Hypoglycemic Activity of Rice Resistant-Starch Metabolites: A Mechanistic Network Pharmacology and In Vitro Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Screening of Differential Metabolites and Predicting Targets of RRS
2.3. Screening of Common Targets of Metabolites and T2DM
2.4. Construction of Protein–Protein Interaction (PPI) Network and Screening of Key Targets
2.5. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis
2.6. Construction of Component–Target–Pathway–Disease
2.7. Molecular Docking of Core Metabolites of RRS and Key Targets
2.8. Molecular Docking of Core Metabolites of RRS, α-Glucosidase, and α-Amylase
2.9. Assay of Inhibition of α-Glucosidase by Core Metabolites
2.10. Assay of Inhibition of α-Amylase by Core Metabolites
2.11. Statistical Analysis
3. Results and Discussion
3.1. Metabolites and Potential Targets of RRS
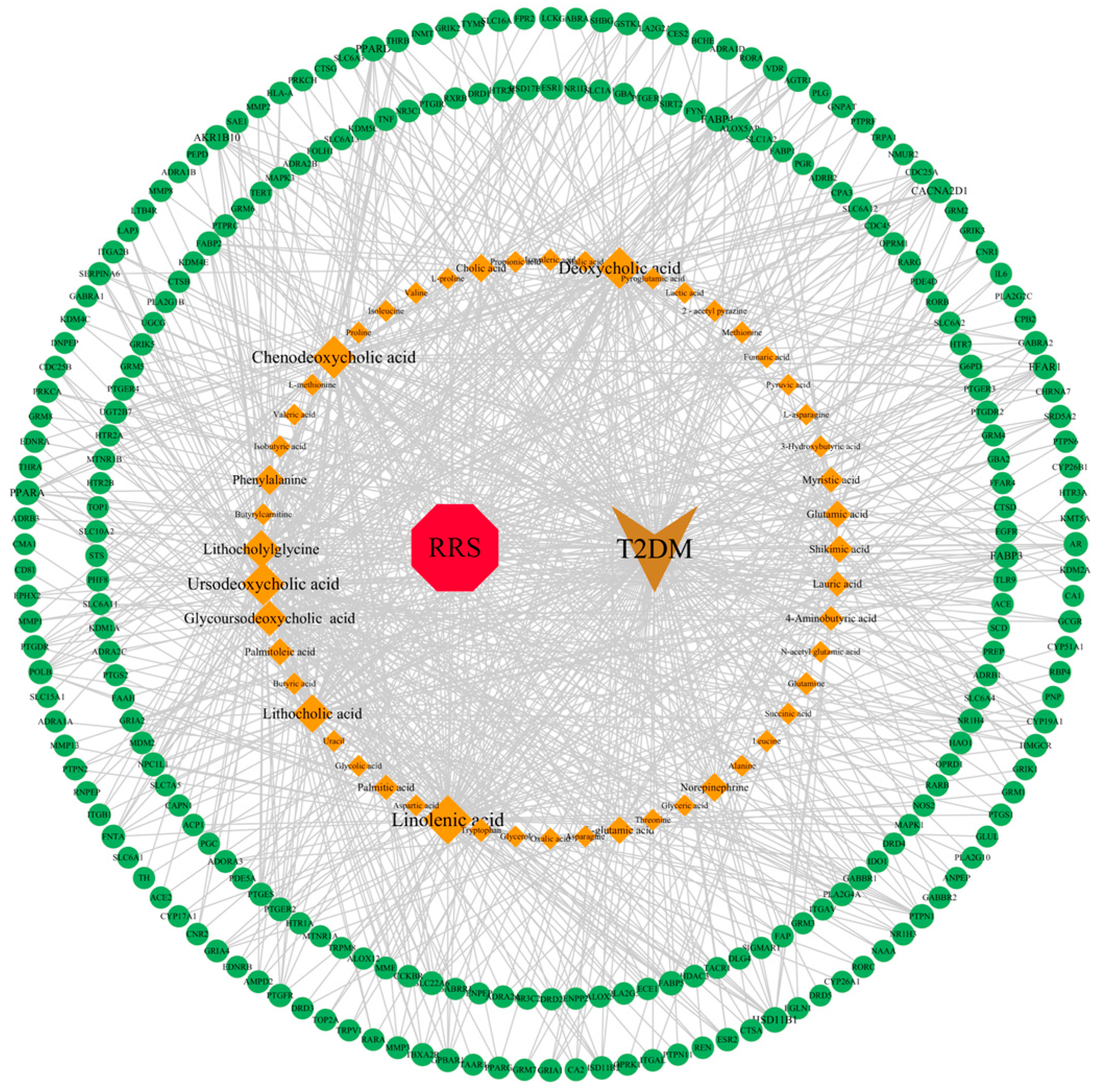
3.2. Screening of Intersection Targets between T2DM and Metabolites of RRS
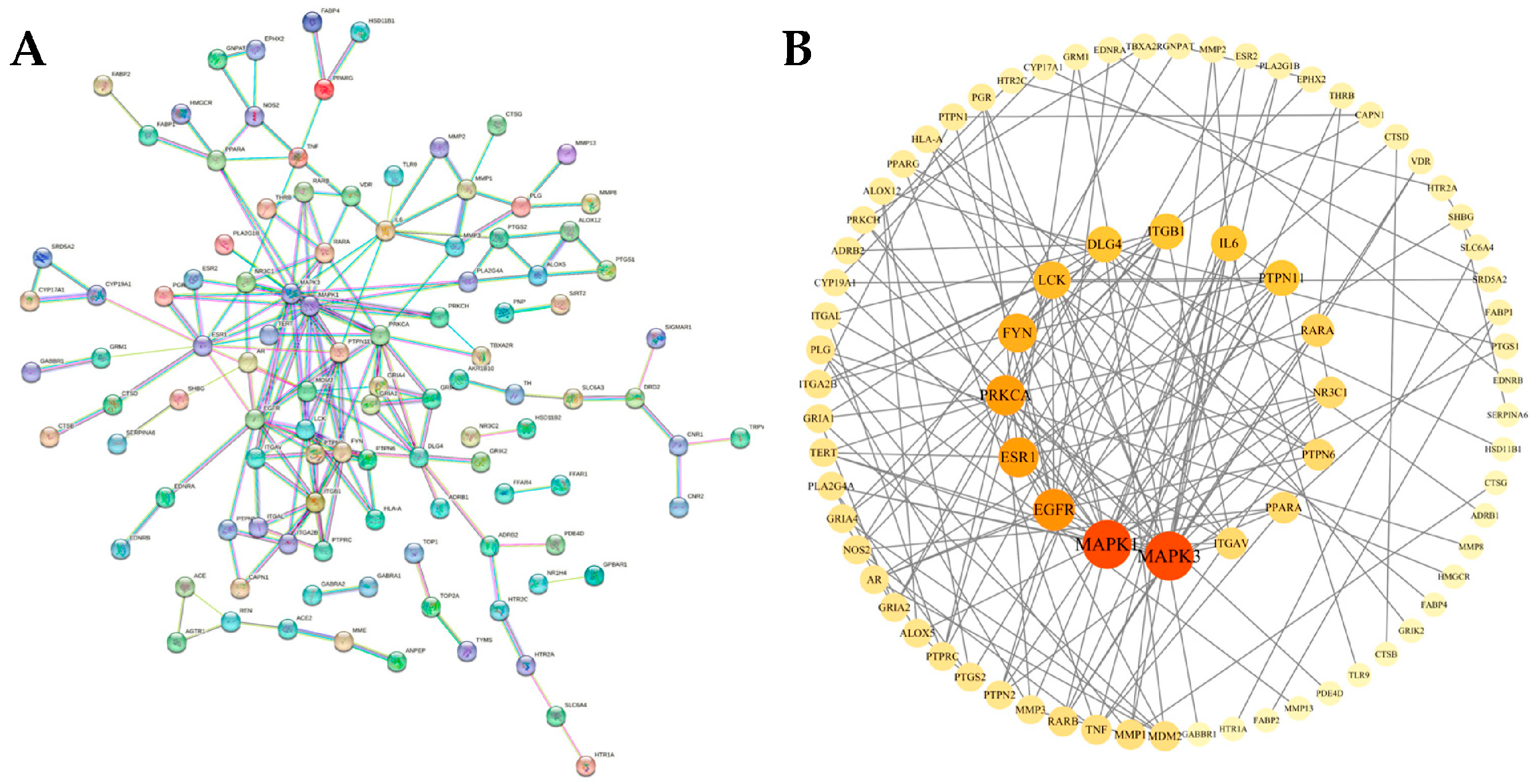
3.3. Construction and Analysis of PPI Protein Interaction Network
3.4. GO Functional Annotation and KEGG Pathway Enrichment Analysis
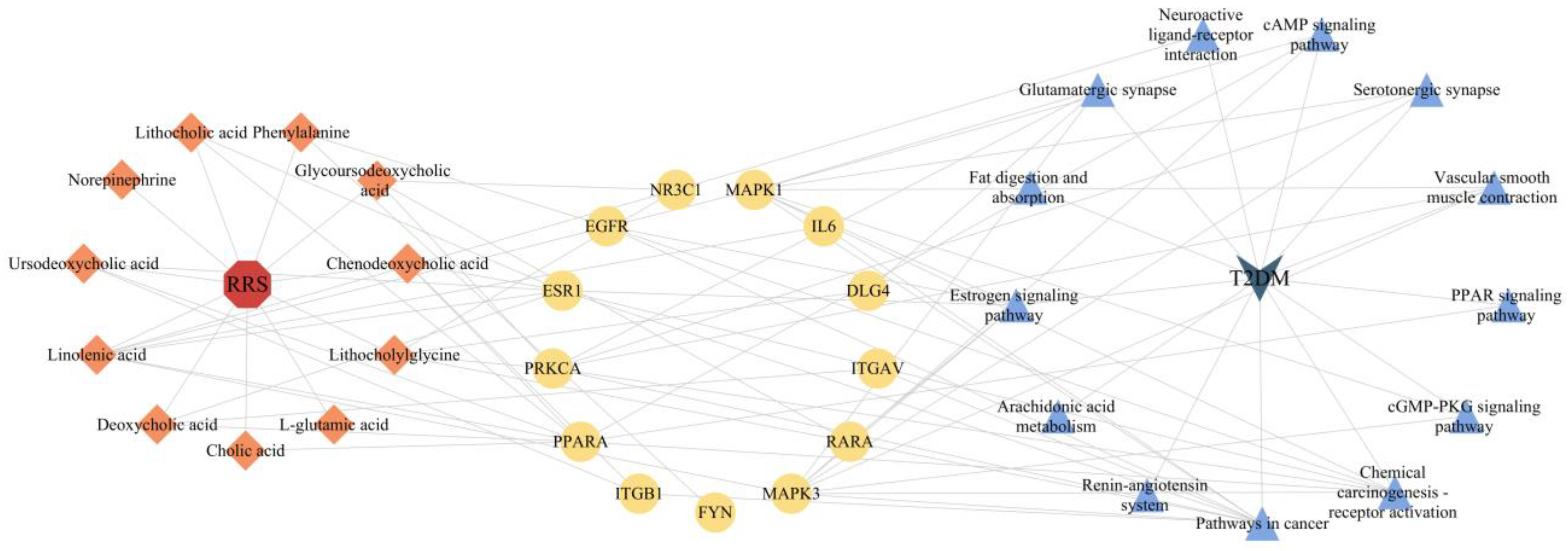
3.5. Construction of RRS-Metabolite-Target-Pathway-T2DM Network
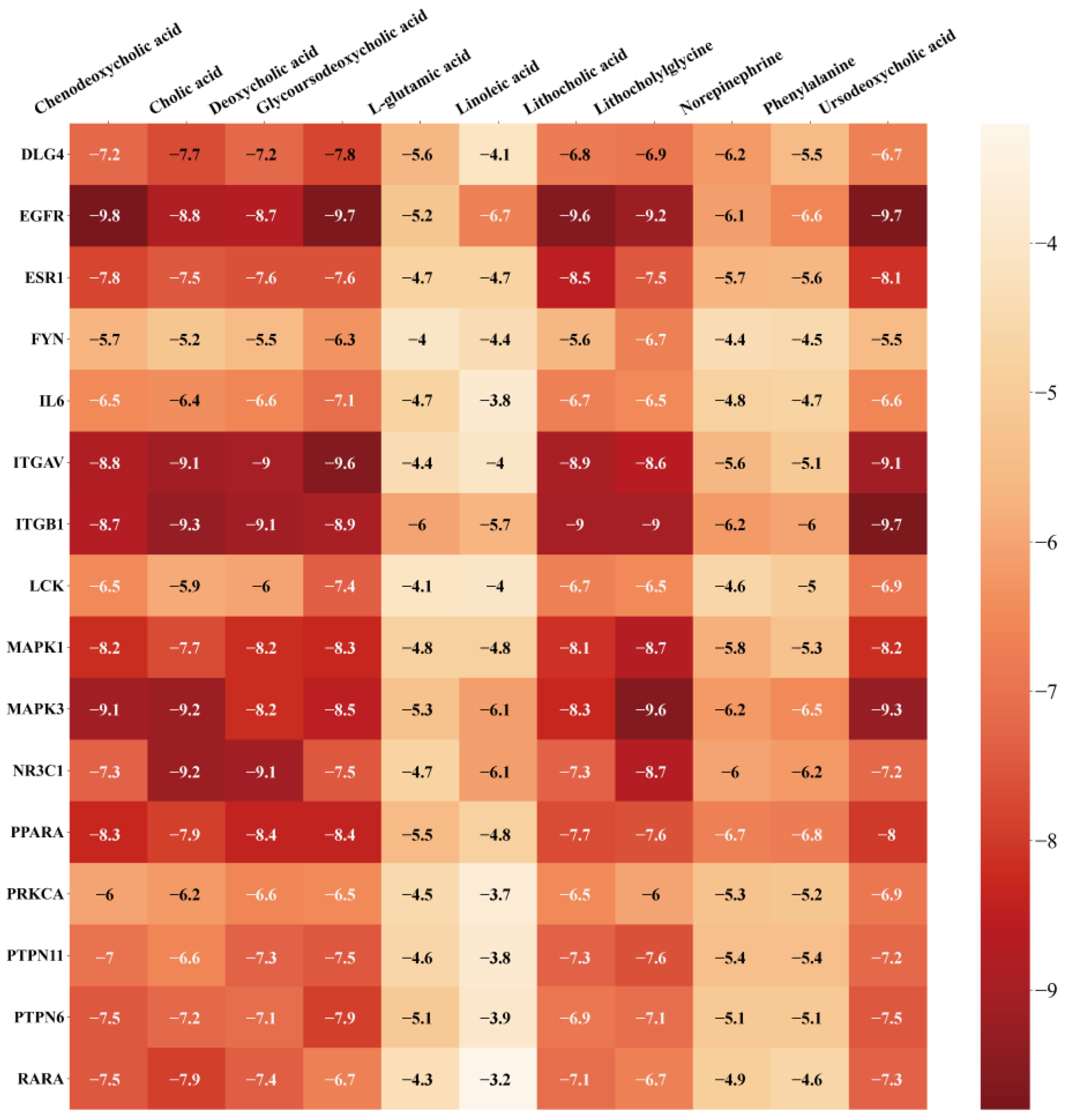
3.6. Verification of Molecule Docking between Core Metabolites of RRS and Targets
3.7. Molecular Docking Validation of Key RRS Metabolites with α-Glucosidase and α-Amylase
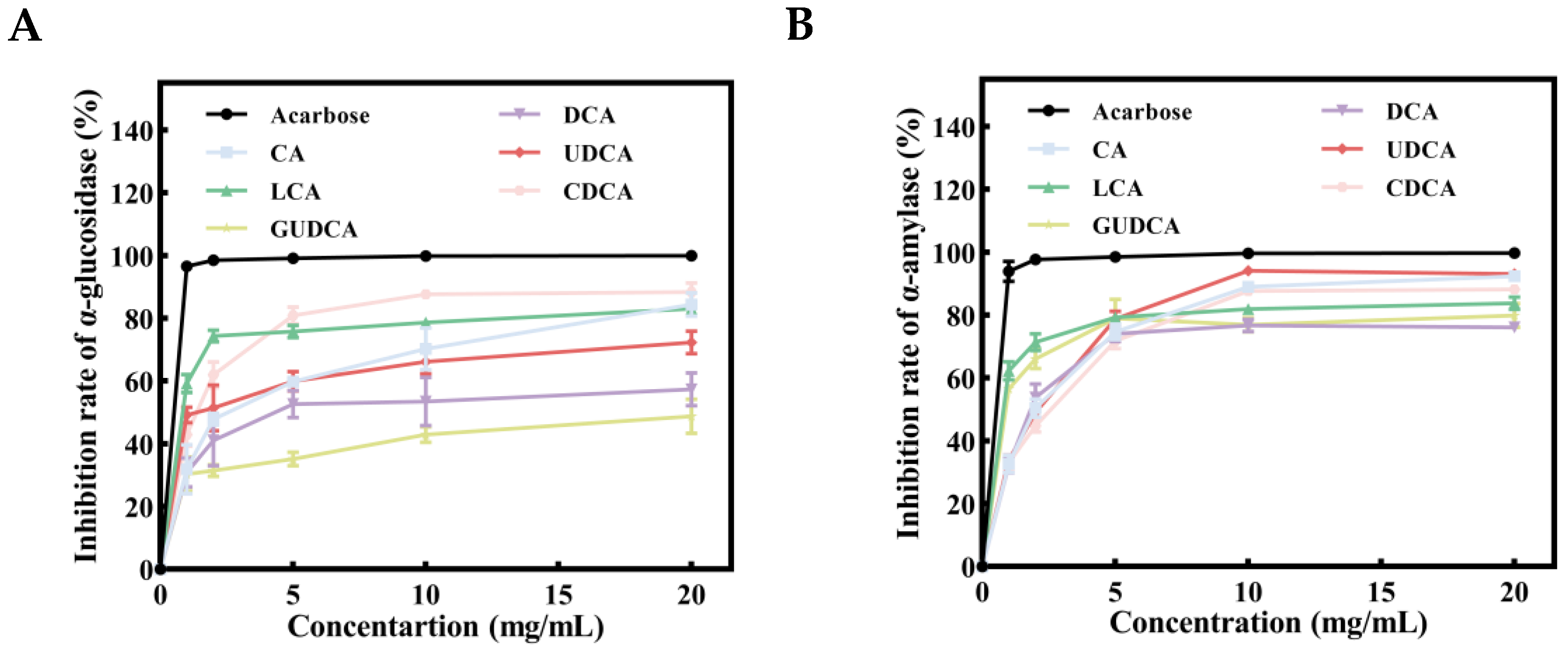
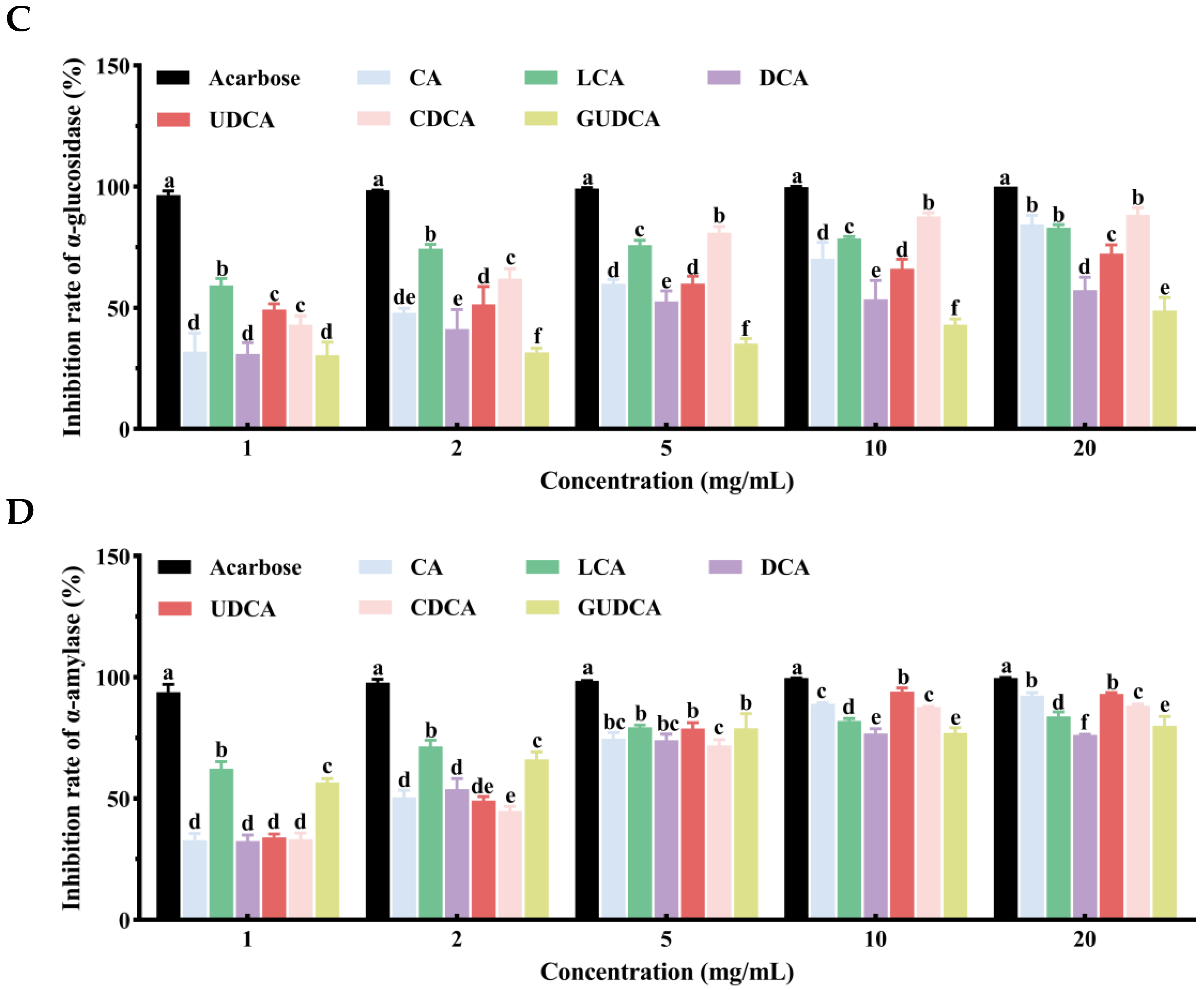
3.8. Inhibitory Effects of Bile Acids on α-Glucosidase and α-Amylase
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Poznyak, A.; Grechko, A.V.; Poggio, P.; Myasoedova, V.A.; Alfieri, V.; Orekhov, A.N. The Diabetes Mellitus-Atherosclerosis Connection: The Role of Lipid and Glucose Metabolism and Chronic Inflammation. Int. J. Mol. Sci. 2020, 21, 1835. [Google Scholar] [CrossRef] [PubMed]
- Stumvoll, M.; Goldstein, B.J.; Van Haeften, T.W. Type 2 diabetes: Principles of pathogenesis and therapy. Lancet 2005, 365, 1333–1346. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, E.; Lim, S.; Lamptey, R.; Webb, D.R.; Davies, M.J. Type 2 diabetes. Lancet 2022, 400, 1803–1820. [Google Scholar] [CrossRef] [PubMed]
- Kolb, H.; Martin, S. Environmental/lifestyle factors in the pathogenesis and prevention of type 2 diabetes. BMC Med. 2017, 15, 131. [Google Scholar] [CrossRef] [PubMed]
- Rakesh, K.C.; Narola, A.K.; Chaurasia, K.; Vigyan, K. Nutritional and health benefits of Millets: A review. Pharma Innov. 2023, 12, 3360–3363. [Google Scholar] [CrossRef]
- Meng, J.; Zhu, Y.; Ma, H.; Wang, X.; Zhao, Q. The role of traditional Chinese medicine in the treatment of cognitive dysfunction in type 2 diabetes. J. Ethnopharmacol. 2021, 280, 114464. [Google Scholar] [CrossRef] [PubMed]
- Venkatakrishnan, K.; Chiu, H.-F.; Wang, C.-K. Popular functional foods and herbs for the management of type-2-diabetes mellitus: A comprehensive review with special reference to clinical trials and its proposed mechanism. J. Funct. Foods 2019, 57, 425–438. [Google Scholar] [CrossRef]
- Tester, R.F.; Karkalas, J.; Qi, X. Starch—Composition, fine structure and architecture. J. Cereal Sci. 2004, 39, 151–165. [Google Scholar] [CrossRef]
- Kaimal, A.M.; Mujumdar, A.S.; Thorat, B.N. Resistant starch from millets: Recent developments and applications in food industries. Trends Food Sci. Technol. 2021, 111, 563–580. [Google Scholar] [CrossRef]
- Liu, J.; Lu, W.; Liang, Y.; Wang, L.; Jin, N.; Zhao, H.; Fan, B.; Wang, F. Research Progress on Hypoglycemic Mechanisms of Resistant Starch: A Review. Molecules 2022, 27, 7111. [Google Scholar] [CrossRef]
- Jiang, F.; Du, C.; Jiang, W.; Wang, L.; Du, S.-k. The preparation, formation, fermentability, and applications of resistant starch. Int. J. Biol. Macromol. 2020, 150, 1155–1161. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, M.; Ma, Q.; Tian, B.; Nie, C.; Chen, Z.; Li, J. Health beneficial effects of resistant starch on diabetes and obesity via regulation of gut microbiota: A review. Food Funct. 2020, 11, 5749–5767. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Wang, F.; Ren, X.; Wang, Y.; Blanchard, C. Resistant starch manipulated hyperglycemia/hyperlipidemia and related genes expression in diabetic rats. Int. J. Biol. Macromol. 2015, 75, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, B.; Kumari, A.; Sarkhel, S.; Jha, S.; Mukherjee, A.; Jain, M.; Mohan, A.; Roy, A. Rice Starch Phase Transition and Detection During Resistant Starch Formation. Food Rev. Int. 2024, 40, 158–184. [Google Scholar] [CrossRef]
- Kim, Y.J.; Kim, J.G.; Lee, W.K.; So, K.M.; Kim, J.K. Trial data of the anti-obesity potential of a high resistant starch diet for canines using Dodamssal rice and the identification of discriminating markers in feces for metabolic profiling. Metabolomics 2019, 15, 21. [Google Scholar] [CrossRef] [PubMed]
- Wan, J. Effects of Rice with Different Amounts of Resistant Starch on Mice Fed A High-fat Diet: Regulation of Lipid Metabolism and Gut Microbiota. Ph.D. Thesis, Huazhong Agricultural University, Wuhan, China, 2021. [Google Scholar] [CrossRef]
- Gao, P.; Zhang, Y.; Gao, C.; Xiang, X.; Zhang, X.; Wang, Z. Effects of brown rice on metabolomics related to glucose and lipid in normal rats. Wei Sheng Yan Jiu 2021, 50, 600–608. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Gao, W.; Zhang, Y.; Liu, C.; Wang, G.; Wu, H.; Gao, W. Integrated Phytochemical Analysis Based on UPLC-MS and Network Pharmacology Approaches to Explore the Quality Control Markers for the Quality Assessment of Trifolium pratense L. Molecules 2020, 25, 3787. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Ding, Q. New progress of interdisciplinary research between network toxicology, quality markers and TCM network pharmacology. Chinese Herbal. Med. 2019, 11, 347–348. [Google Scholar] [CrossRef]
- Xiao, Q.Y.; Ye, T.Y.; Wang, X.L.; Han, L.; Wang, T.X.; Qi, D.M.; Cheng, X.R.; Wang, S.Q. A network pharmacology-based study on key pharmacological pathways and targets of Qi Fu Yin acting on Alzheimer’s disease. Exp. Gerontol. 2021, 149, 111336. [Google Scholar] [CrossRef]
- Tian, J.L.; Si, X.; Wang, Y.H.; Gong, E.S.; Xie, X.; Zhang, Y.; Li, B.; Shu, C. Bioactive flavonoids from Rubus corchorifolius inhibit α-glucosidase and α-amylase to improve postprandial hyperglycemia. Food Chem. 2021, 341, 128149. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, B.; Xiao, J.; Huang, Q.; Li, C.; Fu, X. Physicochemical, functional, and biological properties of water-soluble polysaccharides from Rosa roxburghii Tratt fruit. Food Chem. 2018, 249, 127–135. [Google Scholar] [CrossRef]
- Qian, L. Diagnosis of Non-Alcoholic Fatty Liver Disease and Clinical Studies of Resistant Starch Intervention. Ph.D. Thesis, Shanghai Jiaotong University, Shanghai, China, 2022. [Google Scholar] [CrossRef]
- Segal, M.S.; Gollub, E.; Johnson, R.J. Is the fructose index more relevant with regards to cardiovascular disease than the glycemic index? Eur. J. Nutr. 2007, 46, 406–417. [Google Scholar] [CrossRef]
- Özturan, A.; Arslan, S.; Kocaadam, B.; Elibol, E.; İmamoğlu, İ.; Karadağ, M.G. Effect of inositol and its derivatives on diabetes: A systematic review. Crit. Rev. Food Sci. Nutr. 2019, 59, 1124–1136. [Google Scholar] [CrossRef] [PubMed]
- Lebeck, J. Metabolic impact of the glycerol channels AQP7 and AQP9 in adipose tissue and liver. J. Mol. Endocrinol. 2014, 52, R165–R178. [Google Scholar] [CrossRef] [PubMed]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef] [PubMed]
- Lim, G.E.; Brubaker, P.L. Glucagon-Like Peptide 1 Secretion by the L-Cell: The View from within. Diabetes 2006, 55, S70–S77. [Google Scholar] [CrossRef]
- Zhu, Y.; Dong, L.; Huang, L.; Shi, Z.; Dong, J.; Yao, Y.; Shen, R. Effects of oat β-glucan, oat resistant starch, and the whole oat flour on insulin resistance, inflammation, and gut microbiota in high-fat-diet-induced type 2 diabetic rats. J. Funct. Foods 2020, 69, 103939. [Google Scholar] [CrossRef]
- Nguyen, A.; Bouscarel, B. Bile acids and signal transduction: Role in glucose homeostasis. Cell. Signal. 2008, 20, 2180–2197. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, T.; Jiang, R.; Zhao, A.; Wu, Q.; Kuang, J.; Sun, D.; Ren, Z.; Li, M.; Zhao, M.; et al. Hyocholic acid species improve glucose homeostasis through a distinct TGR5 and FXR signaling mechanism. Cell Metab. 2021, 33, 791–803.e797. [Google Scholar] [CrossRef]
- Saravanan, R.; Pari, L. Succinic acid monoethyl ester prevents oxidative stress in streptozotocin-nicotinamide-induced type2 diabetic rats. J. Basic. Clin. Physiol. Pharmacol. 2006, 17, 115–132. [Google Scholar] [CrossRef]
- Hou, X.W.; Wang, Y.; Pan, C.W. Metabolomics in Diabetic Retinopathy: A Systematic Review. Investig. Ophthalmol. Vis. Sci. 2021, 62, 4. [Google Scholar] [CrossRef] [PubMed]
- Tricò, D.; Mengozzi, A.; Nesti, L.; Hatunic, M.; Gabriel Sanchez, R.; Konrad, T.; Lalić, K.; Lalić, N.M.; Mari, A.; Natali, A. Circulating palmitoleic acid is an independent determinant of insulin sensitivity, beta cell function and glucose tolerance in non-diabetic individuals: A longitudinal analysis. Diabetologia 2020, 63, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Xiao, C.; Wu, Q.; Xie, Y.; Chen, X.; Yong, T.; Zhang, R.; Shao, W. Hypoglycemic effect of important edible and medicinal fungi polysaccharides: A review. Microbiol. China 2021, 48, 197–209. [Google Scholar] [CrossRef]
- Aldehaiman, A.; Momin, A.A.; Restouin, A.; Wang, L.; Shi, X.; Aljedani, S.; Opi, S.; Lugari, A.; Shahul Hameed, U.F.; Ponchon, L.; et al. Synergy and allostery in ligand binding by HIV-1 Nef. Biochem. J. 2021, 478, 1525–1545. [Google Scholar] [CrossRef]
- Alicea-Velázquez, N.L.; Jakoncic, J.; Boggon, T.J. Structure-guided studies of the SHP-1/JAK1 interaction provide new insights into phosphatase catalytic domain substrate recognition. J. Struct. Biol. 2013, 181, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Barr, A.J.; Ugochukwu, E.; Lee, W.H.; King, O.N.; Filippakopoulos, P.; Alfano, I.; Savitsky, P.; Burgess-Brown, N.A.; Müller, S.; Knapp, S. Large-scale structural analysis of the classical human protein tyrosine phosphatome. Cell 2009, 136, 352–363. [Google Scholar] [CrossRef] [PubMed]
- Camara-Artigas, A.; Murciano-Calles, J.; Martínez, J.C. Conformational changes in the third PDZ domain of the neuronal postsynaptic density protein 95. Acta Crystallogr. D Struct. Biol. 2019, 75, 381–391. [Google Scholar] [CrossRef]
- Chaikuad, A.; Tacconi, E.M.; Zimmer, J.; Liang, Y.; Gray, N.S.; Tarsounas, M.; Knapp, S. A unique inhibitor binding site in ERK1/2 is associated with slow binding kinetics. Nat. Chem. Biol. 2014, 10, 853–860. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Zhao, B.; Iacob, R.E.; Zhu, J.; Koksal, A.C.; Lu, C.; Engen, J.R.; Springer, T.A. Force interacts with macromolecular structure in activation of TGF-β. Nature 2017, 542, 55–59. [Google Scholar] [CrossRef]
- Elkins, J.M.; Papagrigoriou, E.; Berridge, G.; Yang, X.; Phillips, C.; Gileadi, C.; Savitsky, P.; Doyle, D.A. Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions. Protein Sci. 2007, 16, 683–694. [Google Scholar] [CrossRef]
- He, Y.; Yi, W.; Suino-Powell, K.; Zhou, X.E.; Tolbert, W.D.; Tang, X.; Yang, J.; Yang, H.; Shi, J.; Hou, L.; et al. Structures and mechanism for the design of highly potent glucocorticoids. Cell Res. 2014, 24, 713–726. [Google Scholar] [CrossRef] [PubMed]
- Hosfield, D.J.; Weber, S.; Li, N.S.; Sauvage, M.; Joiner, C.F.; Hancock, G.R.; Sullivan, E.A.; Ndukwe, E.; Han, R.; Cush, S.; et al. Stereospecific lasofoxifene derivatives reveal the interplay between estrogen receptor alpha stability and antagonistic activity in ESR1 mutant breast cancer cells. eLife 2022, 11, e72512. [Google Scholar] [CrossRef] [PubMed]
- Kamata, S.; Oyama, T.; Saito, K.; Honda, A.; Yamamoto, Y.; Suda, K.; Ishikawa, R.; Itoh, T.; Watanabe, Y.; Shibata, T.; et al. PPARα Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates. iScience 2020, 23, 101727. [Google Scholar] [CrossRef] [PubMed]
- le Maire, A.; Teyssier, C.; Erb, C.; Grimaldi, M.; Alvarez, S.; de Lera, A.R.; Balaguer, P.; Gronemeyer, H.; Royer, C.A.; Germain, P.; et al. A unique secondary-structure switch controls constitutive gene repression by retinoic acid receptor. Nat. Struct. Mol. Biol. 2010, 17, 801–807. [Google Scholar] [CrossRef] [PubMed]
- Planken, S.; Behenna, D.C.; Nair, S.K.; Johnson, T.O.; Nagata, A.; Almaden, C.; Bailey, S.; Ballard, T.E.; Bernier, L.; Cheng, H.; et al. Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR. J. Med. Chem. 2017, 60, 3002–3019. [Google Scholar] [CrossRef] [PubMed]
- Reif, A.; Lam, K.; Weidler, S.; Lott, M.; Boos, I.; Lokau, J.; Bretscher, C.; Mönnich, M.; Perkams, L.; Schmälzlein, M.; et al. Natural Glycoforms of Human Interleukin 6 Show Atypical Plasma Clearance. Angew. Chem. Int. Ed. Engl. 2021, 60, 13380–13387. [Google Scholar] [CrossRef] [PubMed]
- Ward, R.A.; Anderton, M.J.; Bethel, P.; Breed, J.; Cook, C.; Davies, E.J.; Dobson, A.; Dong, Z.; Fairley, G.; Farrington, P.; et al. Discovery of a Potent and Selective Oral Inhibitor of ERK1/2 (AZD0364) That Is Efficacious in Both Monotherapy and Combination Therapy in Models of Nonsmall Cell Lung Cancer (NSCLC). J. Med. Chem. 2019, 62, 11004–11018. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Springer, T.A. Metal ion and ligand binding of integrin α5β1. Proc. Natl. Acad. Sci. USA 2014, 111, 17863–17868. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Kim, J.L.; Newcomb, J.R.; Rose, P.E.; Stover, D.R.; Toledo, L.M.; Zhao, H.; Morgenstern, K.A. Structural analysis of the lymphocyte-specific kinase Lck in complex with non-selective and Src family selective kinase inhibitors. Structure 1999, 7, 651–661. [Google Scholar] [CrossRef]
- Istyastono, E.P.; Radifar, M.; Yuniarti, N.; Prasasty, V.D.; Mungkasi, S. PyPLIF HIPPOS: A Molecular Interaction Fingerprinting Tool for Docking Results of AutoDock Vina and PLANTS. J. Chem. Inf. Model. 2020, 60, 3697–3702. [Google Scholar] [CrossRef]
- Yun, C.H.; Boggon, T.J.; Li, Y.; Woo, M.S.; Greulich, H.; Meyerson, M.; Eck, M.J. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: Mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell 2007, 11, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Mo, J.; Dong, S.; Liao, Z.; Zhang, B.; Zhu, P. Integrinβ-1 in disorders and cancers: Molecular mechanisms and therapeutic targets. Cell Commun. Signal. 2024, 22, 71. [Google Scholar] [CrossRef] [PubMed]
- Taherkhani, A.; Khodadadi, P.; Samie, L.; Azadian, Z.; Bayat, Z. Flavonoids as Strong Inhibitors of MAPK3: A Computational Drug Discovery Approach. Int. J. Anal. Chem. 2023, 2023, 8899240. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Deng, Y.; Xie, X.; Ma, J.; Xu, M.; Zhao, X.; Gu, W.; Hong, J.; Wang, W.; Xu, G.; et al. Plasma bile acid changes in type 2 diabetes correlated with insulin secretion in two-step hyperglycemic clamp. J. Diabetes 2018, 10, 874–885. [Google Scholar] [CrossRef] [PubMed]
- Meier, P.J.; Stieger, B. Bile salt transporters. Annu. Rev. Physiol. 2002, 64, 635–661. [Google Scholar] [CrossRef]
- Noel, O.F.; Still, C.D.; Argyropoulos, G.; Edwards, M.; Gerhard, G.S. Bile Acids, FXR, and Metabolic Effects of Bariatric Surgery. J. Obes. 2016, 2016, 4390254. [Google Scholar] [CrossRef]
- Han, S.Y.; Song, H.K.; Cha, J.J.; Han, J.Y.; Kang, Y.S.; Cha, D.R. Farnesoid X receptor (FXR) agonist ameliorates systemic insulin resistance, dysregulation of lipid metabolism, and alterations of various organs in a type 2 diabetic kidney animal model. Acta Diabetol. 2021, 58, 495–503. [Google Scholar] [CrossRef]
- Sun, L.; Xie, C.; Wang, G.; Wu, Y.; Wu, Q.; Wang, X.; Liu, J.; Deng, Y.; Xia, J.; Chen, B.; et al. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat. Med. 2018, 24, 1919–1929. [Google Scholar] [CrossRef] [PubMed]
- Ðanić, M.; Stanimirov, B.; Pavlović, N.; Goločorbin-Kon, S.; Al-Salami, H.; Stankov, K.; Mikov, M. Pharmacological Applications of Bile Acids and Their Derivatives in the Treatment of Metabolic Syndrome. Front. Pharmacol. 2018, 9, 1382. [Google Scholar] [CrossRef]
- Castellanos-Jankiewicz, A.; Guzmán-Quevedo, O.; Fénelon, V.S.; Zizzari, P.; Quarta, C.; Bellocchio, L.; Tailleux, A.; Charton, J.; Fernandois, D.; Henricsson, M.; et al. Hypothalamic bile acid-TGR5 signaling protects from obesity. Cell Metab. 2021, 33, 1483–1492.e1410. [Google Scholar] [CrossRef]
- Huang, S.; Ma, S.; Ning, M.; Yang, W.; Ye, Y.; Zhang, L.; Shen, J.; Leng, Y. TGR5 agonist ameliorates insulin resistance in the skeletal muscles and improves glucose homeostasis in diabetic mice. Metabolism 2019, 99, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Kuhre, R.E.; Holst, J.J.; Kappe, C. The regulation of function, growth and survival of GLP-1-producing L-cells. Clin. Sci. 2016, 130, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Xie, S.; Chi, Z.; Zhang, J.; Liu, Y.; Zhang, L.; Zheng, M.; Zhang, X.; Xia, D.; Ke, Y.; et al. Bile Acids Control Inflammation and Metabolic Disorder through Inhibition of NLRP3 Inflammasome. Immunity 2016, 45, 802–816. [Google Scholar] [CrossRef]
- Renga, B.; Mencarelli, A.; Cipriani, S.; D‘Amore, C.; Carino, A.; Bruno, A.; Francisci, D.; Zampella, A.; Distrutti, E.; Fiorucci, S. The bile acid sensor FXR is required for immune-regulatory activities of TLR-9 in intestinal inflammation. PLoS ONE 2013, 8, e54472. [Google Scholar] [CrossRef] [PubMed]
- Hosseinkhani, F.; Heinken, A.; Thiele, I.; Lindenburg, P.W.; Harms, A.C.; Hankemeier, T. The contribution of gut bacterial metabolites in the human immune signaling pathway of non-communicable diseases. Gut Microbes 2021, 13, 1882927. [Google Scholar] [CrossRef] [PubMed]
- Normanno, N.; De Luca, A.; Bianco, C.; Strizzi, L.; Mancino, M.; Maiello, M.R.; Carotenuto, A.; De Feo, G.; Caponigro, F.; Salomon, D.S. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 2006, 366, 2–16. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yao, S.; Ding, J.M.; Chen, W.; Guo, Y. Enhancer-Gene Interaction Analyses Identified the Epidermal Growth Factor Receptor as a Susceptibility Gene for Type 2 Diabetes Mellitus. Diabetes Metab. J. 2021, 45, 241–250. [Google Scholar] [CrossRef]
- Farah, R.I.; Al-Sabbagh, M.Q.; Momani, M.S.; Albtoosh, A.; Arabiat, M.; Abdulraheem, A.M.; Aljabiri, H.; Abufaraj, M. Diabetic kidney disease in patients with type 2 diabetes mellitus: A cross-sectional study. BMC Nephrol. 2021, 22, 223. [Google Scholar] [CrossRef]
- Shi, R.; Zhao, W.M.; Zhu, L.; Wang, R.F.; Wang, D.G. Identification of basement membrane markers in diabetic kidney disease and immune infiltration by using bioinformatics analysis and experimental verification. IET Syst. Biol. 2023, 17, 316–326. [Google Scholar] [CrossRef]
- Weng, S.; Zemany, L.; Standley, K.N.; Novack, D.V.; La Regina, M.; Bernal-Mizrachi, C.; Coleman, T.; Semenkovich, C.F. β3 integrin deficiency promotes atherosclerosis and pulmonary inflammation in high-fat-fed, hyperlipidemic mice. Proc. Natl. Acad. Sci. USA 2003, 100, 6730–6735. [Google Scholar] [CrossRef]
- Cheng, R.; Ma, J.-x. Angiogenesis in diabetes and obesity. Rev. Endocr. Metab. Disord. 2015, 16, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zhu, X.; Liu, Y.; Jiang, F.; Chen, M.; Cheng, L.; Cheng, X. Gene Expression Profiling of Type 2 Diabetes Mellitus by Bioinformatics Analysis. Comput. Math. Methods Med. 2020, 2020, 9602016. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Wang, J.; Wang, H.; Zhou, J.; Qi, H.; Naji, Y.; Zhao, L.; Tang, Y.; Dai, Y. Knockdown of NR3C1 inhibits the proliferation and migration of clear cell renal cell carcinoma through activating endoplasmic reticulum stress–mitophagy. J. Transl. Med. 2023, 21, 701. [Google Scholar] [CrossRef] [PubMed]
- Rafacho, A.; Ortsäter, H.; Nadal, A.; Quesada, I. Glucocorticoid treatment and endocrine pancreas function: Implications for glucose homeostasis, insulin resistance and diabetes. J. Endocrinol. 2014, 223, R49–R62. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, R.; Kluwe, B.; Odei, J.B.; Echouffo Tcheugui, J.B.; Sims, M.; Kalyani, R.R.; Bertoni, A.G.; Golden, S.H.; Joseph, J.J. The association of morning serum cortisol with glucose metabolism and diabetes: The Jackson Heart Study. Psychoneuroendocrinology 2019, 103, 25–32. [Google Scholar] [CrossRef]
- Wei, D.; Liu, X.; Huo, W.; Yu, S.; Li, L.; Wang, C.; Mao, Z. Serum cortisone and glucocorticoid receptor gene (NR3C1) polymorphism in human dysglycemia. Hormones 2020, 19, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Di, S.; Han, L.; An, X.; Kong, R.; Gao, Z.; Yang, Y.; Wang, X.; Zhang, P.; Ding, Q.; Wu, H.; et al. In silico network pharmacology and in vivo analysis of berberine-related mechanisms against type 2 diabetes mellitus and its complications. J. Ethnopharmacol. 2021, 276, 114180. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Deng, X.L.; Aleteng, Q.Q.; Li, L.; Zhu, J. Genome-Wide Transcriptome Analysis in Type 2 Diabetes Patients Treated by Sitagliptin. Diabetes Metab. Syndr. Obes. 2022, 15, 1761–1770. [Google Scholar] [CrossRef]
- Noor, F.; Rehman, A.; Ashfaq, U.A.; Saleem, M.H.; Okla, M.K.; Al-Hashimi, A.; AbdElgawad, H.; Aslam, S. Integrating Network Pharmacology and Molecular Docking Approaches to Decipher the Multi-Target Pharmacological Mechanism of Abrus precatorius L. Acting on Diabetes. Pharmaceuticals 2022, 15, 414. [Google Scholar] [CrossRef]
- Wang, A.; Li, Z.; Zhuo, S.; Gao, F.; Zhang, H.; Zhang, Z.; Ren, G.; Ma, X. Mechanisms of Cardiorenal Protection With SGLT2 Inhibitors in Patients With T2DM Based on Network Pharmacology. Front. Cardiovasc. Med. 2022, 9, 857952. [Google Scholar] [CrossRef]
- Yuan, G.; Shi, S.; Jia, Q.; Shi, J.; Shi, S.; Zhang, X.; Shou, X.; Zhu, X.; Hu, Y. Use of Network Pharmacology to Explore the Mechanism of Gegen (Puerariae lobatae Radix) in the Treatment of Type 2 Diabetes Mellitus Associated with Hyperlipidemia. Evid. Based Complement. Altern. Med. 2021, 2021, 6633402. [Google Scholar] [CrossRef] [PubMed]
- Nahoum, V.; Roux, G.; Anton, V.; Rougé, P.; Puigserver, A.; Bischoff, H.; Henrissat, B.; Payan, F. Crystal structures of human pancreatic alpha-amylase in complex with carbohydrate and proteinaceous inhibitors. Biochem. J. 2000, 346, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Tagami, T.; Yamashita, K.; Okuyama, M.; Mori, H.; Yao, M.; Kimura, A. Molecular basis for the recognition of long-chain substrates by plant α-glucosidases. J. Biol. Chem. 2013, 288, 19296–19303. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yang, S.; An, J.; Wang, M.; Li, H.; Liu, X. Statistical Characterization of Food-Derived α-Amylase Inhibitory Peptides: Computer Simulation and Partial Least Squares Regression Analysis. Molecules 2024, 29, 395. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Ding, H.; Hu, X.; Zhang, G.; Gong, D. Galangin inhibits α-glucosidase activity and formation of non-enzymatic glycation products. Food Chem. 2019, 271, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Zaharudin, N.; Salmeán, A.A.; Dragsted, L.O. Inhibitory effects of edible seaweeds, polyphenolics and alginates on the activities of porcine pancreatic α-amylase. Food Chem. 2018, 245, 1196–1203. [Google Scholar] [CrossRef] [PubMed]
- Haeusler, R.A.; Astiarraga, B.; Camastra, S.; Accili, D.; Ferrannini, E. Human insulin resistance is associated with increased plasma levels of 12α-hydroxylated bile acids. Diabetes 2013, 62, 4184–4191. [Google Scholar] [CrossRef] [PubMed]
- Duran-Sandoval, D.; Mautino, G.; Martin, G.; Percevault, F.; Barbier, O.; Fruchart, J.C.; Kuipers, F.; Staels, B. Glucose regulates the expression of the farnesoid X receptor in liver. Diabetes 2004, 53, 890–898. [Google Scholar] [CrossRef] [PubMed]
- Kir, S.; Beddow, S.A.; Samuel, V.T.; Miller, P.; Previs, S.F.; Suino-Powell, K.; Xu, H.E.; Shulman, G.I.; Kliewer, S.A.; Mangelsdorf, D.J. FGF19 as a postprandial, insulin-independent activator of hepatic protein and glycogen synthesis. Science 2011, 331, 1621–1624. [Google Scholar] [CrossRef]
- Katsuma, S.; Hirasawa, A.; Tsujimoto, G. Bile acids promote glucagon-like peptide-1 secretion through TGR5 in a murine enteroendocrine cell line STC-1. Biochem. Biophys. Res. Commun. 2005, 329, 386–390. [Google Scholar] [CrossRef]
- Thomas, C.; Gioiello, A.; Noriega, L.; Strehle, A.; Oury, J.; Rizzo, G.; Macchiarulo, A.; Yamamoto, H.; Mataki, C.; Pruzanski, M.; et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009, 10, 167–177. [Google Scholar] [CrossRef]
- Schauer, P.R.; Bhatt, D.L.; Kirwan, J.P.; Wolski, K.; Aminian, A.; Brethauer, S.A.; Navaneethan, S.D.; Singh, R.P.; Pothier, C.E.; Nissen, S.E.; et al. Bariatric Surgery versus Intensive Medical Therapy for Diabetes—5-Year Outcomes. N. Engl. J. Med. 2017, 376, 641–651. [Google Scholar] [CrossRef]


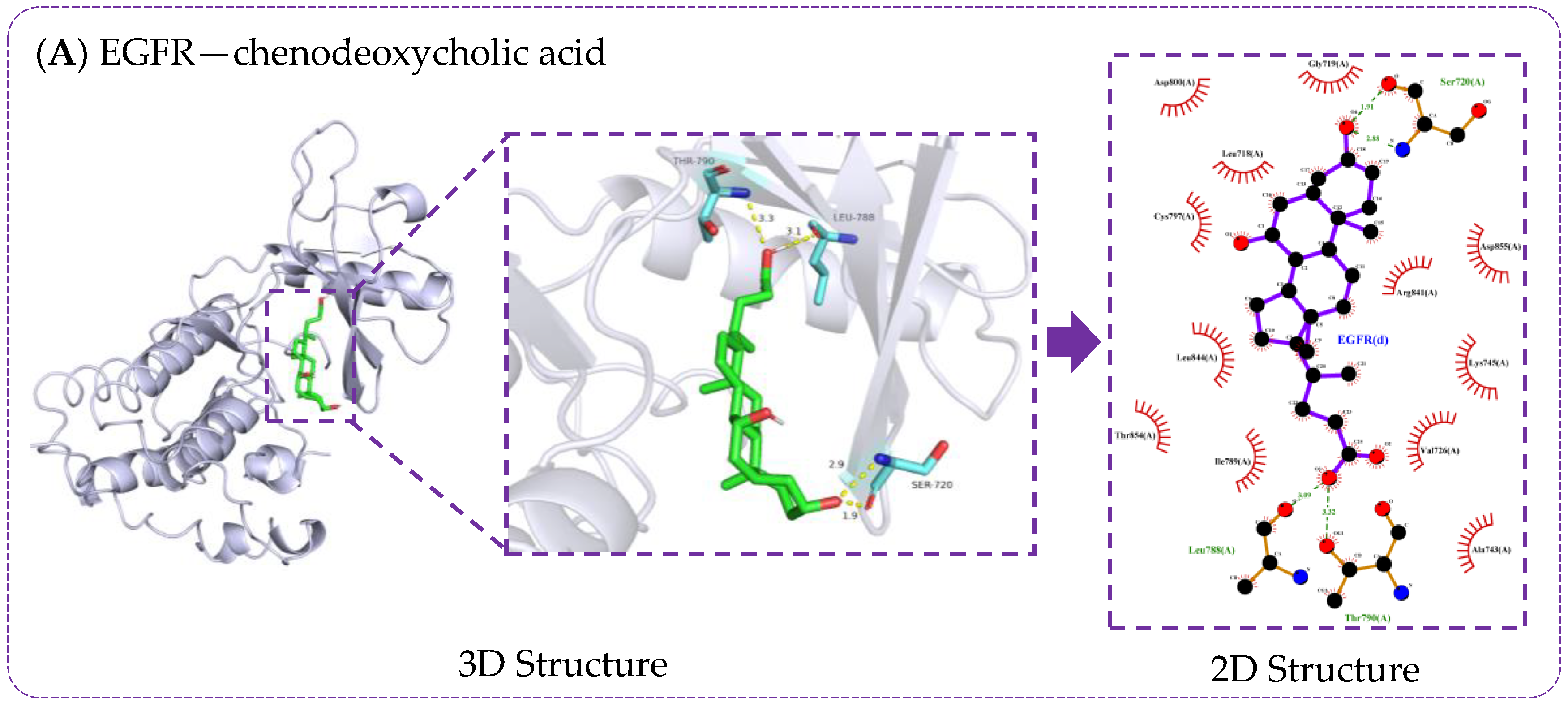
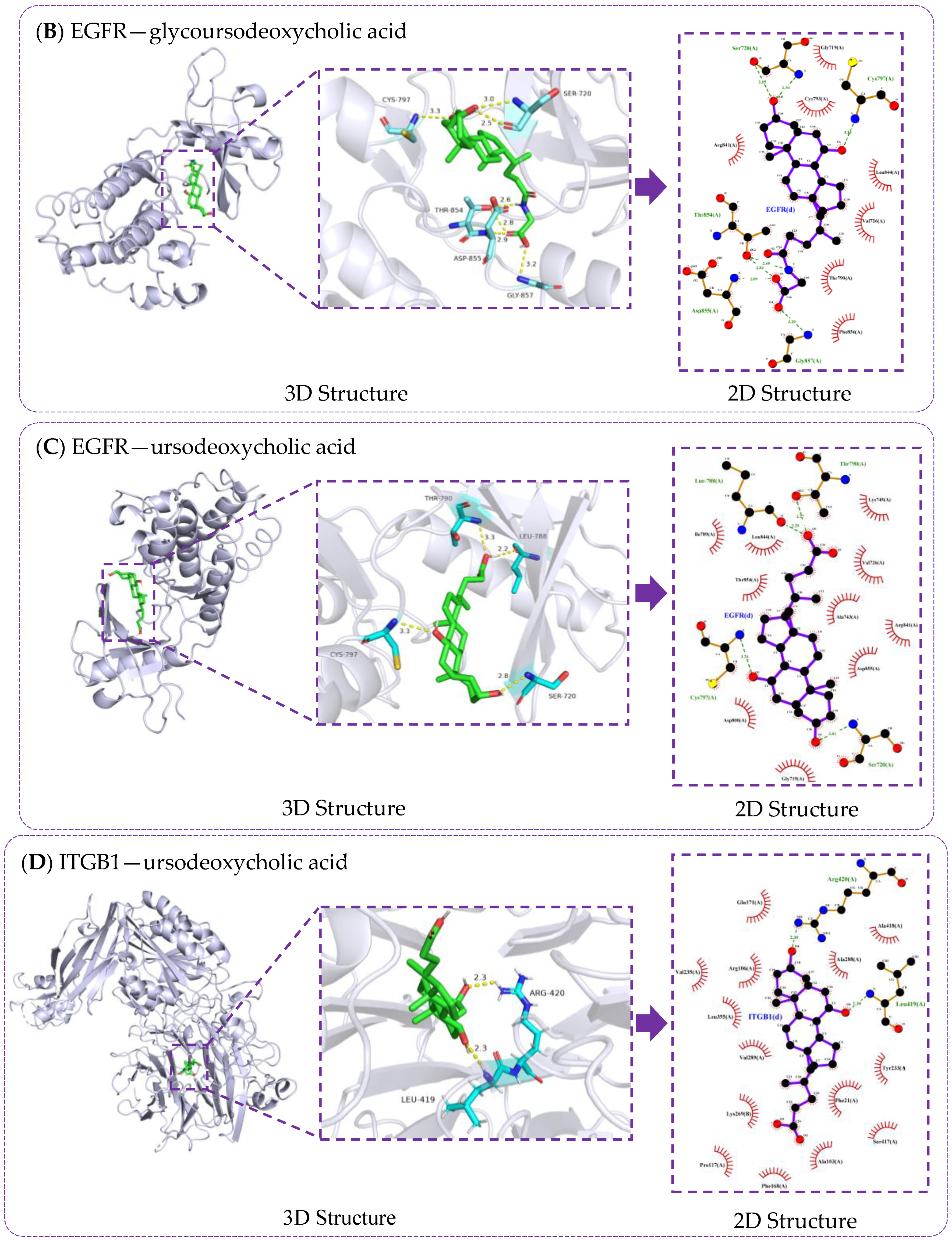
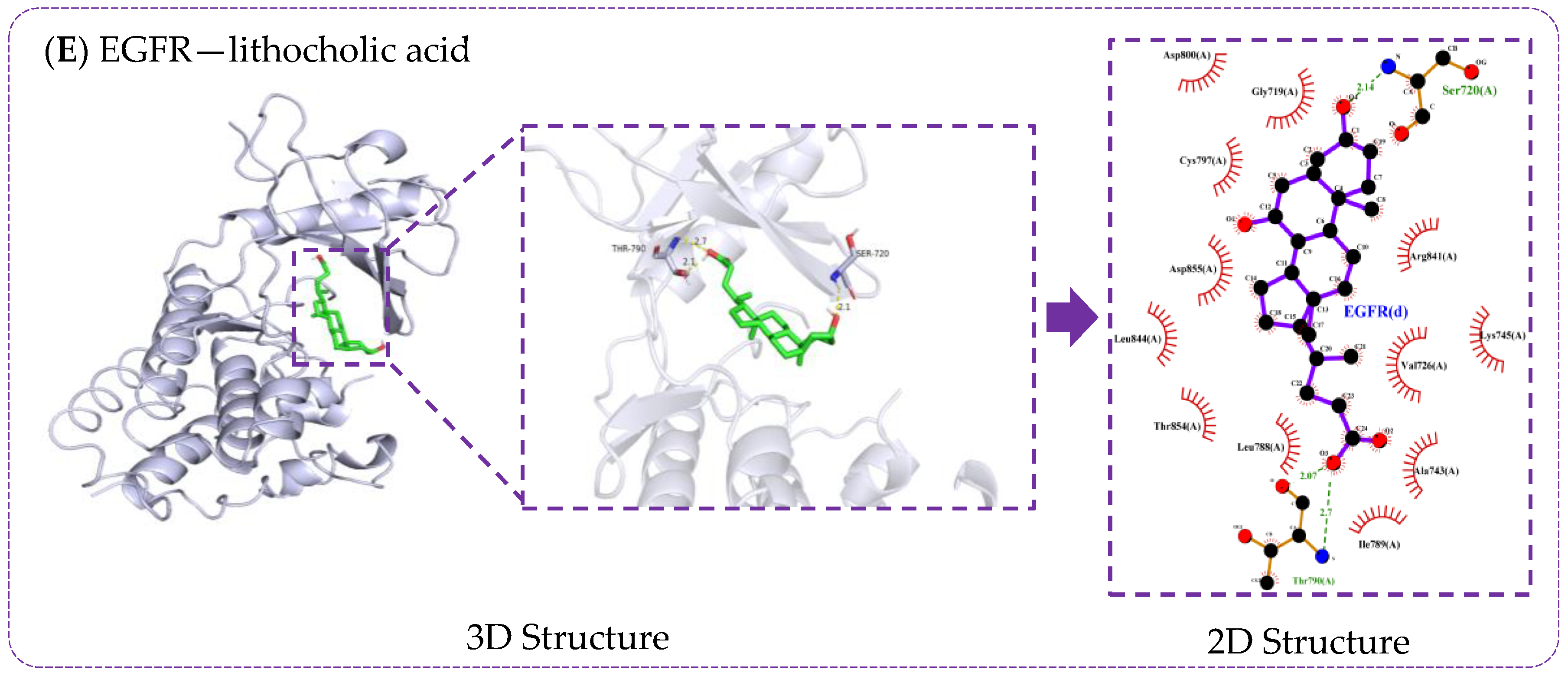
| Metabolites | Principles of Drug-Likeness | Gastrointestinal Absorption Capacity | PubChem CID | ||||
|---|---|---|---|---|---|---|---|
| Lipinski | Ghose | Veber | Egan | Muegge | |||
| 2-acetyl pyrazine | Yes | No | Yes | Yes | No | High | 30914 |
| 3-hydroxybutyric acid | Yes | No | Yes | Yes | No | High | 441 |
| 4-aminobutyric acid | Yes | No | Yes | Yes | No | High | 119 |
| Alanine | Yes | No | Yes | Yes | No | High | 5950 |
| Asparagine | Yes | No | Yes | Yes | No | High | 6267 |
| Aspartic acid | Yes | No | Yes | Yes | No | High | 5960 |
| Butyric acid | Yes | No | Yes | Yes | No | High | 264 |
| Butyrylcarnitine | Yes | No | Yes | Yes | Yes | High | 213114 |
| Chenodeoxycholic acid | Yes | Yes | Yes | Yes | Yes | High | 10133 |
| Cholic acid | Yes | Yes | Yes | Yes | Yes | High | 221493 |
| Deoxycholic acid | Yes | Yes | Yes | Yes | Yes | High | 222528 |
| Fumaric acid | Yes | No | Yes | Yes | No | High | 444972 |
| Glutamic acid | Yes | No | Yes | Yes | No | High | 33032 |
| Glutamine | Yes | No | Yes | Yes | No | High | 5961 |
| Glyceric acid | Yes | No | Yes | Yes | No | High | 752 |
| Glycerol | Yes | No | Yes | Yes | No | High | 753 |
| Glycolic acid | Yes | No | Yes | Yes | No | High | 757 |
| Glycoursodeoxycholic acid | Yes | No | Yes | Yes | Yes | High | 12310288 |
| Isobutyric acid | Yes | No | Yes | Yes | No | High | 6590 |
| Isoleucine | Yes | No | Yes | Yes | No | High | 6306 |
| Isovaleric acid | Yes | No | Yes | Yes | No | High | 10430 |
| Lactic acid | Yes | No | Yes | Yes | No | High | 612 |
| L-asparagine | Yes | No | Yes | Yes | No | High | 6267 |
| Lauric acid | Yes | Yes | Yes | Yes | Yes | High | 3893 |
| Leucine | Yes | No | Yes | Yes | No | High | 6106 |
| L-glutamic acid | Yes | No | Yes | Yes | No | High | 33032 |
| Linolenic acid | Yes | No | No | Yes | No | High | 5280934 |
| Lithocholic acid | Yes | Yes | Yes | Yes | Yes | High | 9903 |
| Lithocholylglycine | Yes | No | Yes | Yes | Yes | High | 115245 |
| L-methionine | Yes | No | Yes | Yes | No | High | 6137 |
| L-proline | Yes | No | Yes | Yes | No | High | 145742 |
| Malic acid | Yes | No | Yes | Yes | No | High | 525 |
| Methionine | Yes | No | Yes | Yes | No | High | 6137 |
| Myristic acid | Yes | Yes | No | Yes | No | High | 11005 |
| N-acetyl glutamic acid | Yes | No | Yes | Yes | No | High | 70914 |
| Norepinephrine | Yes | Yes | Yes | Yes | No | High | 439260 |
| Oxalic acid | Yes | No | Yes | Yes | No | High | 971 |
| Palmitic acid | Yes | Yes | No | Yes | No | High | 985 |
| Palmitoleic acid | Yes | Yes | No | Yes | No | High | 445638 |
| Phenylalanine | Yes | Yes | Yes | Yes | No | High | 6140 |
| Proline | Yes | No | Yes | Yes | No | High | 145742 |
| Propionic acid | Yes | No | Yes | Yes | No | High | 1032 |
| Pyroglutamic acid | Yes | No | Yes | Yes | No | High | 7405 |
| Pyruvic acid | Yes | No | Yes | Yes | No | High | 1060 |
| Shikimic acid | Yes | No | Yes | Yes | No | High | 8742 |
| Succinic acid | Yes | No | Yes | Yes | No | High | 1110 |
| Threonine | Yes | No | Yes | Yes | No | High | 6288 |
| Tryptophan | Yes | Yes | Yes | Yes | Yes | High | 6305 |
| Uracil | Yes | No | Yes | Yes | No | High | 1174 |
| Ursodeoxycholic acid | Yes | Yes | Yes | Yes | Yes | High | 31401 |
| Valeric acid | Yes | No | Yes | Yes | No | High | 7991 |
| Valine | Yes | No | Yes | Yes | No | High | 6287 |
| Target | Betweenness Centrality | Closeness Centrality | Topological Coefficient | Degree |
|---|---|---|---|---|
| MAPK3 | 0.223813617 | 0.428571429 | 0.155555556 | 18 |
| MAPK1 | 0.199221844 | 0.428571429 | 0.145555556 | 18 |
| EGFR | 0.140099305 | 0.384236453 | 0.226824458 | 13 |
| ESR1 | 0.194078415 | 0.371428571 | 0.204545455 | 12 |
| PRKCA | 0.212868248 | 0.39 | 0.191056911 | 12 |
| FYN | 0.086891474 | 0.366197183 | 0.227272727 | 11 |
| LCK | 0.032375537 | 0.345132743 | 0.306666667 | 10 |
| DLG4 | 0.19912985 | 0.331914894 | 0.189814815 | 9 |
| ITGB1 | 0.042968916 | 0.320987654 | 0.247863248 | 9 |
| IL6 | 0.169924934 | 0.352941176 | 0.233333333 | 9 |
| PTPN11 | 0.044866981 | 0.378640777 | 0.287749288 | 9 |
| RARA | 0.040924344 | 0.348214286 | 0.3 | 7 |
| NR3C1 | 0.064554392 | 0.342105263 | 0.369047619 | 6 |
| PTPN6 | 0.003184922 | 0.310756972 | 0.438596491 | 6 |
| PPARA | 0.135433492 | 0.329113924 | 0.306666667 | 6 |
| ITGAV | 0.017906128 | 0.331914894 | 0.410714286 | 6 |
| KEGG ID | Pathway | Gene Ratio (%) | p Value | Counts |
|---|---|---|---|---|
| hsa04080 | Neuroactive ligand–receptor interaction | 24.50331126 | 6.08 × 10−19 | 37 |
| hsa05200 | Pathways in cancer | 17.8807947 | 3.93 × 10−7 | 27 |
| hsa00590 | Arachidonic acid metabolism | 6.622516556 | 5.68 × 10−7 | 10 |
| hsa04614 | Renin–angiotensin system | 4.635761589 | 1.42 × 10−6 | 7 |
| hsa04924 | Renin secretion | 6.622516556 | 1.66 × 10−6 | 10 |
| hsa04726 | Serotonergic synapse | 7.947019868 | 2.64 × 10−6 | 12 |
| hsa05207 | Chemical carcinogenesis–receptor activation | 9.933774834 | 9.29 × 10−6 | 15 |
| hsa04915 | Estrogen signaling pathway | 7.947019868 | 1.55 × 10−5 | 12 |
| hsa04724 | Glutamatergic synapse | 7.284768212 | 1.66 × 10−5 | 11 |
| hsa03320 | PPAR signaling pathway | 5.960264901 | 2.91 × 10−5 | 9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, J.; Dai, J.; Chen, Y.; Wang, Z.; Sha, R.; Mao, J.; Mao, Y. Hypoglycemic Activity of Rice Resistant-Starch Metabolites: A Mechanistic Network Pharmacology and In Vitro Approach. Metabolites 2024, 14, 224. https://doi.org/10.3390/metabo14040224
Ren J, Dai J, Chen Y, Wang Z, Sha R, Mao J, Mao Y. Hypoglycemic Activity of Rice Resistant-Starch Metabolites: A Mechanistic Network Pharmacology and In Vitro Approach. Metabolites. 2024; 14(4):224. https://doi.org/10.3390/metabo14040224
Chicago/Turabian StyleRen, Jianing, Jing Dai, Yue Chen, Zhenzhen Wang, Ruyi Sha, Jianwei Mao, and Yangchen Mao. 2024. "Hypoglycemic Activity of Rice Resistant-Starch Metabolites: A Mechanistic Network Pharmacology and In Vitro Approach" Metabolites 14, no. 4: 224. https://doi.org/10.3390/metabo14040224
APA StyleRen, J., Dai, J., Chen, Y., Wang, Z., Sha, R., Mao, J., & Mao, Y. (2024). Hypoglycemic Activity of Rice Resistant-Starch Metabolites: A Mechanistic Network Pharmacology and In Vitro Approach. Metabolites, 14(4), 224. https://doi.org/10.3390/metabo14040224







