Cryoprotectant-Mediated Cold Stress Mitigation in Litchi Flower Development: Transcriptomic and Metabolomic Perspectives
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Experimental Design
2.2. Biochemical Characterization of the Flower Spikes
2.2.1. Determination of Soluble Protein and Sugars
2.2.2. Estimation of Malondialdehyde (MDA) and Proline
2.2.3. Analysis of Antioxidant Enzymes (CAT, SOD, and POD)
2.2.4. Estimation of the Activities of Key Proline Enzymes (P5CS, P5CR, ProDH, and δ-OAT)
2.3. Transcriptome Analysis
2.4. Metabolome Analysis
2.5. Statistical Analysis
3. Results
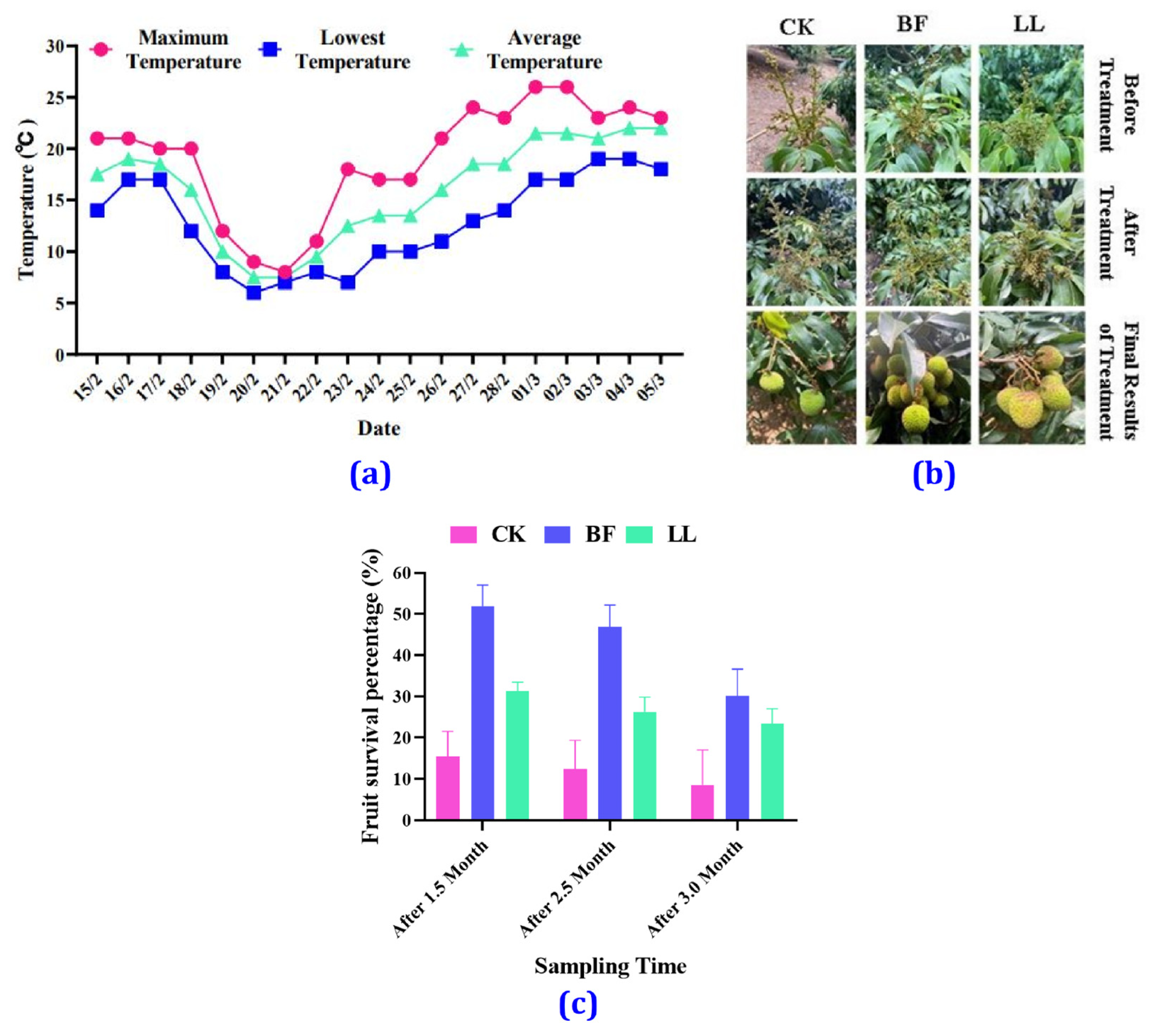
3.1. Temperature Variation
3.2. Morphological Changes
3.3. Biochemical Characterization on Inflorescence Growth
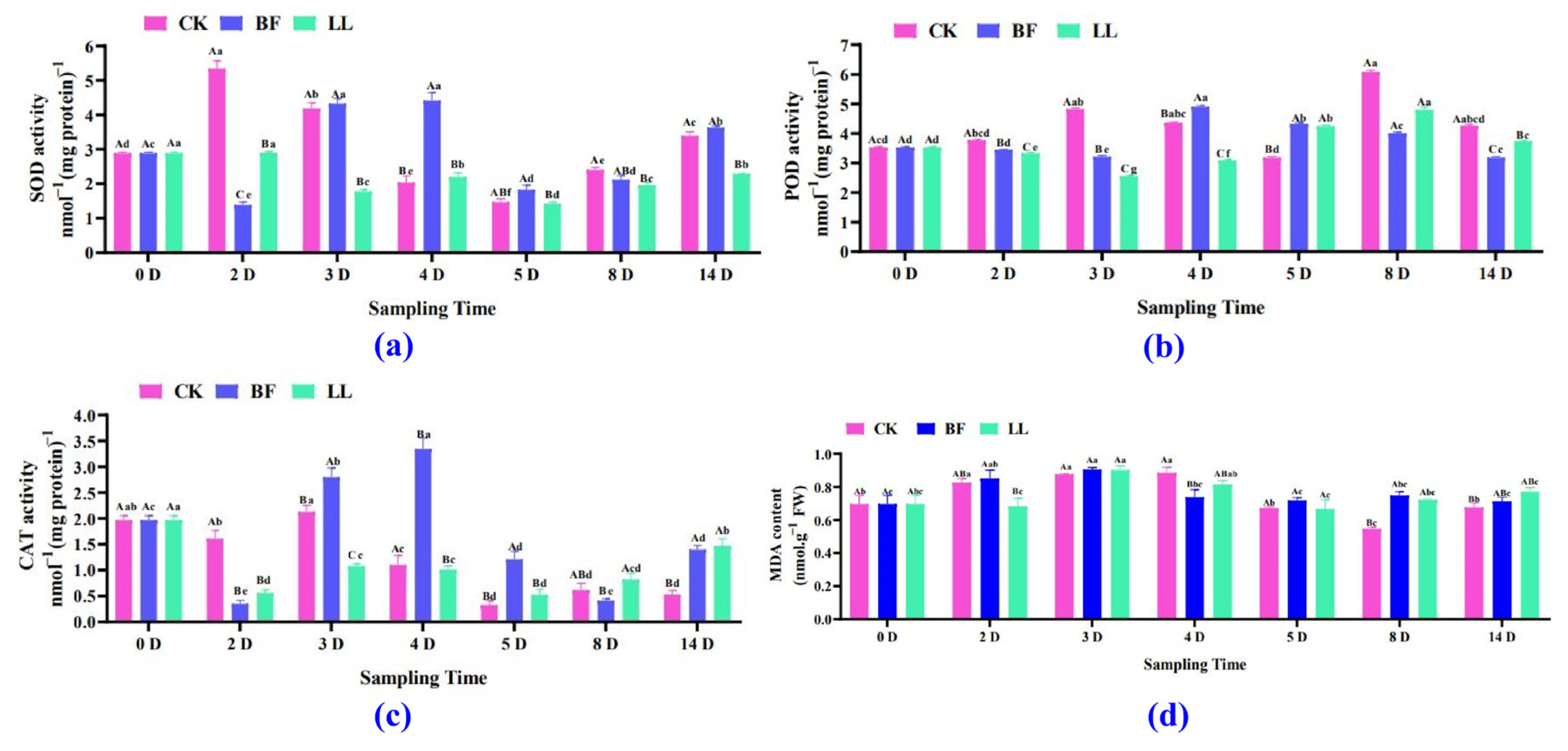
3.3.1. Antioxidant Enzyme and Malondialdehyde (MDA) Analysis
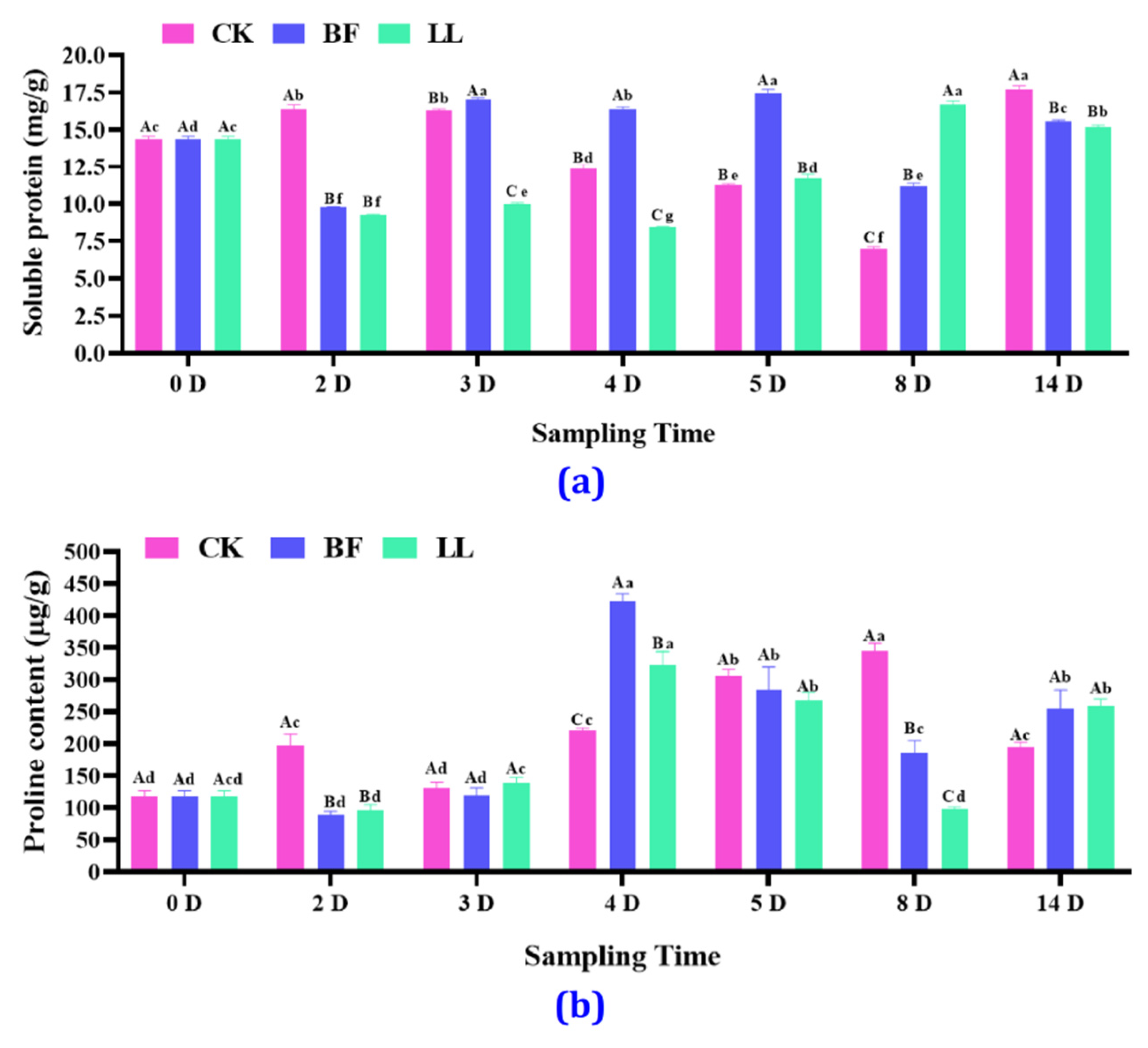
3.3.2. Soluble Protein and Proline Content
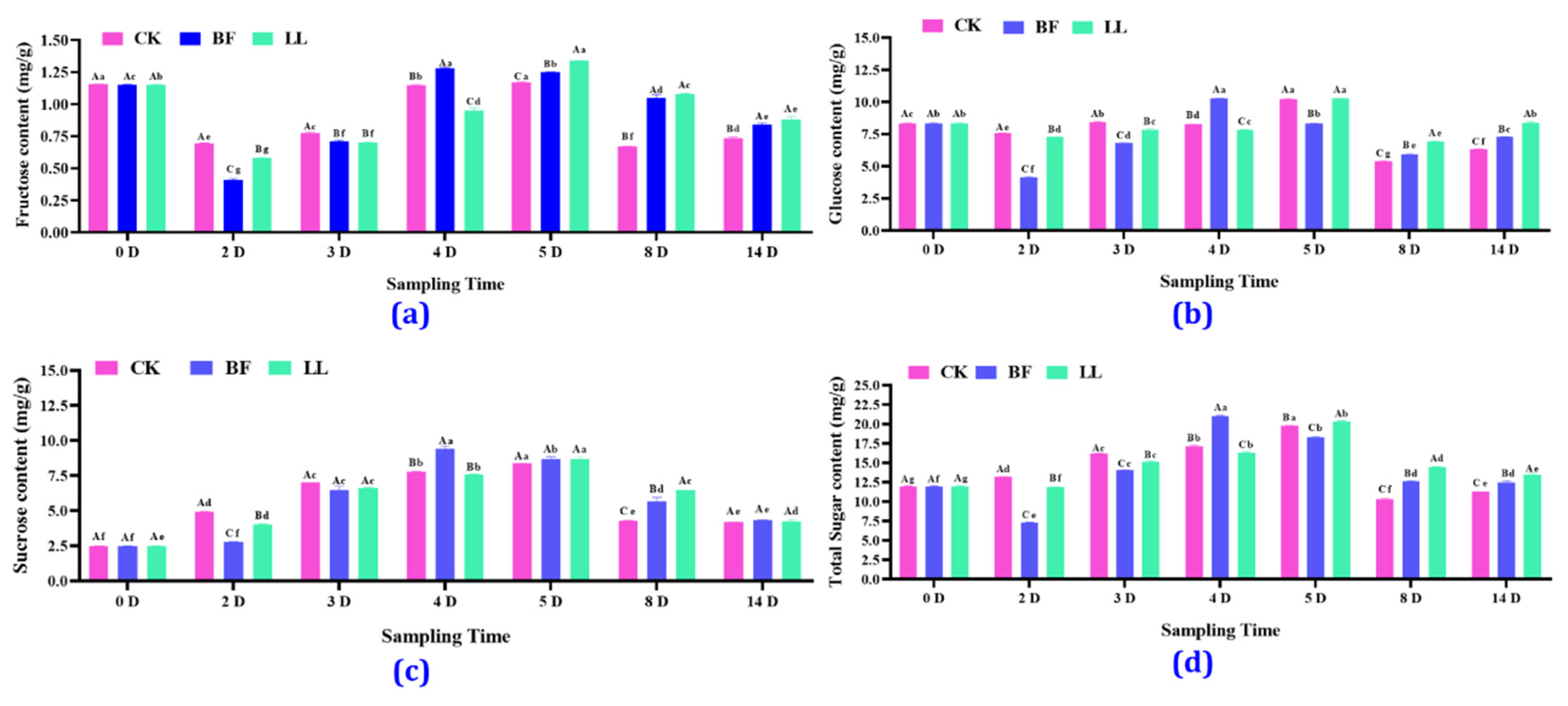
3.3.3. Different Soluble Sugars’ Content
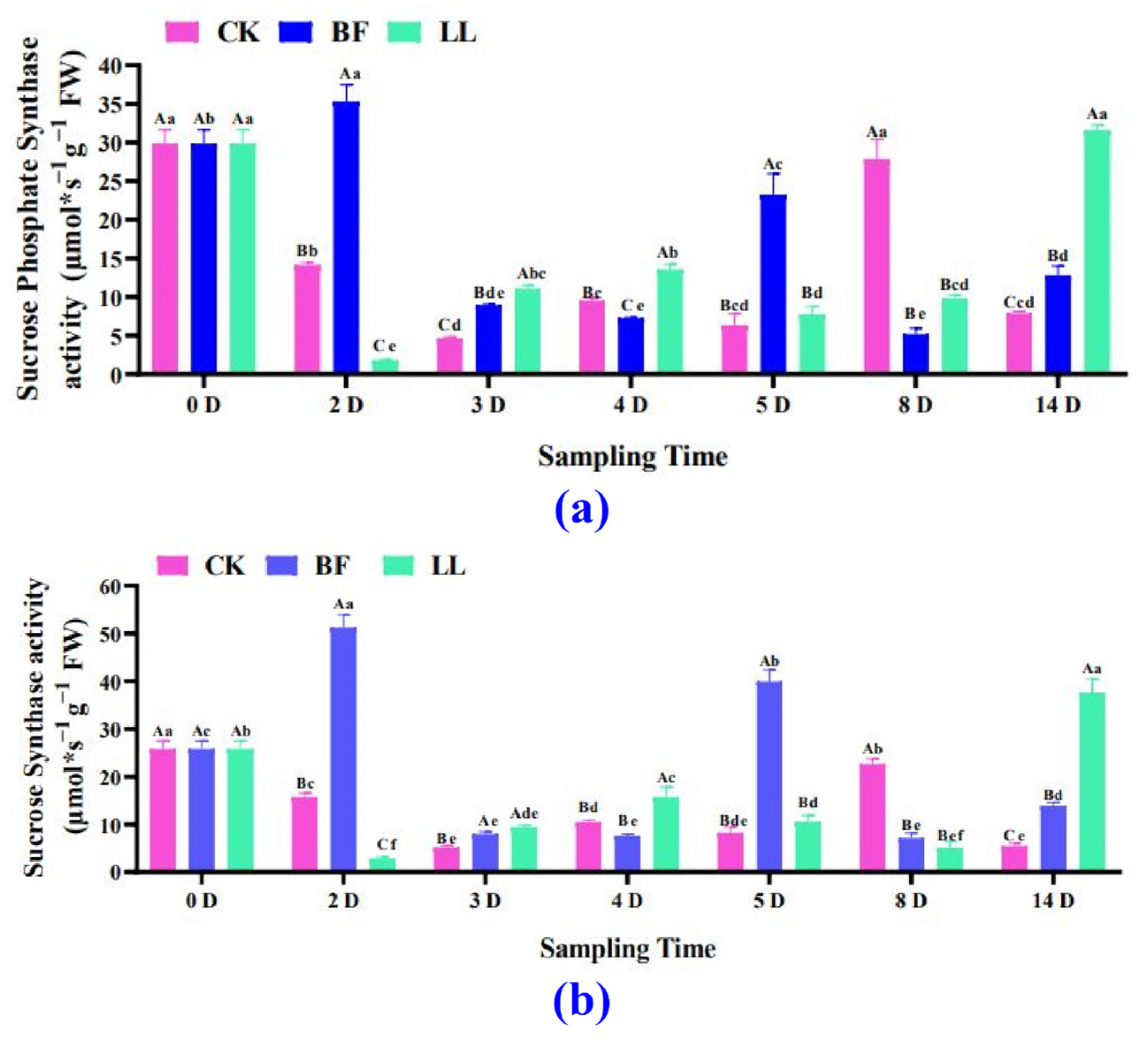
3.3.4. Sucrose Synthetase and Sucrose Phosphate Synthetase Analysis
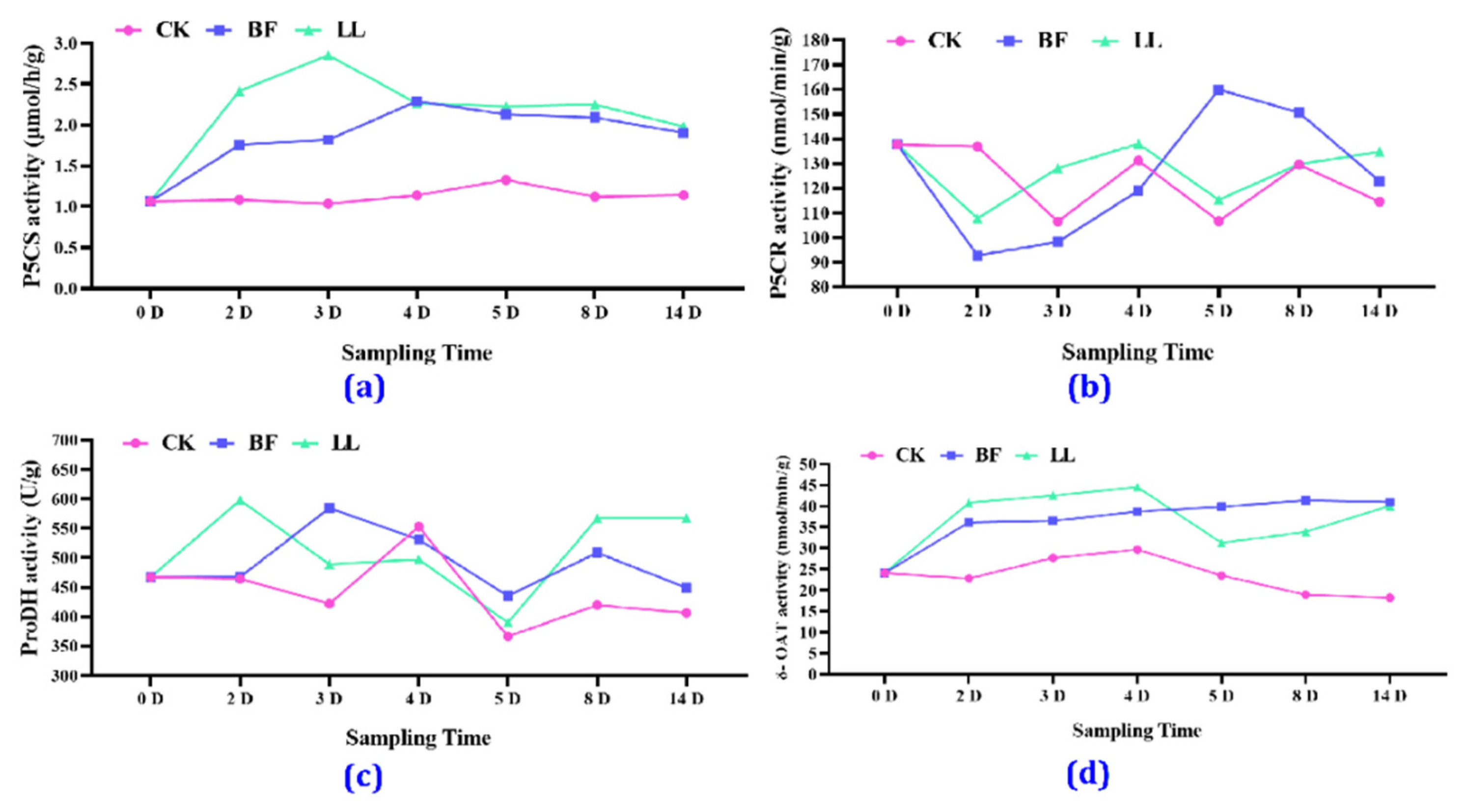
3.3.5. Key Proline-Related Enzyme Activities
3.4. Transcriptome Analysis
3.4.1. Transcriptome Sequencing Data Analysis
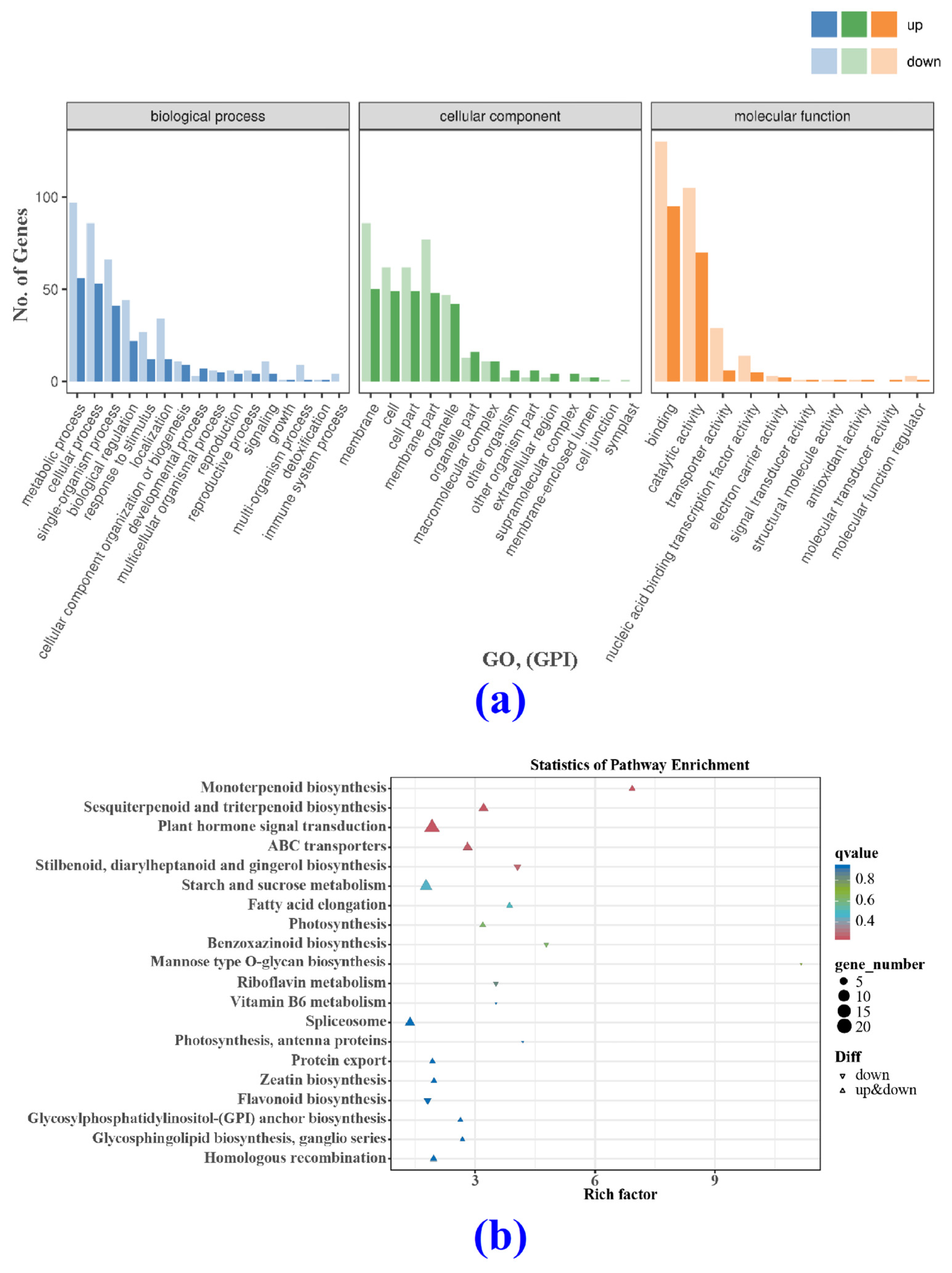
3.4.2. Differentially Expressed Genes (DEGs) and Gene Ontology (GO)
3.4.3. KEGG Analysis
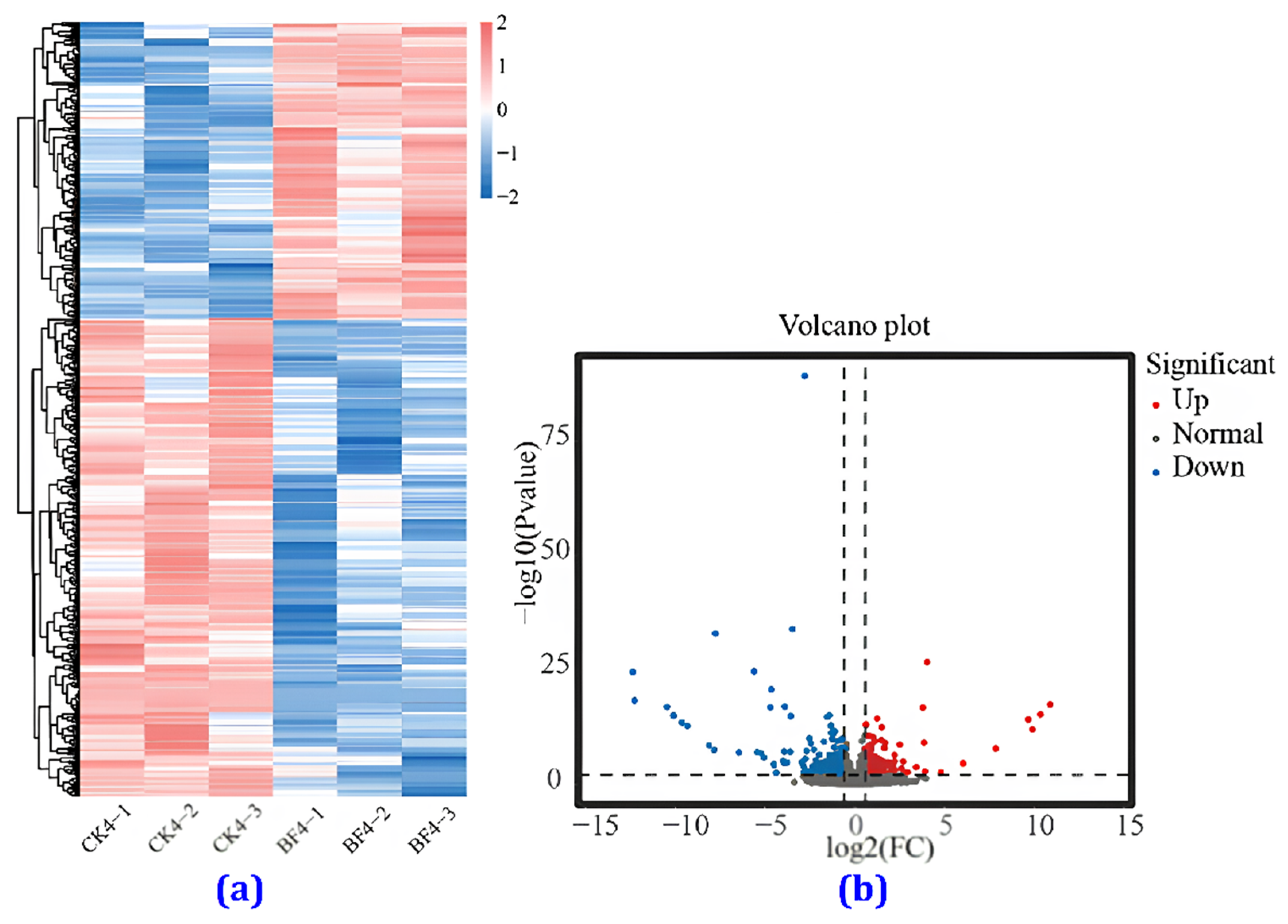
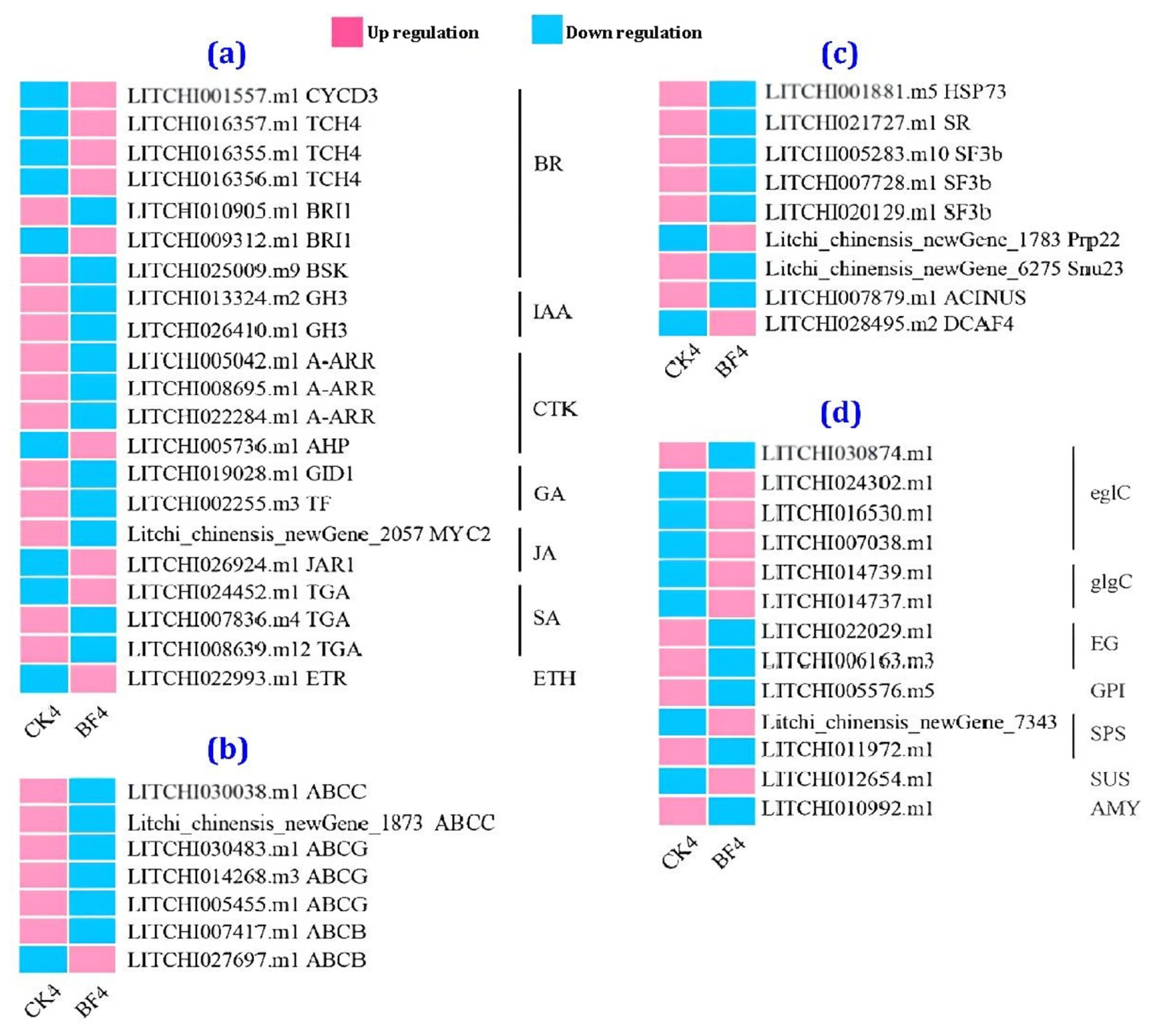
3.4.4. Heat Map Analysis
3.4.5. Comparison of the Transcriptome between the CK and the BF Groups
3.5. Metabolome Analysis
3.5.1. Differentially Accumulated Metabolite (DAM) Analysis
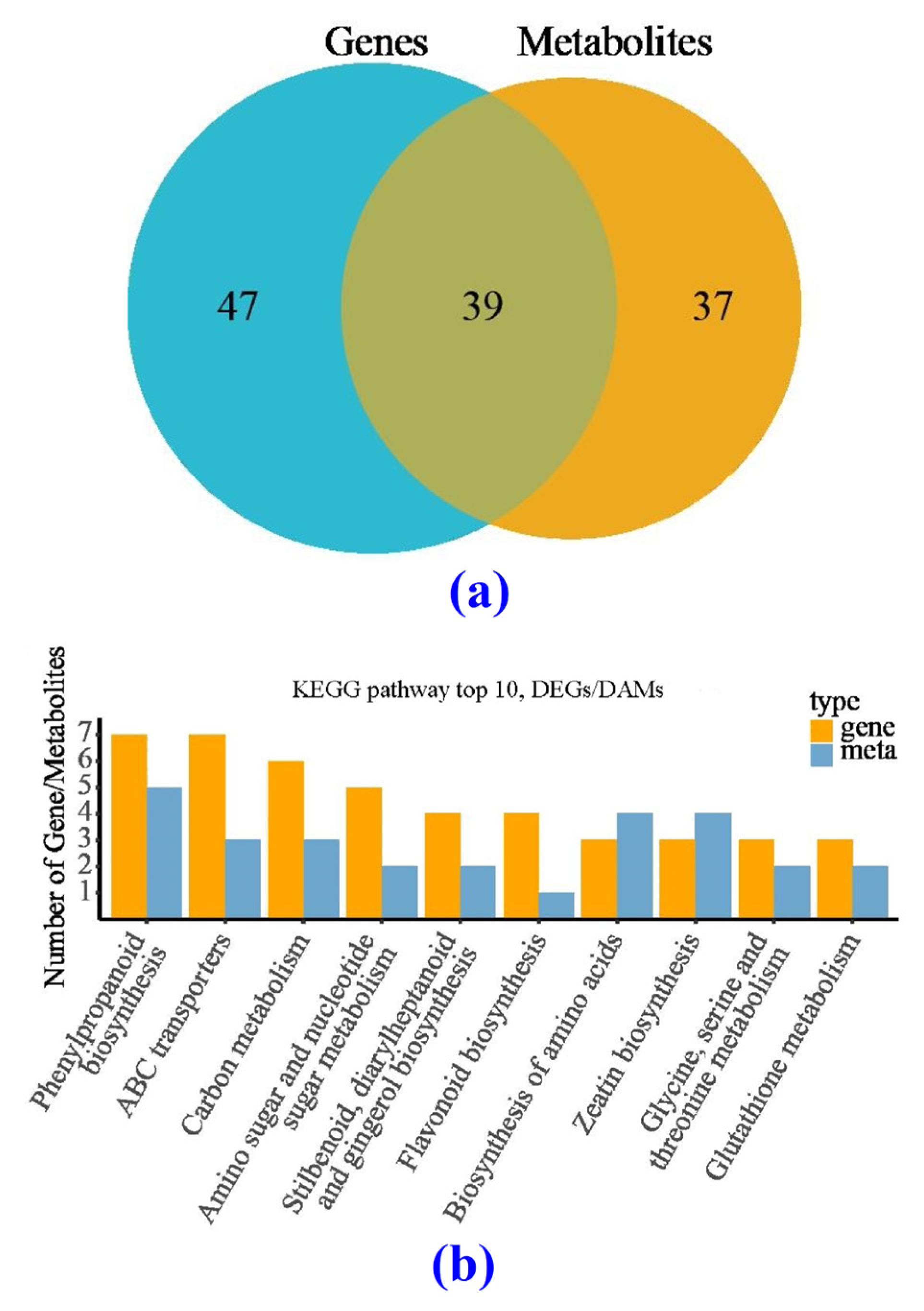
3.5.2. KEGG Analysis
3.5.3. Comparison of the Metabolome between the CK and BF Groups
3.6. Correlation Analysis between Transcription and Metabolism for the CK and BF Groups
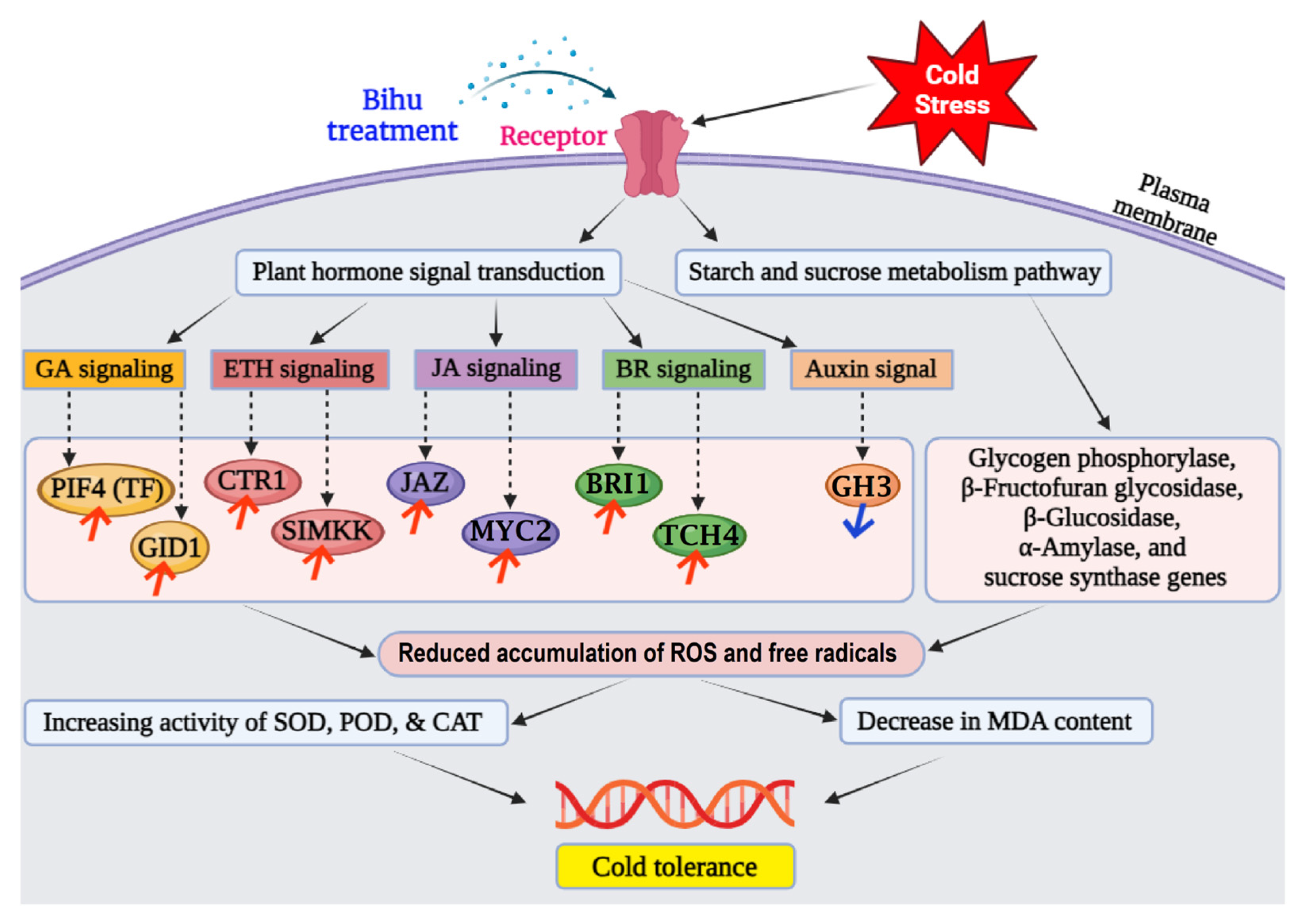
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, H.; Huang, D.; Ma, Q.; Qi, W.; Li, H. Factors Influencing the Technology Adoption Behaviours of Litchi Farmers in China. Sustainability 2020, 12, 271. [Google Scholar] [CrossRef]
- Shan, X.; Yang, Y.; Wei, S.; Wang, C.; Shen, W.; Chen, H.; Shen, J. Involvement of CBF in the Fine-Tuning of Litchi Flowering Time and Cold and Drought Stresses. Front. Plant Sci. 2023, 14, 1167458. [Google Scholar] [CrossRef]
- Menzel, C.M.; Simpson, D.R. Temperatures above 20DegCC Reduce Flowering in Lychee (Litchi Chinensis Sonn.). J. Hortic. Sci. 1995, 70, 981–987. [Google Scholar] [CrossRef]
- Bentz, B.J.; Rgnire, J.; Fettig, C.J.; Hansen, E.M.; Hayes, J.L.; Hicke, J.A.; Kelsey, R.G.; Negron, J.F.; Seybold, S.J. Climate Change and Bark Beetles of the Western United States and Canada: Direct and Indirect Effects. Bioscience 2010, 60, 602–613. [Google Scholar] [CrossRef]
- Vinocur, B.; Altman, A. Recent Advances in Engineering Plant Tolerance to Abiotic Stress: Achievements and Limitations. Curr. Opin. Biotechnol. 2005, 16, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Xiao, Q.; Qiu, H.; Chen, C.; Chen, H. Integrative Effect of Drought and Low Temperature on Litchi (Litchi Chinensis Sonn.) Floral Initiation Revealed by Dynamic Genome-Wide Transcriptome Analysis. Sci. Rep. 2016, 6, 32005. [Google Scholar] [CrossRef]
- O’Hare, T.J. Interaction of Temperature and Vegetative Flush Maturity Influences Shoot Structure and Development of Lychee (Litchi Chinensis Sonn.). Sci. Hortic. 2002, 95, 203–211. [Google Scholar] [CrossRef]
- O’Hare, T.J. Impact of Root and Shoot Temperature on Bud Dormancy and Floral Induction in Lychee (Litchi Chinensis Sonn.). Sci. Hortic. 2004, 99, 21–28. [Google Scholar] [CrossRef]
- Mitra, S.K.; Thingreingam Irenus, K.S. Elucidation of the Factors Affecting Poor Flowering in Litchi (Litchi Chinensis Sonn.) and Measures to Overcome. Acta Hortic. 2018, 1229, 143–149. [Google Scholar] [CrossRef]
- Stern, R.A.; Meron, M.; Naor, A.; Wallach, R.; Bravdo, B.; Gazit, S. Effect of Fall Irrigation Level in “Mauritius” and “Floridian” Lychee on Soil and Plant Water Status, Flowering Intensity, and Yield. J. Am. Soc. Hortic. Sci. 1998, 123, 150–155. [Google Scholar] [CrossRef]
- Carr, M.K.V.; Menzel, C.M. The Water Relations and Irrigation Requirements of Lychee (Litchi Chinensis Sonn.): A Review. Exp. Agric. 2014, 50, 481–497. [Google Scholar] [CrossRef][Green Version]
- Airaki, M.; Leterrier, M.; Mateos, R.M.; Valderrama, R.; Chaki, M.; Barroso, J.B.; del Río, L.A.; Palma, J.M.; Corpas, F.J. Metabolism of Reactive Oxygen Species and Reactive Nitrogen Species in Pepper (Capsicum Annuum L.) Plants under Low Temperature Stress. Plant Cell Env. 2012, 35, 281–295. [Google Scholar] [CrossRef] [PubMed]
- Gull, A.; Lone, A.A.; Wani, N.U.I. Biotic and Abiotic Stresses in Plants. Abiotic Biot. Stress. Plants 2016, 11, 13. [Google Scholar]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive Oxygen Species, Oxidative Damage, and Antioxidative Defense Mechanism in Plants under Stressful Conditions. J. Bot. 2012, 2012, 1–26. [Google Scholar] [CrossRef]
- Bhattacharjee, S. Reactive Oxygen Species and Oxidative Burst: Roles in Stress, Senescence and Signal Transduction in Plants. Curr. Sci. 2005, 89, 1113–1121. [Google Scholar]
- Holaday, A.S.; Martindale, W.; Aired, R.; Brooks, A.L.; Leegood, R.C. Changes in Activities of Enzymes of Carbon Metabolism in Leaves during Exposure of Plants to Low Temperature. Plant Physiol. 1992, 98, 1105–1114. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Wang, S.; Xie, J.; Tan, P.; Han, L. Discontinuous Low Temperature Stress and Plant Growth Regulators during the Germination Period Promote Roots Growth in Alfalfa (Medicago Sativa L.). Plant Physiol. Biochem. 2023, 197, 107624. [Google Scholar] [CrossRef] [PubMed]
- Castonguay, Y.; Nadeau, P.; Lechasseur, P.; Chouinard, L. Differential Accumulation of Carbohydrates in Alfalfa Cultivars of Contrasting Winterhardiness. Crop Sci. 1995, 35, 509–516. [Google Scholar] [CrossRef]
- Li, W.C.; Wu, J.Y.; Zhang, H.N.; Shi, S.Y.; Liu, L.Q.; Shu, B.; Liang, Q.Z.; Xie, J.H.; Wei, Y.Z. De Novo Assembly and Characterization of Pericarp Transcriptome and Identification of Candidate Genes Mediating Fruit Cracking in Litchi Chinensis Sonn. Int. J. Mol. Sci. 2014, 15, 17667–17685. [Google Scholar] [CrossRef]
- Zhang, C.; Shi, S. Physiological and Proteomic Responses of Contrasting Alfalfa (Medicago Sativa L.) Varieties to Peg-Induced Osmotic Stress. Front. Plant Sci. 2018, 9, 333359. [Google Scholar] [CrossRef]
- Chen, H.; Huang, H. China Litchi Industry: Development, Achievements and Problems. Acta Hortic. 2001, 558, 31–39. [Google Scholar] [CrossRef]
- Xu, Q.; Dong, Y.; Wang, Y.; Yang, R.; Xu, C. Determinants and Identification of the Northern Boundary of China’s Tropical Zone. J. Geogr. Sci. 2018, 28, 31–45. [Google Scholar] [CrossRef]
- Neuhoff, V.; Stamm, R.; Pardowitz, I.; Arold, N.; Ehrhardt, W.; Taube, D. Essential Problems in Quantification of Proteins Following Colloidal Staining with Coomassie Brilliant Blue Dyes in Polyacrylamide Gels, and Their Solution. Electrophoresis 1990, 11, 101–117. [Google Scholar] [CrossRef] [PubMed]
- Medlicott, A.P.; Thompson, A.K. Analysis of Sugars and Organic Acids in Ripening Mango Fruits (Mangifera Indica L. Var Keitt) by High Performance Liquid Chromatography. J. Sci. Food Agric. 1985, 36, 561–566. [Google Scholar] [CrossRef]
- Dhindsa, R.S.; Matowe, W. Drought Tolerance in Two Mosses: Correlated with Enzymatic Defence against Lipid Peroxidation. J. Exp. Bot. 1981, 32, 79–91. [Google Scholar] [CrossRef]
- Abrahám, E.; Hourton-Cabassa, C.; Erdei, L.; Szabados, L. Methods for Determination of Proline in Plants. Methods Mol. Biol. 2010, 639, 317–331. [Google Scholar] [CrossRef] [PubMed]
- Shah, K.; Kumar, R.G.; Verma, S.; Dubey, R.S. Effect of Cadmium on Lipid Peroxidation, Superoxide Anion Generation and Activities of Antioxidant Enzymes in Growing Rice Seedlings. Plant Sci. 2001, 161, 1135–1144. [Google Scholar] [CrossRef]
- Sun, M.; Zigman, S. An Improved Spectrophotometric Assay for Superoxide Dismutase Based on Epinephrine Autoxidation. Anal. Biochem. 1978, 90, 81–89. [Google Scholar] [CrossRef]
- Verma, S.; Dubey, R.S. Lead Toxicity Induces Lipid Peroxidation and Alters the Activities of Antioxidant Enzymes in Growing Rice Plants. Plant Sci. 2017, 3, 879–882. [Google Scholar] [CrossRef]
- Yang, S.L.; Lan, S.S.; Gong, M. Hydrogen Peroxide-Induced Proline and Metabolic Pathway of Its Accumulation in Maize Seedlings. J. Plant Physiol. 2009, 166, 1694–1699. [Google Scholar] [CrossRef]
- Winfield, M.O.; Lu, C.; Wilson, I.D.; Coghill, J.A.; Edwards, K.J. Plant Responses to Cold: Transcriptome Analysis of Wheat. Plant Biotechnol. J. 2010, 8, 749–771. [Google Scholar] [CrossRef]
- Tang, H.; Zhang, X.; Gong, B.; Yan, Y.; Shi, Q. Proteomics and Metabolomics Analysis of Tomato Fruit at Different Maturity Stages and under Salt Treatment. Food Chem. 2020, 311, 126009. [Google Scholar] [CrossRef]
- Paul, G.K.; Mahmud, S.; Dutta, A.K.; Sarkar, S.; Laboni, A.A.; Hossain, M.S.; Nagata, A.; Karmaker, P.; Razu, M.H.; Kazi, T.; et al. Volatile Compounds of Bacillus Pseudomycoides Induce Growth and Drought Tolerance in Wheat (Triticum Aestivum L.). Sci. Rep. 2022, 12, 19137. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Yao, Y.; Zeng, H.; Zhang, X. Integrated Metabolome and Transcriptome Analysis Reveals a Potential Mechanism for Water Accumulation Mediated Translucency in Pineapple (Ananas Comosus (L.) Merr.) Fruit. Int. J. Mol. Sci. 2023, 24, 7199. [Google Scholar] [CrossRef]
- Zhao, Y.; Song, C.; Brummell, D.A.; Qi, S.; Lin, Q.; Bi, J.; Duan, Y. Salicylic Acid Treatment Mitigates Chilling Injury in Peach Fruit by Regulation of Sucrose Metabolism and Soluble Sugar Content. Food Chem. 2021, 358, 129867. [Google Scholar] [CrossRef] [PubMed]
- Geng, J.; Wei, T.; Wang, Y.; Huang, X.; Liu, J.H. Overexpression of PtrbHLH, a Basic Helix-Loop-Helix Transcription Factor from Poncirus Trifoliata, Confers Enhanced Cold Tolerance in Pummelo (Citrus Grandis) by Modulation of H2O2 Level via Regulating a CAT Gene. Tree Physiol. 2019, 39, 2045–2054. [Google Scholar] [CrossRef]
- Thomashow, M.F. Molecular Basis of Plant Cold Acclimation: Insights Gained from Studying the CBF Cold Response Pathway. Plant Physiol. 2010, 154, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional Regulatory Networks in Cellular Responses and Tolerance to Dehydration and Cold Stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef]
- Kidokoro, S.; Maruyama, K.; Nakashima, K.; Imura, Y.; Narusaka, Y.; Shinwari, Z.K.; Osakabe, Y.; Fujita, Y.; Mizoi, J.; Shinozaki, K.; et al. The Phytochrome-Interacting Factor PIF7 Negatively Regulates Dreb1 Expression under Circadian Control in Arabidopsis. Plant Physiol. 2009, 151, 2046–2057. [Google Scholar] [CrossRef]
- Lee, C.M.; Thomashow, M.F. Photoperiodic Regulation of the C-Repeat Binding Factor (CBF) Cold Acclimation Pathway and Freezing Tolerance in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2012, 109, 15054–15059. [Google Scholar] [CrossRef]
- Jiang, B.; Shi, Y.; Zhang, X.; Xin, X.; Qi, L.; Guo, H.; Li, J.; Yang, S. PIF3 Is a Negative Regulator of the CBF Pathway and Freezing Tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E6695–E6702. [Google Scholar] [CrossRef] [PubMed]
- Lantzouni, O.; Alkofer, A.; Falter-Braun, P.; Schwechheimer, C. Growth-Regulating Factors Interact with Dellas and Regulate Growth in Cold Stress. Plant Cell 2020, 32, 1018–1034. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Lu, S.; Li, Z.; Cheng, J.; Hu, P.; Zhu, T.; Wang, X.; Jin, M.; Wang, X.; Li, L.; et al. CYCLIC NUCLEOTIDE-GATED ION CHANNELs 14 and 16 Promote Tolerance to Heat and Chilling in Rice1[OPEN]. Plant Physiol. 2020, 183, 1794–1808. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Chen, L.; Li, S.; Zhang, Y.; Xu, R.; Liu, Z.; Liu, W.; Kong, J.; Huang, X.; Wang, Y.; et al. BRI1 and BAK1 Interact with G Proteins and Regulate Sugar-Responsive Growth and Development in Arabidopsis. Nat. Commun. 2018, 9, 1522. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, D.; Zhou, R.; Wang, X.; Dossa, K.; Wang, L.; Zhang, Y.; Yu, J.; Gong, H.; Zhang, X.; et al. Transcriptome and Metabolome Analyses of Two Contrasting Sesame Genotypes Reveal the Crucial Biological Pathways Involved in Rapid Adaptive Response to Salt Stress. BMC Plant Biol. 2019, 19, 66. [Google Scholar] [CrossRef] [PubMed]
- Vitalini, S.; Cas, M.D.; Rubino, F.M.; Vigentini, I.; Foschino, R.; Iriti, M.; Paroni, R. LC-MS/MS-Based Profiling of Tryptophan-Related Metabolites in Healthy Plant Foods. Molecules 2020, 25, 311. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.C.; Song, R.F.; Zheng, S.Q.; Li, T.T.; Zhang, B.L.; Gao, X.; Lu, Y.T. Coordination of Plant Growth and Abiotic Stress Responses by Tryptophan Synthase β Subunit 1 through Modulation of Tryptophan and ABA Homeostasis in Arabidopsis. Mol. Plant 2022, 15, 973–990. [Google Scholar] [CrossRef] [PubMed]
- Schwechheimer, C. Gibberellin Signaling in Plants—The Extended Version. Front. Plant Sci. 2012, 2, 107. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Gong, F.; Cheminant, S.; Alioua, M.; Hedden, P.; Genschika, P. The Cold-Inducible CBF1 Factor-Dependent Signaling Pathway Modulates the Accumulation of the Growth-Repressing DELLA Proteins via Its Effect on Gibberellin Metabolism. Plant Cell 2008, 20, 2117–2129. [Google Scholar] [CrossRef]
- Richter, R.; Bastakis, E.; Schwechheimer, C. Cross-Repressive Interactions between SOC1 and the GATAs GNC and GNL/CGA1 in the Control of Greening, Cold Tolerance, and Flowering Time in Arabidopsis. Plant Physiol. 2013, 162, 1992–2004. [Google Scholar] [CrossRef]
- Hsieh, T.H.; Lee, J.T.; Yang, P.T.; Chiu, L.H.; Charng, Y.Y.; Wang, Y.C.; Chan, M.T. Heterology Expression of the Arabidopsis C-Repeat/Dehydration Response Element Binding Factor 1 Gene Confers Elevated Tolerance to Chilling and Oxidative Stresses in Transgenic Tomato. Plant Physiol. 2002, 129, 1086–1094. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Xu, M.; Wu, L.; Shen, C.; Ma, H.; Lin, J. CbCBF from Capsella Bursa-Pastoris Enhances Cold Tolerance and Restrains Growth in Nicotiana Tabacum by Antagonizing with Gibberellin and Affecting Cell Cycle Signaling. Plant Mol. Biol. 2014, 85, 259–275. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Yang, Y.; Guo, Y.; Wu, C.; Yang, G.; Gao, Z.; Zheng, C. Cotton GhDREB1 Increases Plant Tolerance to Low Temperature and Is Negatively Regulated by Gibberellic Acid. New Phytol. 2006, 176, 70–81. [Google Scholar]
- Richter, R.; Behringer, C.; Müller, I.K.; Schwechheimer, C. The GATA-Type Transcription Factors GNC and GNL/CGA1 Repress Gibberellin Signaling Downstream from DELLA Proteins and Phytochrome-Interacting Factors. Genes. Dev. 2010, 24, 2093–2104. [Google Scholar] [CrossRef] [PubMed]
- Franklin, K.A.; Lee, S.H.; Patel, D.; Kumar, S.V.; Spartz, A.K.; Gu, C.; Ye, S.; Yu, P.; Breen, G.; Cohen, J.D.; et al. Phytochrome-Interacting Factor 4 (PIF4) Regulates Auxin Biosynthesis at High Temperature. Proc. Natl. Acad. Sci. USA 2011, 108, 20231–20235. [Google Scholar] [CrossRef]
- Bernardo-García, S.; de Lucas, M.; Martínez, C.; Espinosa-Ruiz, A.; Davière, J.M.; Prat, S. BR-Dependent Phosphorylation Modulates PIF4 Transcriptional Activity and Shapes Diurnal Hypocotyl Growth. Genes. Dev. 2014, 28, 1681–1694. [Google Scholar] [CrossRef] [PubMed]
- Clouse, S.D. Brassinosteroid Signal Transduction: From Receptor Kinase Activation to Transcriptional Networks Regulating Plant Development. Plant Cell 2011, 23, 1219–1230. [Google Scholar] [CrossRef]
- Xia, X.J.; Wang, Y.J.; Zhou, Y.H.; Tao, Y.; Mao, W.H.; Shi, K.; Asami, T.; Chen, Z.; Yu, J.Q. Reactive Oxygen Species Are Involved in Brassinosteroid-Induced Stress Tolerance in Cucumber1[w]. Plant Physiol. 2009, 150, 801–814. [Google Scholar] [CrossRef] [PubMed]
- Singh, I.; Kumar, U.; Singh, S.K.; Gupta, C.; Singh, M.; Kushwaha, S.R. Physiological and Biochemical Effect of 24-Epibrassinoslide on Cold Tolerance in Maize Seedlings. Physiol. Mol. Biol. Plants 2012, 18, 229–236. [Google Scholar] [CrossRef]
- Jiang, Y.P.; Huang, L.F.; Cheng, F.; Zhou, Y.H.; Xia, X.J.; Mao, W.H.; Shi, K.; Yu, J.Q. Brassinosteroids Accelerate Recovery of Photosynthetic Apparatus from Cold Stress by Balancing the Electron Partitioning, Carboxylation and Redox Homeostasis in Cucumber. Physiol. Plant 2013, 148, 133–145. [Google Scholar] [CrossRef]
- Kagale, S.; Divi, U.K.; Krochko, J.E.; Keller, W.A.; Krishna, P. Brassinosteroid Confers Tolerance in Arabidopsis Thaliana and Brassica Napus to a Range of Abiotic Stresses. Planta 2007, 225, 353–364. [Google Scholar] [CrossRef]
- Hannah, M.A.; Heyer, A.G.; Hincha, D.K. A Global Survey of Gene Regulation during Cold Acclimation in Arabidopsis Thaliana. PLoS Genet. 2005, 1, 0179–0196. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Khurana, J.P. Transcript Profiling Reveals Diverse Roles of Auxin-Responsive Genes during Reproductive Development and Abiotic Stress in Rice. FEBS J. 2009, 276, 3148–3162. [Google Scholar] [CrossRef] [PubMed]
- Gavelienė, V.; Novickienė, L.; Pakalniškytė, L. Effect of Auxin Physiological Analogues on Rapeseed (Brassica Napus) Cold Hardening, Seed Yield and Quality. J. Plant Res. 2013, 126, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, K.; Urano, K.; Yoshiwara, K.; Morishita, Y.; Sakurai, N.; Suzuki, H.; Kojima, M.; Sakakibara, H.; Shibata, D.; Saito, K.; et al. Integrated Analysis of the Effects of Cold and Dehydration on Rice Metabolites, Phytohormones, and Gene Transcripts. Plant Physiol. 2014, 164, 1759–1771. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Liu, H.; Xiong, L. Endogenous Auxin and Jasmonic Acid Levels Are Differentially Modulated by Abiotic Stresses in Rice. Front. Plant Sci. 2013, 4, 397. [Google Scholar] [CrossRef]
- Munro, D.; Treberg, J.R. A Radical Shift in Perspective: Mitochondria as Regulators of Reactive Oxygen Species. J. Exp. Biol. 2017, 220, 1170–1180. [Google Scholar] [CrossRef]
- Cui, X.; Wang, Y.; Wu, J.; Han, X.; Gu, X.; Lu, T.; Zhang, Z. The RNA Editing Factor DUA1 Is Crucial to Chloroplast Development at Low Temperature in Rice. New Phytol. 2019, 221, 834–849. [Google Scholar] [CrossRef] [PubMed]
- Nie, W.F.; Wang, M.M.; Xia, X.J.; Zhou, Y.H.; Shi, K.; Chen, Z.; Yu, J.Q. Silencing of Tomato RBOH1 and MPK2 Abolishes Brassinosteroid-Induced H2O2 Generation and Stress Tolerance. Plant Cell Env. 2013, 36, 789–803. [Google Scholar] [CrossRef]
- Baier, M.; Bittner, A.; Prescher, A.; van Buer, J. Preparing Plants for Improved Cold Tolerance by Priming. Plant Cell Env. 2019, 42, 782–800. [Google Scholar] [CrossRef]
- Ding, Y.; Shi, Y.; Yang, S. Advances and Challenges in Uncovering Cold Tolerance Regulatory Mechanisms in Plants. New Phytol. 2019, 222, 1690–1704. [Google Scholar] [CrossRef] [PubMed]
- Kiran, A.; Sharma, P.N.; Awasthi, R.; Nayyar, H.; Seth, R.; Chandel, S.S.; Siddique, K.H.M.; Zinta, G.; Sharma, K.D. Disruption of Carbohydrate and Proline Metabolism in Anthers under Low Temperature Causes Pollen Sterility in Chickpea. Env. Exp. Bot. 2021, 188, 104500. [Google Scholar] [CrossRef]
- Parthasarathy, A.; Savka, M.A.; Hudson, A.O. The Synthesis and Role of β-Alanine in Plants. Front. Plant Sci. 2019, 10, 921. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, Y.; Yuan, L.; Zhou, F.; Gao, Y.; Kang, Z.; Li, T.; Hu, X. Exogenous 5-Aminolevulinic Acid Alleviates Low-Temperature Injury by Regulating Glutathione Metabolism and β-Alanine Metabolism in Tomato Seedling Roots. Ecotoxicol. Env. Saf. 2022, 245, 83–93. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, X.-W.; Cao, X.-Y.; Jiang, W.-H.; Xu, G.-Z.; Liang, Q.-Z.; Yang, Z.-Y. Cryoprotectant-Mediated Cold Stress Mitigation in Litchi Flower Development: Transcriptomic and Metabolomic Perspectives. Metabolites 2024, 14, 223. https://doi.org/10.3390/metabo14040223
Zheng X-W, Cao X-Y, Jiang W-H, Xu G-Z, Liang Q-Z, Yang Z-Y. Cryoprotectant-Mediated Cold Stress Mitigation in Litchi Flower Development: Transcriptomic and Metabolomic Perspectives. Metabolites. 2024; 14(4):223. https://doi.org/10.3390/metabo14040223
Chicago/Turabian StyleZheng, Xue-Wen, Xin-Yue Cao, Wen-Hao Jiang, Guang-Zhao Xu, Qing-Zhi Liang, and Zhuan-Ying Yang. 2024. "Cryoprotectant-Mediated Cold Stress Mitigation in Litchi Flower Development: Transcriptomic and Metabolomic Perspectives" Metabolites 14, no. 4: 223. https://doi.org/10.3390/metabo14040223
APA StyleZheng, X.-W., Cao, X.-Y., Jiang, W.-H., Xu, G.-Z., Liang, Q.-Z., & Yang, Z.-Y. (2024). Cryoprotectant-Mediated Cold Stress Mitigation in Litchi Flower Development: Transcriptomic and Metabolomic Perspectives. Metabolites, 14(4), 223. https://doi.org/10.3390/metabo14040223



