muscle. Cell Metab. 2012, 15, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Wieckowski, M.R.; Wojtczak, L. Fatty acid-induced uncoupling of oxidative phosphorylation is partly due to opening of the mitochondrial permeability transition pore. FEBS Lett. 1998, 423, 339–342. [Google Scholar] [CrossRef]
- Rottenberg, H.; Hoek, J.B. The path from mitochondrial ROS to aging runs through the mitochondrial permeability transition pore. Aging Cell 2017, 16, 943–955. [Google Scholar] [CrossRef]
- Furuno, T.; Kanno, T.; Arita, K.; Asami, M.; Utsumi, T.; Doi, Y.; Inoue, M.; Utsumi, K. Roles of long chain fatty acids and carnitine in mitochondrial membrane permeability transition. Biochem. Pharmacol. 2001, 62, 1037–1046. [Google Scholar] [CrossRef] [PubMed]
- Berezhnov, A.V.; Fedotova, E.I.; Nenov, M.N.; Kasymov, V.A.; Pimenov, O.Y.; Dynnik, V.V. Dissecting cellular mechanisms of long-chain acylcarnitines-driven cardiotoxicity: Disturbance of calcium homeostasis, activation of Ca2+-dependent phospholipases, and mitochondrial energetics collapse. Int. J. Mol. Sci. 2020, 21, 7461. [Google Scholar] [CrossRef] [PubMed]
- Babich, H.; Schuck, A.G.; Weisburg, J.H.; Zuckerbraun, H.L. Research strategies in the study of the pro-oxidant nature of polyphenol nutraceuticals. J. Toxicol. 2011, 2011, 467305. [Google Scholar] [CrossRef]
- Song, B.; Wang, W.; Tang, X.; Goh, R.M.W.J.; Thuya, W.L.; Ho, P.C.L.; Chen, L.; Wang, L. Inhibitory Potential of Resveratrol in Cancer Metastasis: From Biology to Therapy. Cancers 2023, 15, 2758. [Google Scholar] [CrossRef]
- McCann, M.R.; De la Rosa, M.V.G.; Rosania, G.R.; Stringer, K.A. L-carnitine and acylcarnitines: Mitochondrial biomarkers for precision medicine. Metabolites 2021, 11, 51. [Google Scholar] [CrossRef]
- Miller, M.J.; Cusmano-Ozog, K.; Oglesbee, D.; Young, S. Laboratory analysis of acylcarnitines, 2020 update: A technical standard of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2021, 23, 249–258. [Google Scholar] [CrossRef]
- Silva, P.; Sureda, A.; Tur, J.A.; Andreoletti, P.; Cherkaoui-Malki, M.; Latruffe, N. How efficient is resveratrol as an antioxidant of the Mediterranean diet, towards alterations during the aging process? Free Radic. Res. 2019, 53, 1101–1112. [Google Scholar] [CrossRef]
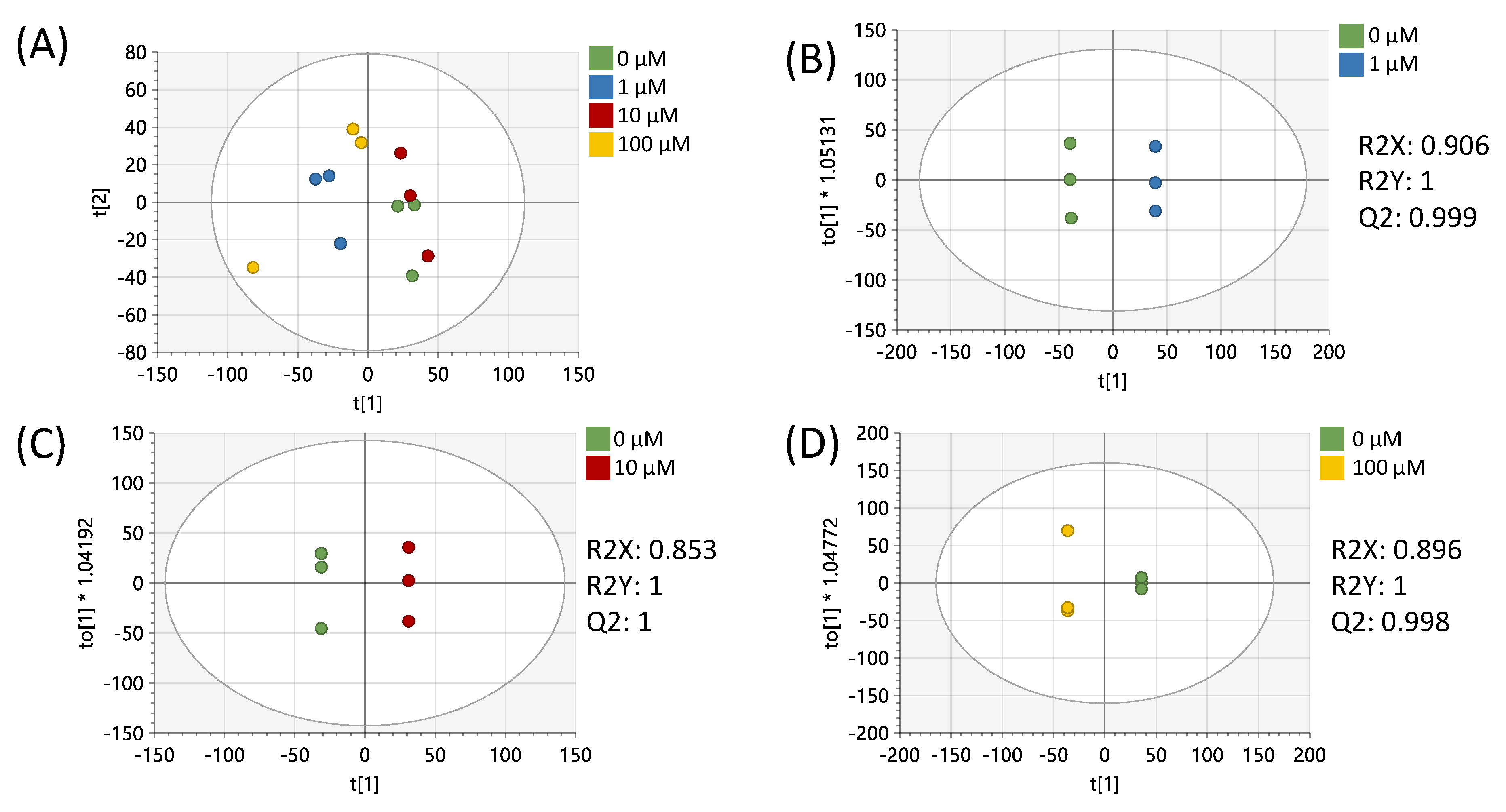

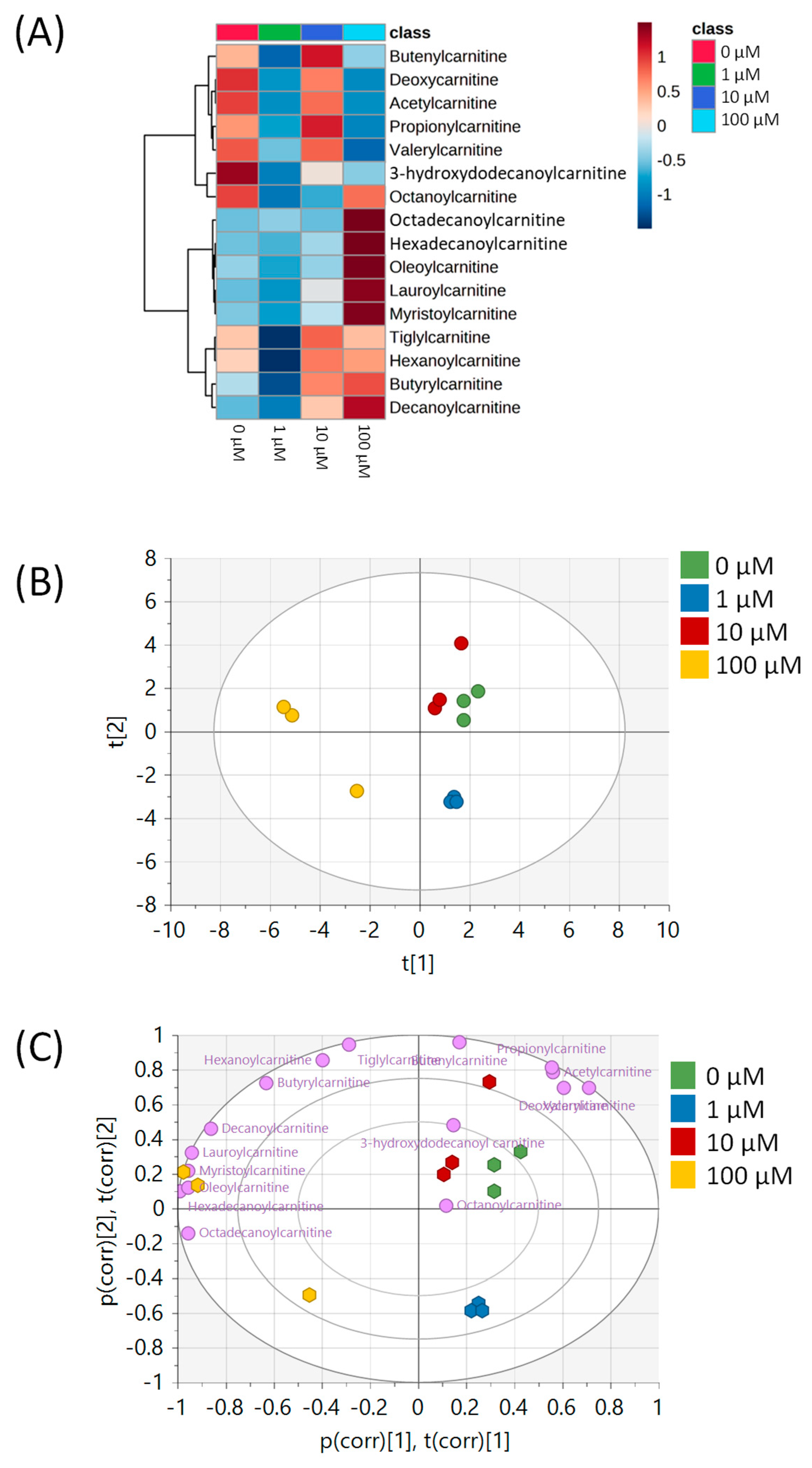


| Pathway Name | Pathway Total a | Hits.total b | Hits.sig c | FET d |
|---|---|---|---|---|
| Bile acid biosynthesis | 82 | 35 | 27 | 0.0036 |
| Carnitine shuttle | 72 | 18 | 14 | 0.0051 |
| Urea cycle/amino group metabolism | 85 | 25 | 14 | 0.0104 |
| Aspartate and asparagine metabolism | 114 | 45 | 20 | 0.0128 |
| Putative anti-inflammatory metabolite formation from EPA | 27 | 2 | 2 | 0.0159 |
| Glutathione metabolism | 19 | 9 | 6 | 0.0243 |
| Purine metabolism | 80 | 19 | 12 | 0.0288 |
| Lysine metabolism | 52 | 16 | 7 | 0.0406 |
| Fold Change | p-Value | VIP | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Metabolite Name | Ontology Level | 1 µM/ 0 µM | 10 µM/ 0 µM | 100 µM/ 0 µM | 0 µM vs. 1 µM | 0 µM vs. 10 µM | 0 µM vs. 100 µM | 0 µM vs. 1 µM | 0 µM vs. 10 µM | 0 µM vs. 100 µM |
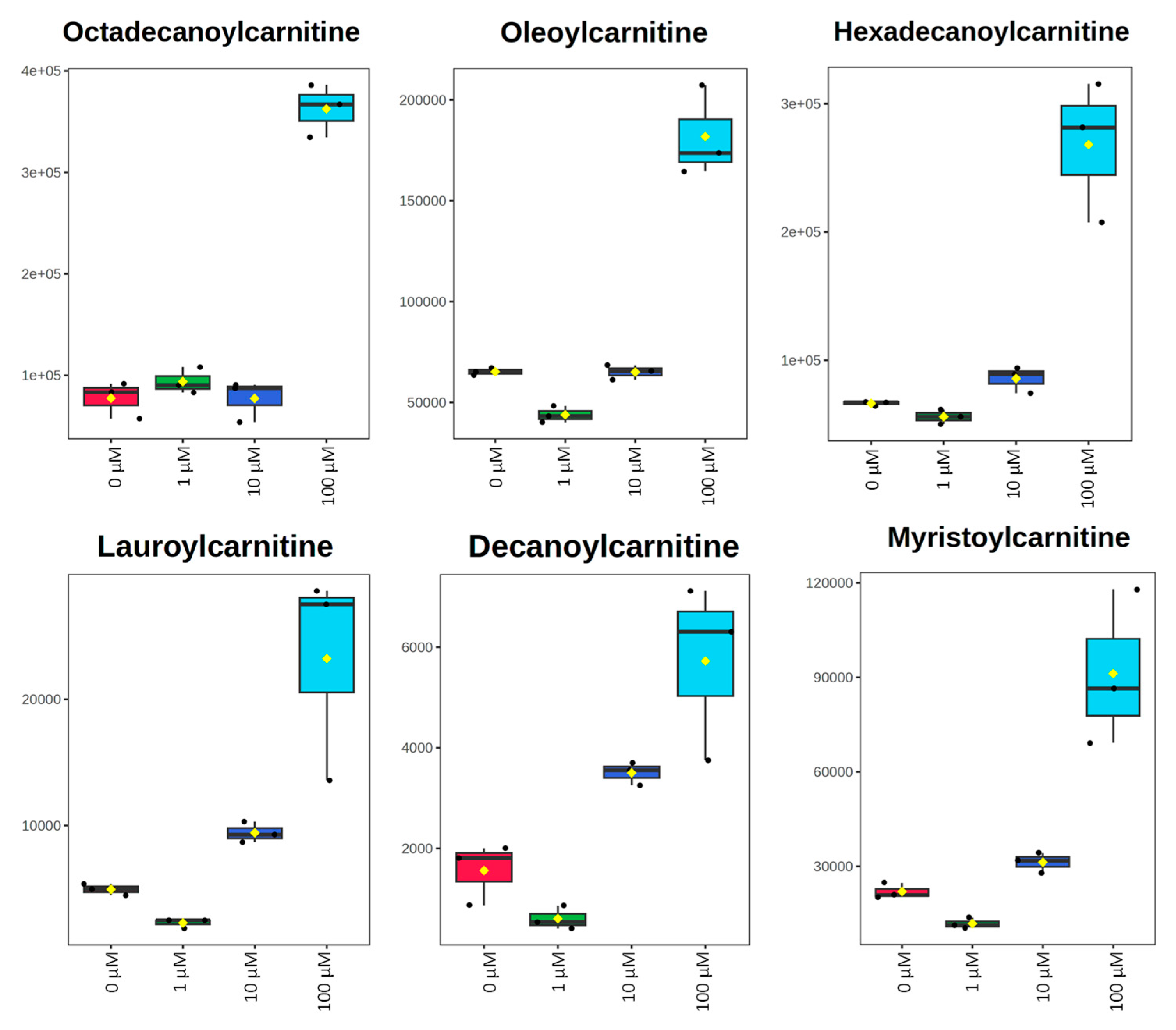
| Lauroylcarnitine | OL2b | 0.46 | 1.91 | 4.70 | 1.3 × 10−3 | 1.2 × 10−3 | 0.020 | 1.33 | 1.58 | 1.32 |
| Octadecanoylcarnitine | OL2b | 1.21 | 1.00 | 4.67 | 0.27 | 0.99 | 9.8 × 10−5 | 0.90 | 0.77 | 1.43 |
| Myristoylcarnitine | OL2b | 0.54 | 1.42 | 4.15 | 3.8 × 10−3 | 0.015 | 8.5 × 10−3 | 1.31 | 1.46 | 1.35 |
| Hexadecanoylcarnitine | OL2b | 0.84 | 1.29 | 4.04 | 0.040 | 0.031 | 3.2 × 10−3 | 1.14 | 1.43 | 1.39 |
| Decanoylcarnitine | OL2b | 0.39 | 2.24 | 3.67 | 0.063 | 6.5 × 10−3 | 0.018 | 1.16 | 1.53 | 1.33 |
| Oleoylcarnitine | OL2b | 0.67 | 1.00 | 2.79 | 1.2 × 10−3 | 0.95 | 8.6 × 10−4 | 1.33 | 0.77 | 1.41 |
| Butyrylcarnitine | OL2a | 0.47 | 1.47 | 1.60 | 2.1 × 10−3 | 3.4 × 10−3 | 0.29 | 1.32 | 1.56 | 0.99 |
| Hexanoylcarnitine | OL2a | 0.33 | 1.19 | 1.12 | 0.012 | 0.35 | 0.76 | 1.27 | 1.01 | 0.76 |
| Tiglylcarnitine | OL1 | 0.38 | 1.18 | 1.01 | 1.3 × 10−3 | 0.25 | 0.96 | 1.33 | 1.10 | 0.73 |
| Octanoylcarnitine | OL2a | 0.43 | 0.53 | 0.94 | 0.61 | 0.68 | 0.97 | 0.80 | 0.81 | 0.75 |
| Butenylcarnitine | OL2a | 0.36 | 1.31 | 0.67 | 2.0 × 10−3 | 0.30 | 0.16 | 1.32 | 1.04 | 1.10 |
| Acetylcarnitine | OL1 | 0.44 | 1.22 | 0.57 | 3.9 × 10−3 | 0.30 | 0.035 | 1.31 | 1.05 | 1.28 |
| Deoxycarnitine | OL1 | 0.57 | 0.92 | 0.56 | 0.010 | 0.57 | 0.015 | 1.27 | 0.87 | 1.33 |
| 3-OH-dodecanoylcarnitine | OL2a | 0.21 | 0.54 | 0.39 | 0.17 | 0.45 | 0.31 | 1.03 | 0.65 | 0.96 |
| Propionylcarnitine | OL1 | 0.38 | 1.26 | 0.29 | 1.7 × 10−3 | 0.30 | 2.0 × 10−3 | 1.32 | 1.04 | 1.40 |
| Valerylcarnitine | OL1 | 0.48 | 0.98 | 0.24 | 1.4 × 10−3 | 0.87 | 8.1 × 10−4 | 1.33 | 0.77 | 1.42 |
| Fold Change | p-Value | VIP | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Metabolite Name | Ontology Level | 25 µM/ 0 µM | 50 µM/ 0 µM | 100 µM/ 0 µM | 0 µM vs. 25 µM | 0 µM vs. 50 µM | 0 µM vs. 100 µM | 0 µM vs. 25 µM | 0 µM vs. 50 µM | 0 µM vs. 100 µM |
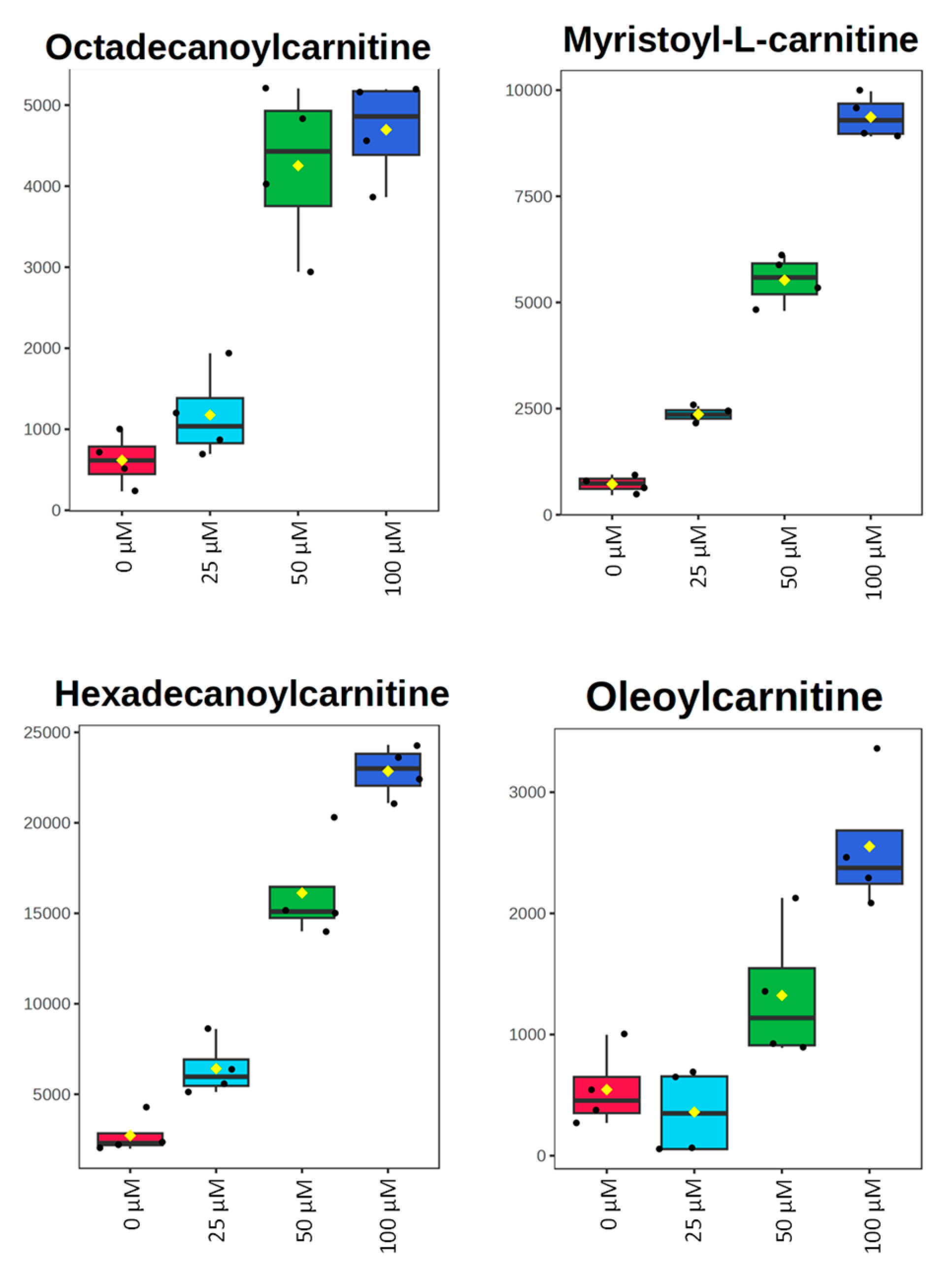
| Octadecanoylcarnitine | OL1 | 1.91 | 6.89 | 7.61 | 0.13 | 4.6 × 10−4 | 2.5 × 10−5 | 0.98 | 1.43 | 1.42 |
| Oleoylcarnitine | OL1 | 0.61 | 2.43 | 4.68 | 0.43 | 0.057 | 7.9 × 10−4 | 0.87 | 1.18 | 1.37 |
| Hexadecanoylcarnitine | OL1 | 2.36 | 5.93 | 8.40 | 7.4 × 10−3 | 1.2 × 10−4 | 4.6 × 10−7 | 1.37 | 1.47 | 1.45 |
| Myristoylcarnitine | OL1 | 3.27 | 7.65 | 12.96 | 1.7 × 10−5 | 4.5 × 10−6 | 6.7 × 10−8 | 1.52 | 1.49 | 1.45 |
| Butenylcarnitine | OL2a | 1.00 | 0.91 | 1.27 | 0.96 | 0.45 | 5.9 × 10−3 | 0.64 | 0.89 | 1.27 |
| Valerylcarnitine | OL1 | 0.69 | 0.49 | 0.33 | 3.5 × 10−4 | 3.9 × 10−6 | 7.4 × 10−7 | 1.48 | 1.50 | 1.45 |
| Tiglylcarnitine | OL1 | 0.91 | 1.37 | 1.71 | 0.15 | 7.2 × 10−5 | 1.6 × 10−4 | 1.06 | 1.47 | 1.41 |
| Isobutyrylcarnitine | OL1 | 1.21 | 2.20 | 2.45 | 1.2 × 10−3 | 2.2 × 10−7 | 2.8 × 10−7 | 1.45 | 1.51 | 1.45 |
| Propionylcarnitine | OL1 | 0.67 | 0.52 | 0.35 | 1.0 × 10−5 | 8.1 × 10−8 | 2.8 × 10−9 | 1.52 | 1.51 | 1.45 |
| O-Acetylcarnitine | OL1 | 0.72 | 0.72 | 0.66 | 0.015 | 0.011 | 0.017 | 1.28 | 1.30 | 1.19 |
| Carnitine | OL1 | 0.87 | 0.71 | 0.52 | 0.27 | 8.6 × 10−3 | 3.1 × 10−4 | 0.96 | 1.32 | 1.39 |
| Deoxycarnitine | OL1 | 0.90 | 0.91 | 0.75 | 0.11 | 0.66 | 2.1 × 10−2 | 1.06 | 0.58 | 1.16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Falcone, I.G.; Rushing, B.R. Untargeted Metabolomics Reveals Acylcarnitines as Major Metabolic Targets of Resveratrol in Breast Cancer Cells. Metabolites 2025, 15, 250. https://doi.org/10.3390/metabo15040250
Falcone IG, Rushing BR. Untargeted Metabolomics Reveals Acylcarnitines as Major Metabolic Targets of Resveratrol in Breast Cancer Cells. Metabolites. 2025; 15(4):250. https://doi.org/10.3390/metabo15040250
Chicago/Turabian StyleFalcone, Isabella G., and Blake R. Rushing. 2025. "Untargeted Metabolomics Reveals Acylcarnitines as Major Metabolic Targets of Resveratrol in Breast Cancer Cells" Metabolites 15, no. 4: 250. https://doi.org/10.3390/metabo15040250
APA StyleFalcone, I. G., & Rushing, B. R. (2025). Untargeted Metabolomics Reveals Acylcarnitines as Major Metabolic Targets of Resveratrol in Breast Cancer Cells. Metabolites, 15(4), 250. https://doi.org/10.3390/metabo15040250









