Single-Nucleus RNA Sequencing Reveals Muscle-Region-Specific Differences in Fibro-Adipogenic Progenitors Driving Intramuscular Fat Accumulation
Abstract
1. Introduction
2. Materials and Methods
2.1. Muscle Samples
2.2. RNA Preparation from Muscle Tissue
2.3. RNA-Seq Analysis
2.4. Single-Nucleus RNA Sequencing
2.5. Bioinformatics Analysis
2.6. Immunostaining Analysis
2.7. Tissue Staining
3. Results
3.1. Comparison of Gene Expression in Different Muscle Tissues
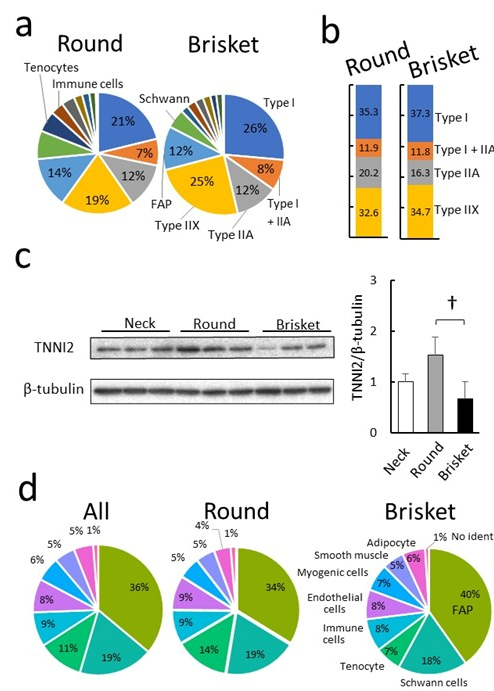
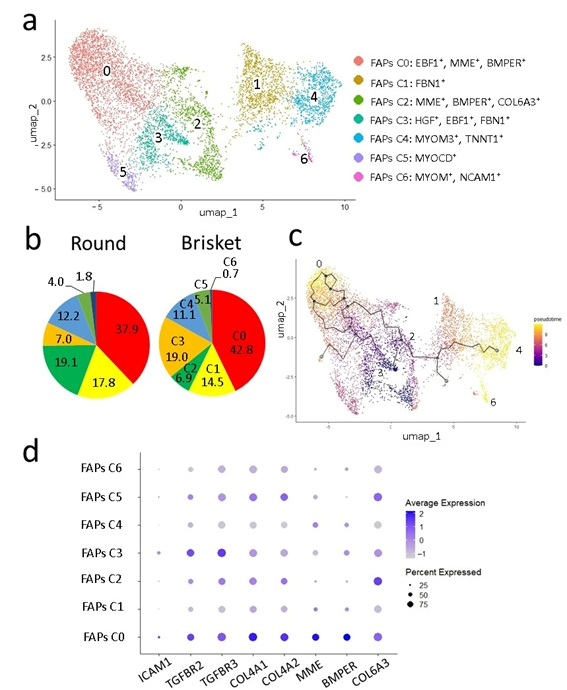
3.2. Comparison of Cells Using Single-Nucleus RNA-Seq Analysis
4. Discussion
4.1. Muscle Tissue Composition
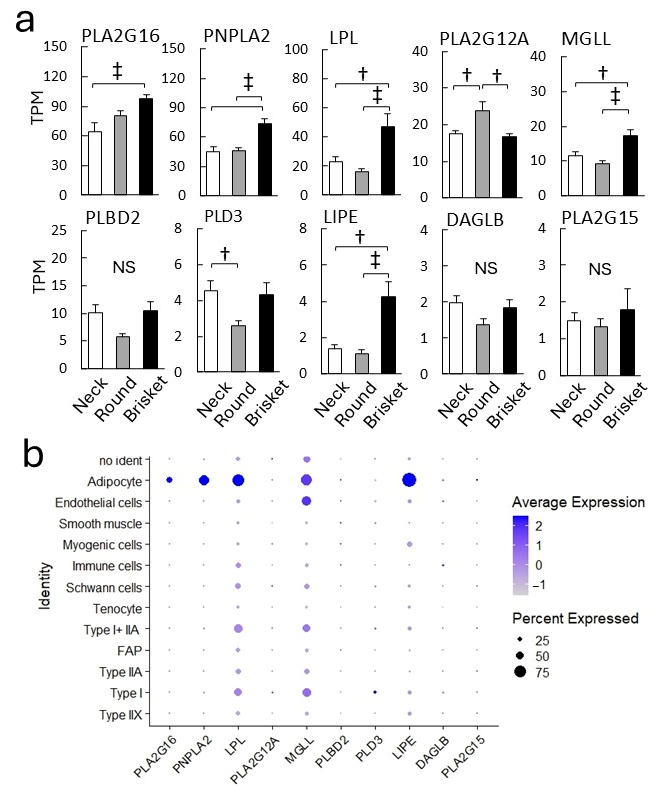
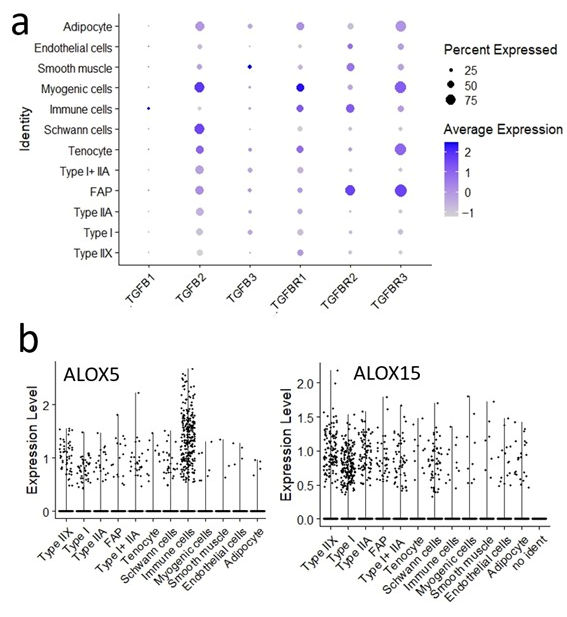
4.2. Lipid Metabolism and Lipoxygenase
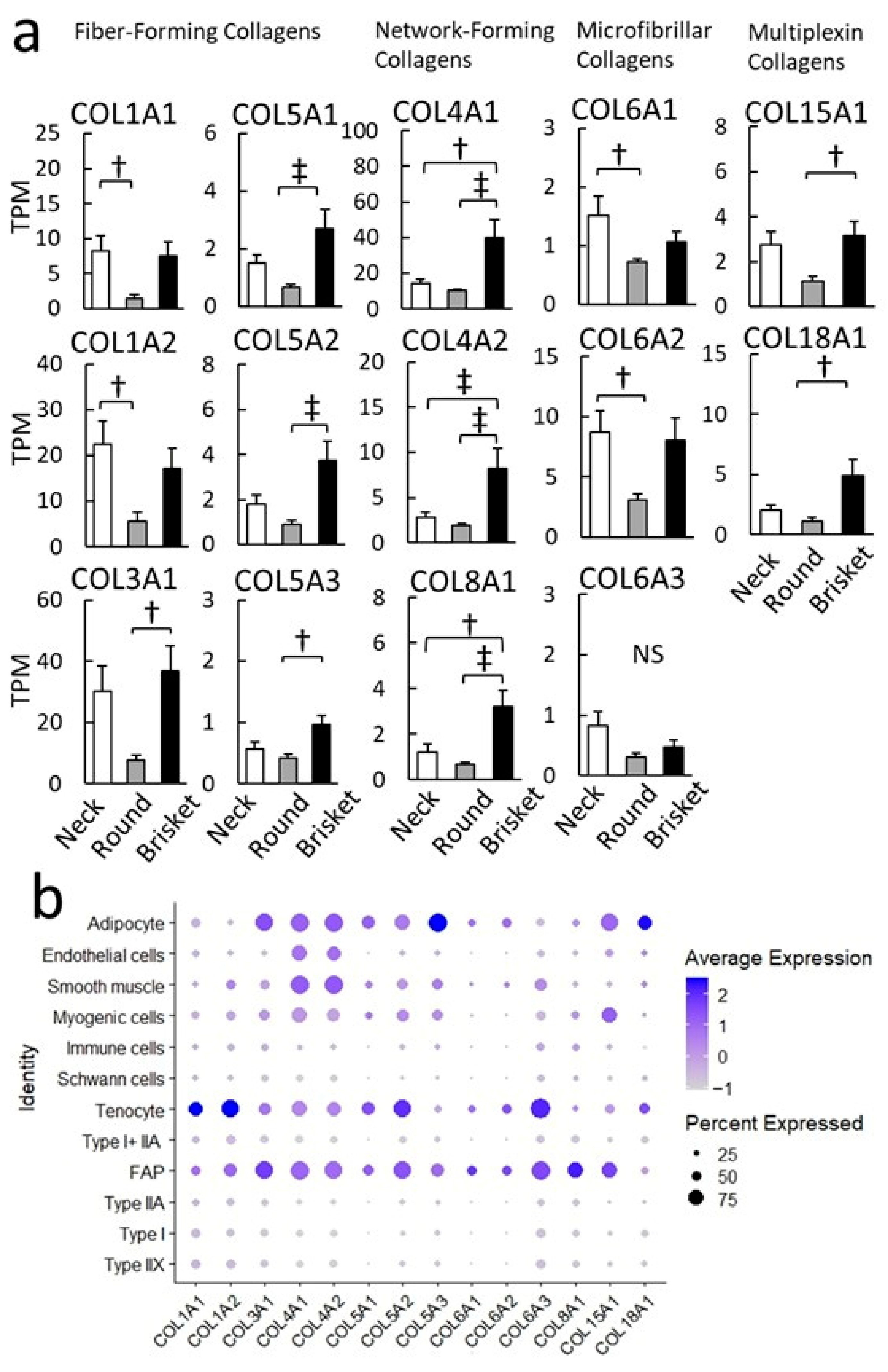
4.3. Collagen and Remodeling
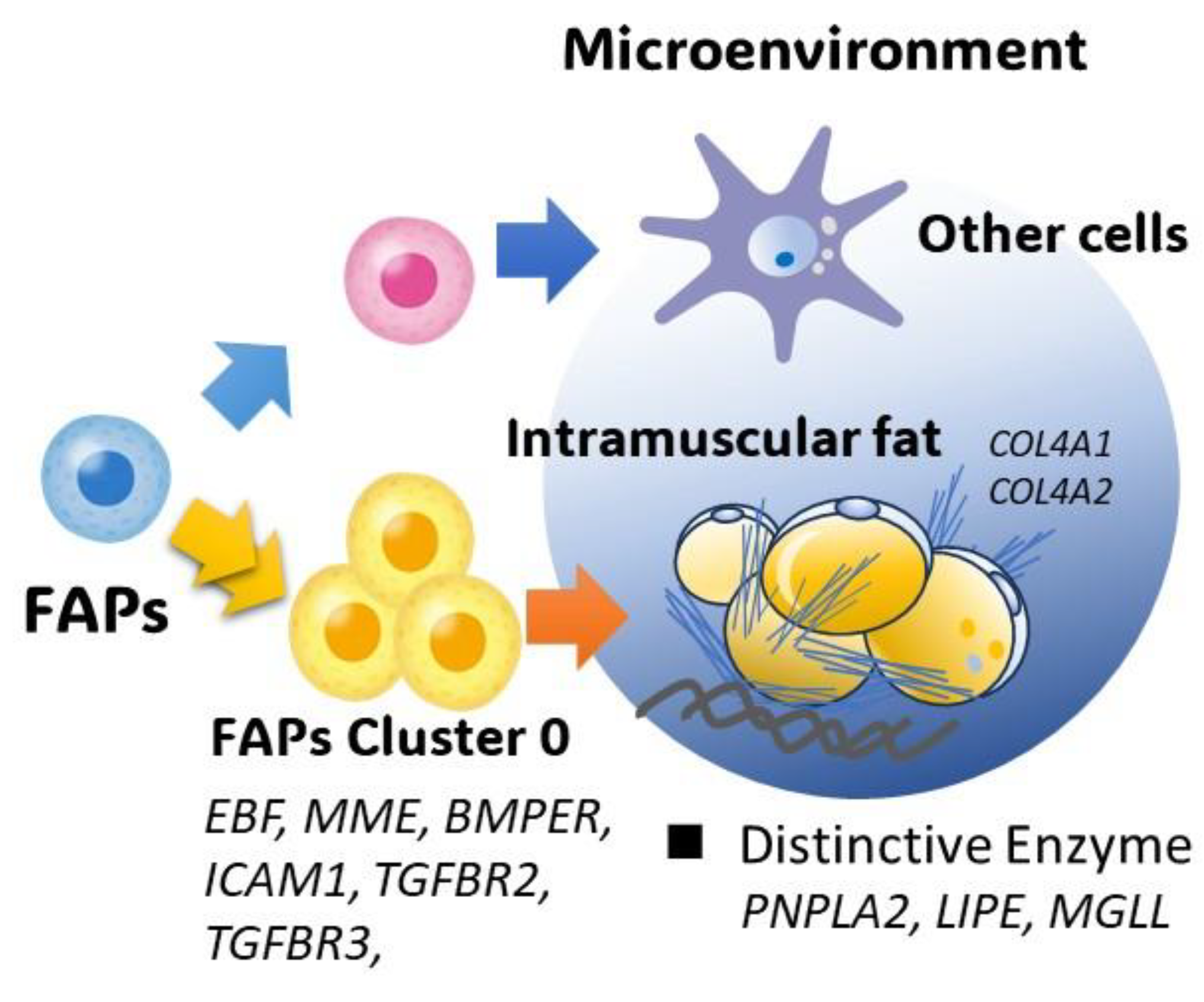
4.4. Fibro–Adipogenic Progenitor Heterogeneity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| IMF | Intramuscular fat |
| scRNA-seq | Single-cell RNA sequencing |
| snRNA-seq | Single-nucleus RNA sequencing |
| NGS | Next-generation sequencing |
| RIN | RNA integrity |
| DEG | Differentially expressed gene |
| TPM | Transcripts per million |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| SE | Standard error |
| NS | Not significant |
| UMAP | Uniform manifold approximation and projection |
References
- Chen, W.; You, W.; Valencak, T.G.; Shan, T. Bidirectional roles of skeletal muscle fibro-adipogenic progenitors in homeostasis and disease. Ageing Res. Rev. 2022, 80, 101682. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Jo, K.; Park, M.K.; Choi, Y.S.; Jung, S. Role of lipids in beef flavor development: A review of research from the past 20 years. Food Chem. 2025, 475, 143310. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Ma, C.; Guan, H.; Shen, J.; Shen, Y.; Li, G.; Sun, G.; Tian, Y.; Kang, X.; Liu, X.; et al. Multi-Omics Insights into Regulatory Mechanisms Underlying Differential Deposition of Intramuscular and Abdominal Fat in Chickens. Biomolecules 2025, 15, 134. [Google Scholar] [CrossRef]
- Su, T.; Fu, Y.; Tan, J.; Gagaoua, M.; Bak, K.H.; Soladoye, O.P.; Zhao, Z.; Zhao, Y.; Wu, W. Effects of intramuscular fat on the flavor of fresh sheep and goat meat: Recent insights into pre-mortem and post-mortem factors. Food Chem. X 2025, 25, 102159. [Google Scholar] [CrossRef]
- Malgwi, I.H.; Halas, V.; Grünvald, P.; Schiavon, S.; Jócsák, I. Genes Related to Fat Metabolism in Pigs and Intramuscular Fat Content of Pork: A Focus on Nutrigenetics and Nutrigenomics. Animals 2022, 12, 150. [Google Scholar] [CrossRef] [PubMed]
- Favaretto, F.; Bettini, S.; Busetto, L.; Milan, G.; Vettor, R. Adipogenic progenitors in different organs: Pathophysiological implications. Rev. Endocr. Metab. Disord. 2022, 23, 71–85. [Google Scholar] [CrossRef]
- Gotoh, T.; Nishimura, T.; Kuchida, K.; Mannen, H. The Japanese Wagyu beef industry: Current situation and future prospects—A review. Asian-Australas. J. Anim. Sci. 2018, 31, 933–950. [Google Scholar] [CrossRef]
- Ueda, S.; Yoshida, Y.; Tateoka, Y.; Kebede, B.; Shinohara, M.; Nakanishi, H.; Fukuda, I.; Shirai, Y. Metabolites and Free Fatty Acids in Japanese Black Beef During Wet Aging. Metabolites 2025, 15, 94. [Google Scholar] [CrossRef]
- Ebrecht, A.C.; Mofokeng, T.M.; Hollmann, F.; Smit, M.S.; Opperman, D.J. Lactones from Unspecific Peroxygenase-Catalyzed In-Chain Hydroxylation of Saturated Fatty Acids. Org. Lett. 2023, 25, 4990–4995. [Google Scholar] [CrossRef]
- Oh, D.K.; Lee, T.E.; Lee, J.; Shin, K.C.; Park, J.B. Biocatalytic oxyfunctionalization of unsaturated fatty acids to oxygenated chemicals via hydroxy fatty acids. Biotechnol. Adv. 2025, 79, 108510. [Google Scholar] [CrossRef]
- Matsuishi, M.; Kume, J.; Itou, Y.; Takahashi, M.; Arai, M.; Nagatomi, H.; Watanabe, K.; Hayase, F.; Okitani, A. Aroma components of Wagyu beef and imported beef. Nihon Chikusan Gakkaiho 2004, 75, 409–415. [Google Scholar] [CrossRef]
- Hirano, T.; Watanabe, T.; Inoue, K.; Sugimoto, Y. Fine-mapping of a marbling trait to a 2.9-cM region on bovine chromosome 7 in Japanese Black cattle. Anim. Genet. 2008, 39, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Sasazaki, S.; Kondo, H.; Moriishi, Y.; Kawaguchi, F.; Oyama, K.; Mannen, H. Comprehensive genotyping analysis of single nucleotide polymorphisms responsible for beef marbling in Japanese Black cattle. BMC Genom. Data 2024, 25, 17. [Google Scholar] [CrossRef] [PubMed]
- Abebe, B.K.; Wang, J.; Guo, J.; Wang, H.; Li, A.; Zan, L. A review of emerging technologies, nutritional practices, and management strategies to improve intramuscular fat composition in beef cattle. Anim. Biotechnol. 2024, 35, 2388704. [Google Scholar] [CrossRef]
- Ueda, S.; Hosoda, M.; Yoshino, K.-I.; Yamanoue, M.; Shirai, Y. Gene Expression Analysis Provides New Insights into the Mechanism of Intramuscular Fat Formation in Japanese Black Cattle. Genes 2021, 12, 1107. [Google Scholar] [CrossRef]
- Wang, X.; Liang, C.; Li, A.; Cheng, G.; Long, F.; Khan, R.; Wang, J.; Zhang, Y.; Wu, S.; Wang, Y.; et al. RNA-Seq and lipidomics reveal different adipogenic processes between bovine perirenal and intramuscular adipocytes. Adipocyte 2022, 11, 448–462. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, X.; Liu, S.; You, W.; Huang, Y.; Zhou, Y.; Chen, W.; Zhang, S.; Wang, J.; Zheng, Q.; et al. Single-nucleus and bulk RNA sequencing reveal cellular and transcriptional mechanisms underlying lipid dynamics in high marbled pork. NPJ Sci. Food 2023, 7, 23. [Google Scholar] [CrossRef]
- Cao, Y.; Xing, Y.; Guan, H.; Ma, C.; Jia, Q.; Tian, W.; Li, G.; Tian, Y.; Kang, X.; Liu, X.; et al. Genomic Insights into Molecular Regulation Mechanisms of Intramuscular Fat Deposition in Chicken. Genes 2023, 14, 2197. [Google Scholar] [CrossRef]
- Wang, L.; Gao, P.; Li, C.; Liu, Q.; Yao, Z.; Li, Y.; Zhang, X.; Sun, J.; Simintiras, C.; Welborn, M.; et al. A single-cell atlas of bovine skeletal muscle reveals mechanisms regulating intramuscular adipogenesis and fibrogenesis. J. Cachexia Sarcopenia Muscle 2023, 14, 2152–2167. [Google Scholar] [CrossRef]
- Chi, Z.; Jia, Q.; Yang, H.; Ren, H.; Jin, C.; He, J.; Wuri, N.; Sui, Z.; Zhang, J.; Mengke, B.; et al. snRNA-seq of adipose tissues reveals the potential cellular and molecular mechanisms of cold and disease resistance in Mongolian cattle. BMC Genom. 2024, 25, 999. [Google Scholar] [CrossRef]
- Li, Z.; Leeming, M.G.; Hastie, M.; Frank, D.; Ha, M.; Warner, R.D. Targeted-metabolomic and untargeted-proteomic approaches reveal the effects of muscle fibre type and postmortem ageing on taste-active compounds in beef. Food Chem. 2024, 460, 140506. [Google Scholar] [CrossRef] [PubMed]
- Bischof, G.; Witte, F.; Terjung, N.; Heinz, V.; Juadjur, A.; Gibis, M. Metabolic, proteomic and microbial changes postmortem and during beef aging. Crit. Rev. Food Sci. Nutr. 2024, 64, 1076–1109. [Google Scholar] [CrossRef]
- Lee, D.Y.; Kim, J.S.; Park, J.; Han, D.; Choi, Y.; Park, J.W.; Lee, J.; Mariano, E., Jr.; Namkung, S.; Hur, S.J. An Investigation of the Status of Commercial Meat Analogs and Their Ingredients: Worldwide and South Korea. Food Sci. Anim. Resour. 2025, 45, 31–61. [Google Scholar] [CrossRef]
- Lee, D.; Lee, S.; Jo, C. Application of Animal Resources into the Maillard Reaction Model System to Improve Meat Flavor. Food Sci. Anim. Resour. 2025, 45, 303–327. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, T.; Yamaguchi, C.; Miyazaki, K.; Chikaraishi, R. The Role of Lipids in Enhancing the Koku Perception of Pork Sausage. J. Oleo Sci. 2024, 73, 503–508. [Google Scholar] [CrossRef]
- Giuliani, G.; Rosina, M.; Reggio, A. Signaling pathways regulating the fate of fibro/adipogenic progenitors (FAPs) in skeletal muscle regeneration and disease. Febs J. 2022, 289, 6484–6517. [Google Scholar] [CrossRef]
- Flores-Opazo, M.; Kopinke, D.; Helmbacher, F.; Fernández-Verdejo, R.; Tuñón-Suárez, M.; Lynch, G.S.; Contreras, O. Fibro-adipogenic progenitors in physiological adipogenesis and intermuscular adipose tissue remodeling. Mol. Asp. Med. 2024, 97, 101277. [Google Scholar] [CrossRef]
- Gallardo, F.S.; Cruz-Soca, M.; Bock-Pereda, A.; Faundez-Contreras, J.; Gutiérrez-Rojas, C.; Gandin, A.; Torresan, V.; Casar, J.C.; Ravasio, A.; Brandan, E. Role of TGF-β/SMAD/YAP/TAZ signaling in skeletal muscle fibrosis. Am. J. Physiol. Cell Physiol. 2025, 328, C1015–C1028. [Google Scholar] [CrossRef]
- Park, S.J.; Beak, S.H.; Jung, D.J.S.; Kim, S.Y.; Jeong, I.H.; Piao, M.Y.; Kang, H.J.; Fassah, D.M.; Na, S.W.; Yoo, S.P.; et al. Genetic, management, and nutritional factors affecting intramuscular fat deposition in beef cattle—A review. Asian-Australas. J. Anim. Sci. 2018, 31, 1043–1061. [Google Scholar] [CrossRef]
- Cao, J.; Spielmann, M.; Qiu, X.; Huang, X.; Ibrahim, D.M.; Hill, A.J.; Zhang, F.; Mundlos, S.; Christiansen, L.; Steemers, F.J.; et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 2019, 566, 496–502. [Google Scholar] [CrossRef]
- Ueda, S.; Hosoda, M.; Kasamatsu, K.; Horiuchi, M.; Nakabayashi, R.; Kang, B.; Shinohara, M.; Nakanishi, H.; Ohto-Nakanishi, T.; Yamanoue, M.; et al. Production of Hydroxy Fatty Acids, Precursors of γ-Hexalactone, Contributes to the Characteristic Sweet Aroma of Beef. Metabolites 2022, 12, 332. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Adiconis, X.; Simmons, S.K.; Kowalczyk, M.S.; Hession, C.C.; Marjanovic, N.D.; Hughes, T.K.; Wadsworth, M.H.; Burks, T.; Nguyen, L.T.; et al. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat. Biotechnol. 2020, 38, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Xu, Z.; You, W.; Zhou, Y.; Nong, Q.; Chen, W.; Shan, T. Lipidomics and single-cell RNA sequencing reveal lipid and cell dynamics of porcine glycerol-injured skeletal muscle regeneration model. Life Sci. 2024, 350, 122742. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.; Li, Q.; Zhu, J.; Cheng, W.; Xie, Y.; Huang, Y.; Zhao, H.; Hao, M.; Wei, H.; Zhao, S. Single-nucleus RNA sequencing and lipidomics reveal characteristics of transcriptional and lipid composition in porcine longissimus dorsi muscle. BMC Genom. 2024, 25, 622. [Google Scholar] [CrossRef]
- Cerk, I.K.; Wechselberger, L.; Oberer, M. Adipose Triglyceride Lipase Regulation: An Overview. Curr. Protein Pept. Sci. 2018, 19, 221–233. [Google Scholar] [CrossRef]
- Pozzi, A.; Yurchenco, P.D.; Iozzo, R.V. The nature and biology of basement membranes. Matrix Biol. 2017, 57–58, 1–11. [Google Scholar] [CrossRef]
- Li, S.N.; Wu, J.F. TGF-β/SMAD signaling regulation of mesenchymal stem cells in adipocyte commitment. Stem Cell Res. Ther. 2020, 11, 41. [Google Scholar] [CrossRef]
- Orafaie, A.; Mousavian, M.; Orafai, H.; Sadeghian, H. An overview of lipoxygenase inhibitors with approach of in vivo studies. Prostaglandins Other Lipid Mediat. 2020, 148, 106411. [Google Scholar] [CrossRef]
- Chen, Z.; Wu, X.; Zheng, D.; Wang, Y.; Chai, J.; Zhang, T.; Wu, P.; Wei, M.; Zhou, T.; Long, K.; et al. Single-Nucleus RNA Sequencing Reveals Cellular Transcriptome Features at Different Growth Stages in Porcine Skeletal Muscle. Cells 2025, 14, 37. [Google Scholar] [CrossRef]
- Xu, Z.; Wu, J.; Li, Y.; Zhou, J.; Zhang, Y.; Qiao, M.; Feng, Y.; Sun, H.; Li, Z.; Li, L.; et al. A Single-Cell Atlas of Porcine Skeletal Muscle Reveals Mechanisms That Regulate Intramuscular Adipogenesis. Int. J. Mol. Sci. 2024, 25, 12935. [Google Scholar] [CrossRef]
- Fitzgerald, G.; Turiel, G.; Gorski, T.; Soro-Arnaiz, I.; Zhang, J.; Casartelli, N.C.; Masschelein, E.; Maffiuletti, N.A.; Sutter, R.; Leunig, M.; et al. MME(+) fibro-adipogenic progenitors are the dominant adipogenic population during fatty infiltration in human skeletal muscle. Commun. Biol. 2023, 6, 111. [Google Scholar] [CrossRef]
- Garritson, J.D.; Zhang, J.; Achenbach, A.; Ferhat, M.; Eich, E.; Stubben, C.J.; Martinez, P.L.; Ibele, A.R.; Hilgendorf, K.I.; Boudina, S. BMPER is a marker of adipose progenitors and adipocytes and a positive modulator of adipogenesis. Commun. Biol. 2023, 6, 638. [Google Scholar] [CrossRef] [PubMed]
- Merrick, D.; Sakers, A.; Irgebay, Z.; Okada, C.; Calvert, C.; Morley, M.P.; Percec, I.; Seale, P. Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science 2019, 364, 6438. [Google Scholar] [CrossRef]
- Qiu, S.; Lin, W.; Zhou, Z.; Hong, Q.; Chen, S.; Li, J.; Zhong, F.; Zhou, Q.; Cui, D. TOX: A potential new immune checkpoint in cancers by pancancer analysis. Discov. Oncol. 2024, 15, 354. [Google Scholar] [CrossRef]
- Rubenstein, A.B.; Smith, G.R.; Raue, U.; Begue, G.; Minchev, K.; Ruf-Zamojski, F.; Nair, V.D.; Wang, X.; Zhou, L.; Zaslavsky, E.; et al. Single-cell transcriptional profiles in human skeletal muscle. Sci. Rep. 2020, 10, 229. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Fu, X.; Yang, G.; Du, M. Review: Enhancing intramuscular fat development via targeting fibro-adipogenic progenitor cells in meat animals. Animal 2020, 14, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Zhu, Q.; Duo, T.; Liang, Z.; Zhang, C.; Cai, S.; Wang, X.; Liu, Y.; Li, Y.; Liu, X.; et al. The Impact of FBN1-α5β1 Axis in Fibro/Adipogenic Progenitor Cells (FAPCD9−) on Intramuscular Fat Content in Pigs. J. Agric. Food Chem. 2024, 72, 9507–9521. [Google Scholar] [CrossRef]
- Kong, L.; Yuan, C.; Guo, T.; Sun, L.; Liu, J.; Lu, Z. Inhibitor of Myom3 inhibits proliferation and promotes differentiation of sheep myoblasts. Genomics 2024, 116, 110921. [Google Scholar] [CrossRef] [PubMed]
- Barruet, E.; Garcia, S.M.; Striedinger, K.; Wu, J.; Lee, S.; Byrnes, L.; Wong, A.; Xuefeng, S.; Tamaki, S.; Brack, A.S.; et al. Functionally heterogeneous human satellite cells identified by single cell RNA sequencing. eLife 2020, 9, e51576. [Google Scholar] [CrossRef]
- Orlich, M.M.; Diéguez-Hurtado, R.; Muehlfriedel, R.; Sothilingam, V.; Wolburg, H.; Oender, C.E.; Woelffing, P.; Betsholtz, C.; Gaengel, K.; Seeliger, M.; et al. Mural Cell SRF Controls Pericyte Migration, Vessel Patterning and Blood Flow. Circ. Res. 2022, 131, 308–327. [Google Scholar] [CrossRef]
- Li, J.; Yang, D.; Chen, C.; Wang, J.; Wang, Z.; Yang, C.; Yu, C.; Li, Z. Single-cell RNA transcriptome uncovers distinct developmental trajectories in the embryonic skeletal muscle of Daheng broiler and Tibetan chicken. BMC Genomics 2025, 26, 187. [Google Scholar] [CrossRef]
- Hogarth, M.W.; Kurukunda, M.P.; Ismat, K.; Uapinyoying, P.; Jaiswal, J.K. Exploring the therapeutic potential of fibroadipogenic progenitors in muscle disease. J. Neuromuscul. Dis. 2025, 12, 22143602241298545. [Google Scholar] [CrossRef]
- Mashima, D.; Oka, Y.; Gotoh, T.; Tomonaga, S.; Sawano, S.; Nakamura, M.; Tatsumi, R.; Mizunoya, W. Correlation between skeletal muscle fiber type and free amino acid levels in Japanese Black steers. Anim. Sci. J. 2019, 90, 604–609. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Mottillo, E.P. Adipocyte lipolysis: From molecular mechanisms of regulation to disease and therapeutics. Biochem. J. 2020, 477, 985–1008. [Google Scholar] [CrossRef]
- He, Z.; Wang, X.; Qi, Y.; Zhu, C.; Zhao, Z.; Zhang, X.; Liu, X.; Li, S.; Zhao, F.; Wang, J.; et al. Long-stranded non-coding RNAs temporal-specific expression profiles reveal longissimus dorsi muscle development and intramuscular fat deposition in Tianzhu white yak. J. Anim. Sci. 2023, 101, skad394. [Google Scholar] [CrossRef]
- Ueda, S.; Yoshida, Y.; Kebede, B.; Kitamura, C.; Sasaki, R.; Shinohara, M.; Fukuda, I.; Shirai, Y. New Implications of Metabolites and Free Fatty Acids in Quality Control of Crossbred Wagyu Beef during Wet Aging Cold Storage. Metabolites 2024, 14, 95. [Google Scholar] [CrossRef]
- Mathavaraj, P.; Muthusamy, V.; Katral, A.; Mandal, P.; Zunjare, R.U.; Hossain, F. Lipoxygenases (LOXs): Will turning off this genetic switch help safeguard the flavor and nutritional quality of stored lipid-rich staple foods? Food Chem. 2025, 470, 142637. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Jiang, X.; Liu, L.; Zhao, Y.; Bai, F.; Wang, J.; Gao, R.; Xu, X. The influence mechanism of phospholipids structure and composition changes caused by oxidation on the formation of flavor substances in sturgeon caviar. Food Chem. 2024, 460, 140585. [Google Scholar] [CrossRef]
- Zhang, K.; Hao, R.; Wang, S.; Zhang, Z.; Li, D.; Li, X.; Zhao, B.; Zhang, S.; Zhao, Y.; Chen, X. Correlation of lipid hydrolysis, oxidation, and molecular transformation with volatile compound revolution in pork during postmortem wet-aging process. Food Chem. 2025, 470, 142656. [Google Scholar] [CrossRef]
- Contreras, O.; Rossi, F.M.V.; Theret, M. Origins, potency, and heterogeneity of skeletal muscle fibro-adipogenic progenitors-time for new definitions. Skelet. Muscle 2021, 11, 16. [Google Scholar] [CrossRef]
- Khoshnoodi, J.; Pedchenko, V.; Hudson, B.G. Mammalian collagen IV. Microsc. Res. Tech. 2008, 71, 357–370. [Google Scholar] [CrossRef]
- Ozdemir, G.; Gulhan, B.; Atayar, E.; Saygılı, S.; Soylemezoglu, O.; Ozcakar, Z.B.; Eroglu, F.K.; Candan, C.; Demir, B.K.; Soylu, A.; et al. COL4A3 mutation is an independent risk factor for poor prognosis in children with Alport syndrome. Pediatr Nephrol 2020, 35, 1941–1952. [Google Scholar] [CrossRef]
- Li, J.; Xu, Y.; Quan, Y.; He, Y.; Lu, F.; Gao, J.; Yao, Y.; Liao, Y. Type IV Collagen Promotes Adipogenic Differentiation of Adipose Stem Cells. Aesthetic Plast. Surg. 2024, 48, 2536–2544. [Google Scholar] [CrossRef]
- Hamrick, M.W.; McGee-Lawrence, M.E.; Frechette, D.M. Fatty Infiltration of Skeletal Muscle: Mechanisms and Comparisons with Bone Marrow Adiposity. Front. Endocrinol. 2016, 7, 69. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, C.; Li, C.; Chen, X.; Xu, R.; Chen, M.; Li, Y.; Liu, Y.; Liu, X.; Chen, Y.; et al. Integrating spatial transcriptomics and single-nucleus RNA-seq revealed the specific inhibitory effects of TGF-β on intramuscular fat deposition. Sci. China Life Sci. 2025, 68, 746–763. [Google Scholar] [CrossRef] [PubMed]
- Yin, K.; Zhang, C.; Deng, Z.; Wei, X.; Xiang, T.; Yang, C.; Chen, C.; Chen, Y.; Luo, F. FAPs orchestrate homeostasis of muscle physiology and pathophysiology. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2024, 38, e70234. [Google Scholar] [CrossRef]
- Stanek, E.; Czamara, K.; Kaczor, A. Increased obesogenic action of palmitic acid during early stage of adipogenesis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2024, 1869, 159525. [Google Scholar] [CrossRef]
- Niu, H.; Wang, H. TOX regulates T lymphocytes differentiation and its function in tumor. Front. Immunol. 2023, 14, 990419. [Google Scholar] [CrossRef]
- Raza, S.H.A.; Pant, S.D.; Wani, A.K.; Mohamed, H.H.; Khalifa, N.E.; Almohaimeed, H.M.; Alshanwani, A.R.; Assiri, R.; Aggad, W.S.; Noreldin, A.E.; et al. Krüppel-like factors family regulation of adipogenic markers genes in bovine cattle adipogenesis. Mol. Cell Probes 2022, 65, 101850. [Google Scholar] [CrossRef]
- Abebe, B.K.; Wang, H.; Li, A.; Zan, L. A review of the role of transcription factors in regulating adipogenesis and lipogenesis in beef cattle. J. Anim. Breed. Genet. 2024, 141, 235–256. [Google Scholar] [CrossRef]
- Zhu, J.; Huang, L.; Zhang, W.; Li, H.; Yang, Y.; Lin, Y.; Zhang, C.; Du, Z.; Xiang, H.; Wang, Y. Single-nucleus transcriptional profiling reveals TCF7L2 as a key regulator in adipogenesis in goat skeletal muscle development. Int J Biol Macromol 2024, 281 Pt 3, 136326. [Google Scholar] [CrossRef]
- Guo, L.; Han, M.; Xu, J.; Zhou, W.; Shi, H.; Chen, S.; Pang, W.; Zhang, X.; Duan, Y.; Yin, Y.; et al. snRNA-Seq and Spatial Transcriptome Reveal Cell-Cell Crosstalk Mediated Metabolic Regulation in Porcine Skeletal Muscle. J. Cachexia Sarcopenia Muscle 2025, 16, e13752. [Google Scholar] [CrossRef] [PubMed]
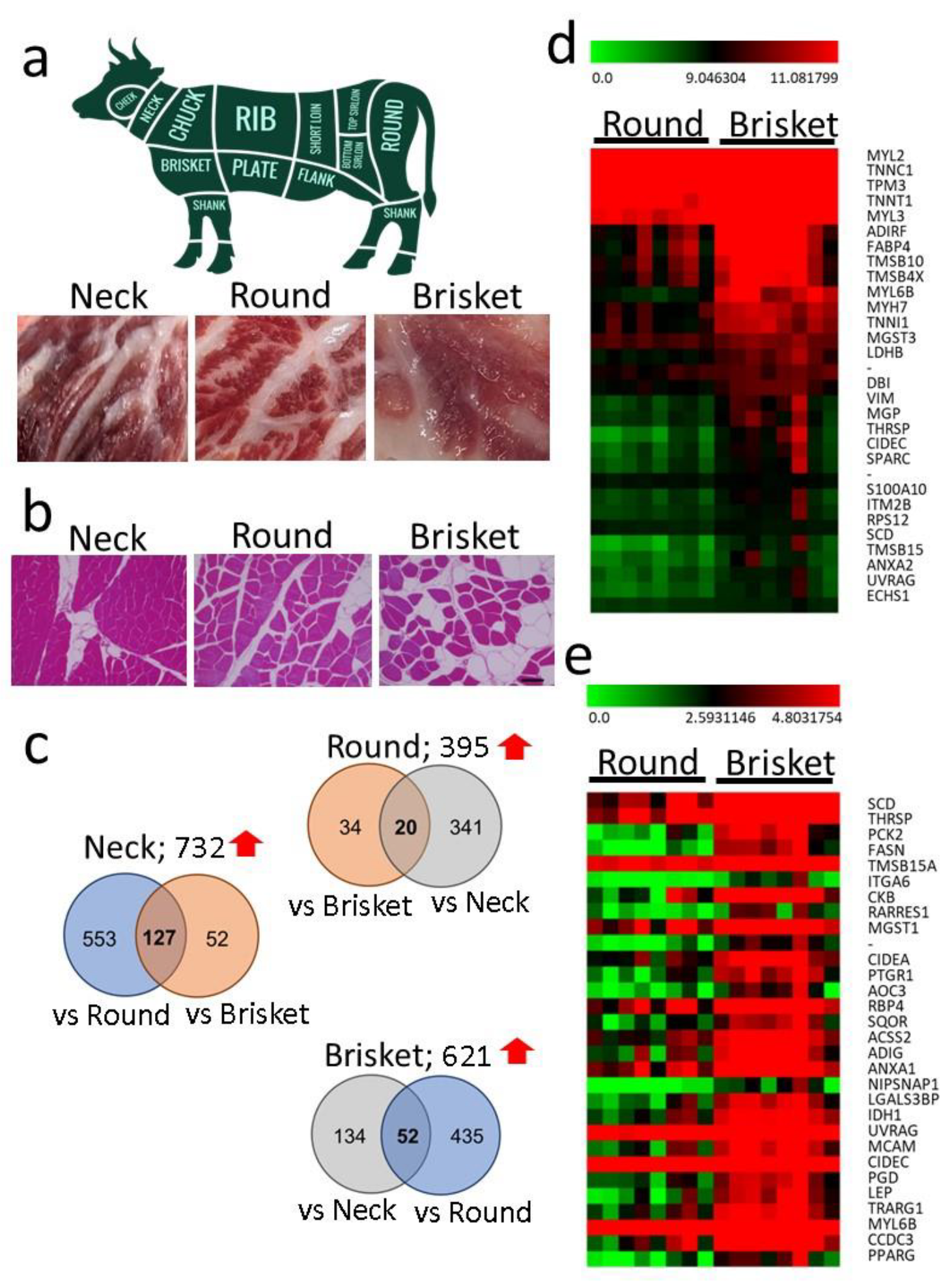
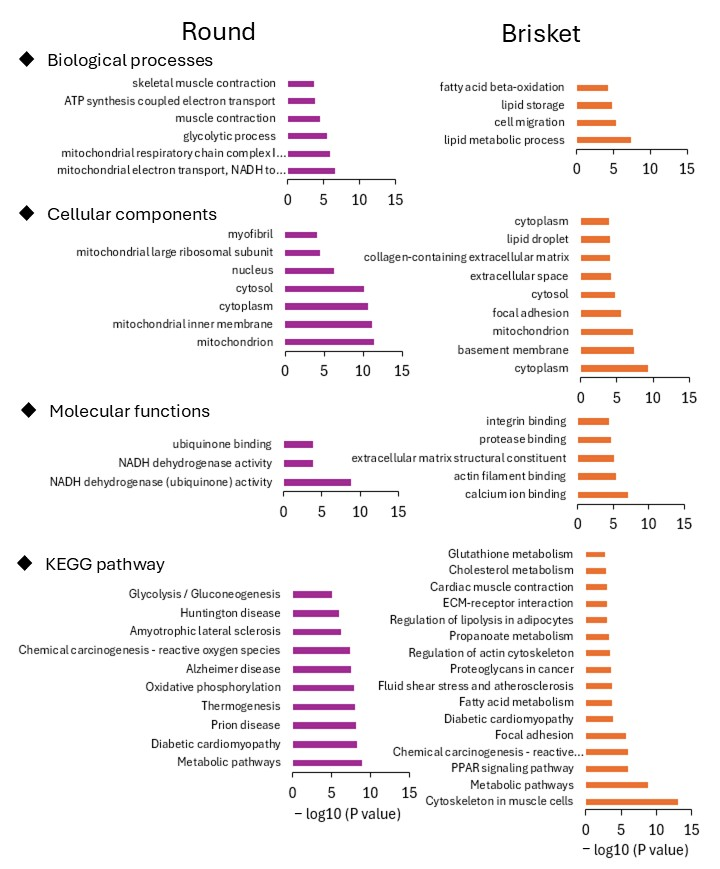
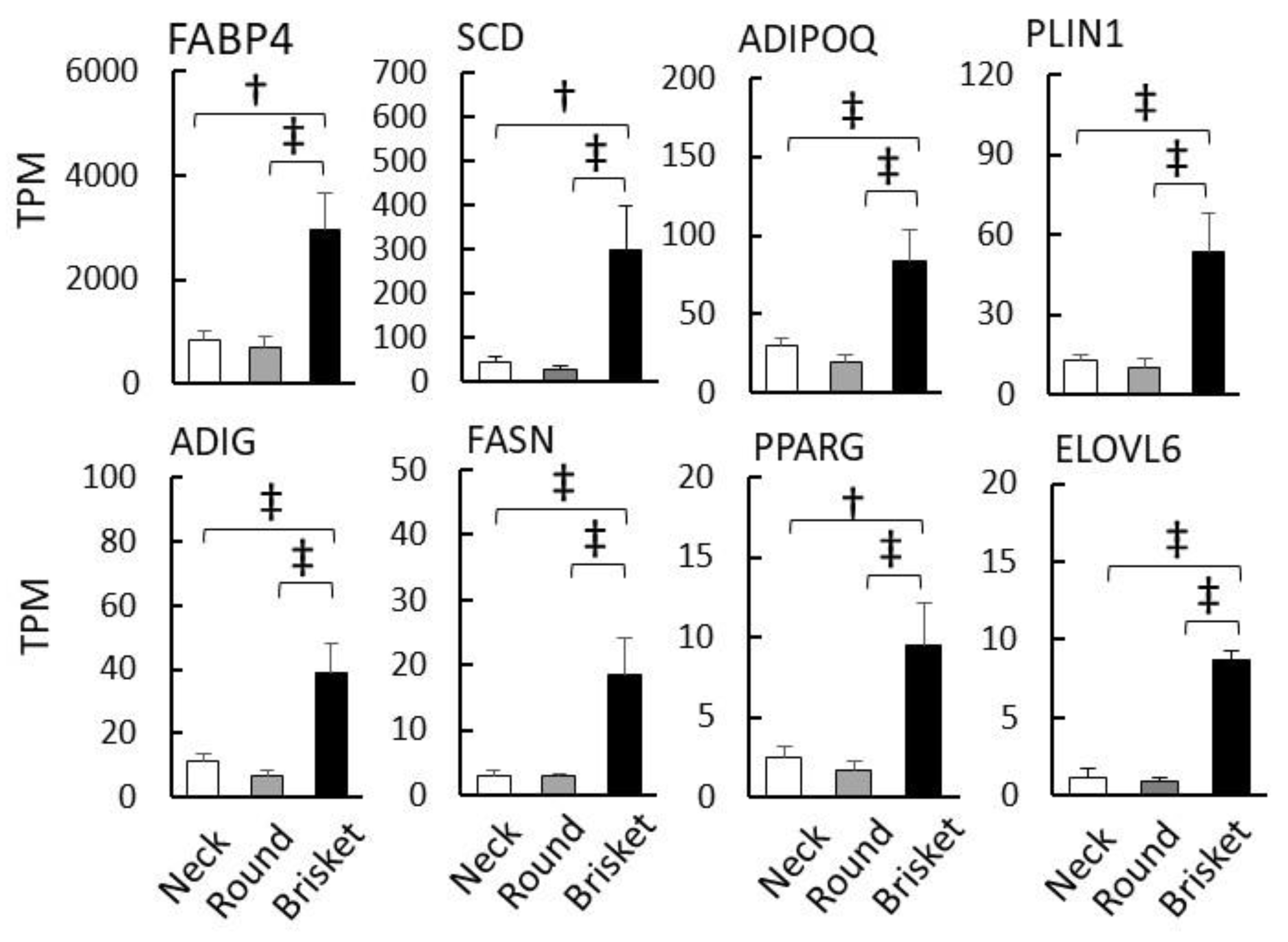
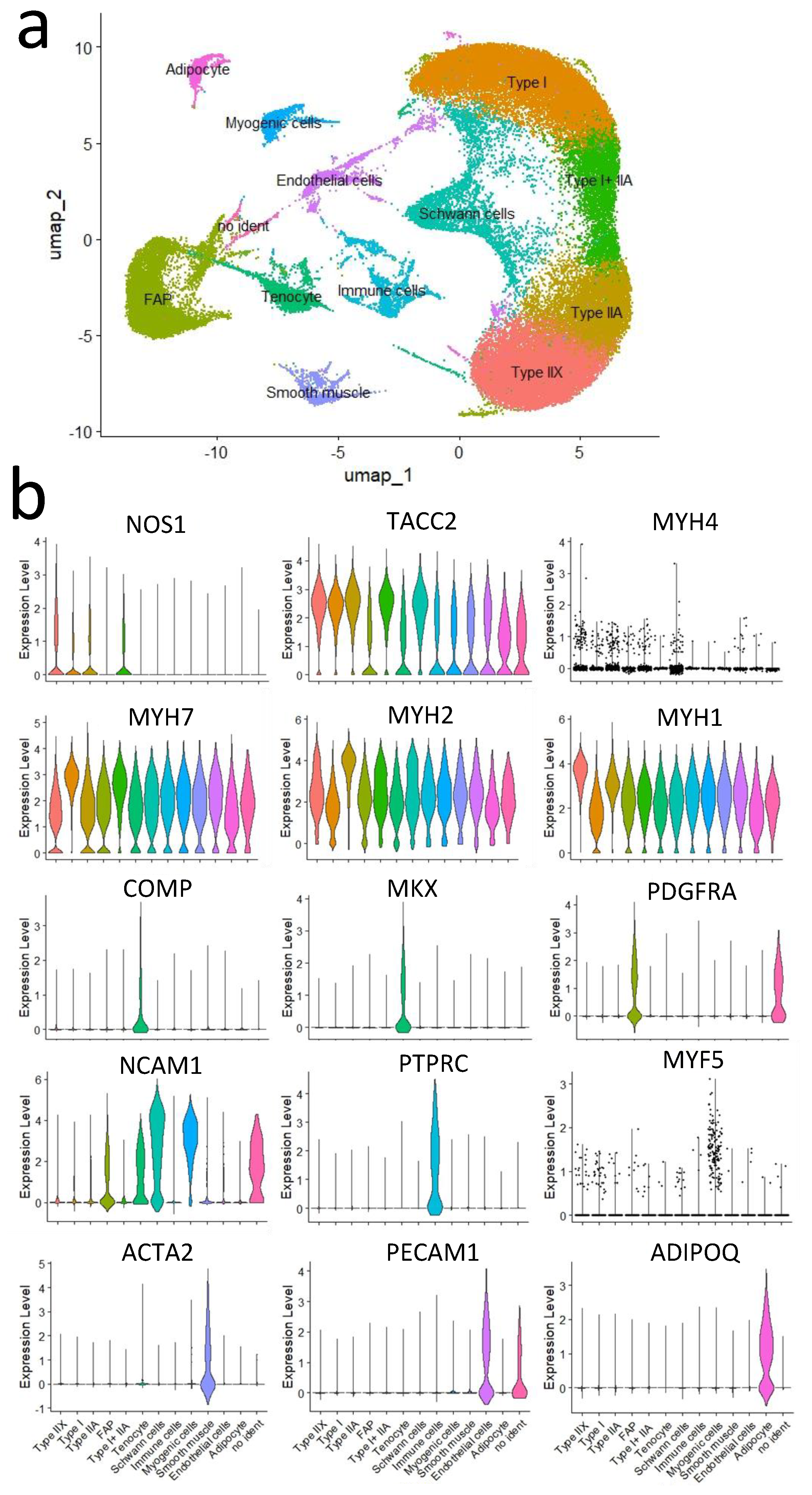
| Muscle Tissue | Total Read (Count) | Mapped Read (Count) | Mapped Read (%) |
|---|---|---|---|
| Neck | 70,838,677 | 70,609,753 | 99.7 |
| Round | 76,025,869 | 75,783,954 | 99.7 |
| Brisket | 70,529,128 | 70,263,281 | 99.6 |
| Muscle Tissue | Estimated Number of Cells | Median Genes Per Cell | Reads Mapped to Genome (%) | Total Genes Detected |
|---|---|---|---|---|
| Brisket | 38,939 | 1654 | 97.10% | 25,580 |
| Round | 51,506 | 1573 | 96.80% | 25,955 |
| Cell Type | Marker Gene |
|---|---|
| Myofibers | Nitric oxide synthase 1 (NOS1) |
| Transforming acidic coiled-coil-containing protein 2 (TACC2) | |
| Myosin heavy chain 1 (MYH1) | |
| Myosin heavy chain 2 (MYH2) | |
| Myosin heavy chain 7 (MYH7) | |
| Fibro–adipogenic progenitors (FAPs) | Platelet-derived growth factor receptor alpha (PDGFRA) |
| Tenocytes | Cartilage oligomeric matrix protein (COMP) |
| Mohawk homeobox (MKX) | |
| Chwann cells | Neural cell adhesion molecule 1 (NCAM1) |
| Immune cells | Protein tyrosine phosphatase receptor type C (PTPRC) |
| Myogenic cells (muscle satellite cells) | Myogenic factor 5 (MYF5) |
| Smooth muscle cells | Actin alpha 2 (ACTA2) |
| Endothelial cells | Platelet and endothelial cell adhesion molecule 1 (PECAM1) |
| Adipocytes | Adiponectin (ADIPOQ) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ueda, S.; Kitamura, C.; Tateoka, Y.; Kanai, A.; Suzuki, Y.; Fukuda, I.; Shirai, Y. Single-Nucleus RNA Sequencing Reveals Muscle-Region-Specific Differences in Fibro-Adipogenic Progenitors Driving Intramuscular Fat Accumulation. Metabolites 2025, 15, 231. https://doi.org/10.3390/metabo15040231
Ueda S, Kitamura C, Tateoka Y, Kanai A, Suzuki Y, Fukuda I, Shirai Y. Single-Nucleus RNA Sequencing Reveals Muscle-Region-Specific Differences in Fibro-Adipogenic Progenitors Driving Intramuscular Fat Accumulation. Metabolites. 2025; 15(4):231. https://doi.org/10.3390/metabo15040231
Chicago/Turabian StyleUeda, Shuji, Chiaki Kitamura, Yuka Tateoka, Akinori Kanai, Yutaka Suzuki, Itsuko Fukuda, and Yasuhito Shirai. 2025. "Single-Nucleus RNA Sequencing Reveals Muscle-Region-Specific Differences in Fibro-Adipogenic Progenitors Driving Intramuscular Fat Accumulation" Metabolites 15, no. 4: 231. https://doi.org/10.3390/metabo15040231
APA StyleUeda, S., Kitamura, C., Tateoka, Y., Kanai, A., Suzuki, Y., Fukuda, I., & Shirai, Y. (2025). Single-Nucleus RNA Sequencing Reveals Muscle-Region-Specific Differences in Fibro-Adipogenic Progenitors Driving Intramuscular Fat Accumulation. Metabolites, 15(4), 231. https://doi.org/10.3390/metabo15040231







