Colonic Dysregulation of Major Metabolic Pathways in Experimental Ulcerative Colitis
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Model
2.2. DSS-Induced Colitis
2.3. Tissue Collection and Protein Extraction
2.4. Sample Preparation for Mass Spectrometry
2.5. Mass Spectrometry Analysis
2.6. Statistical Analysis
3. Results
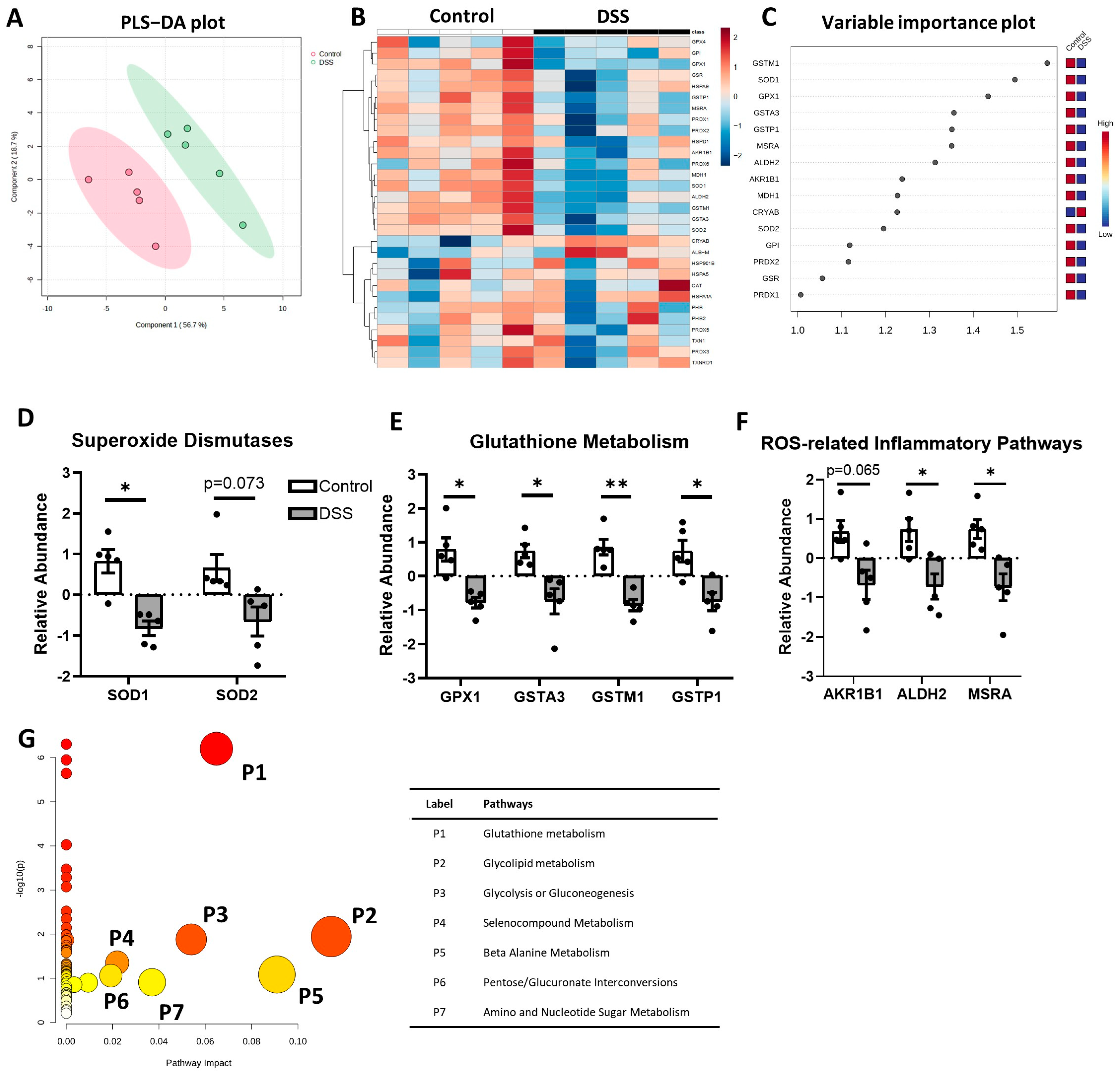
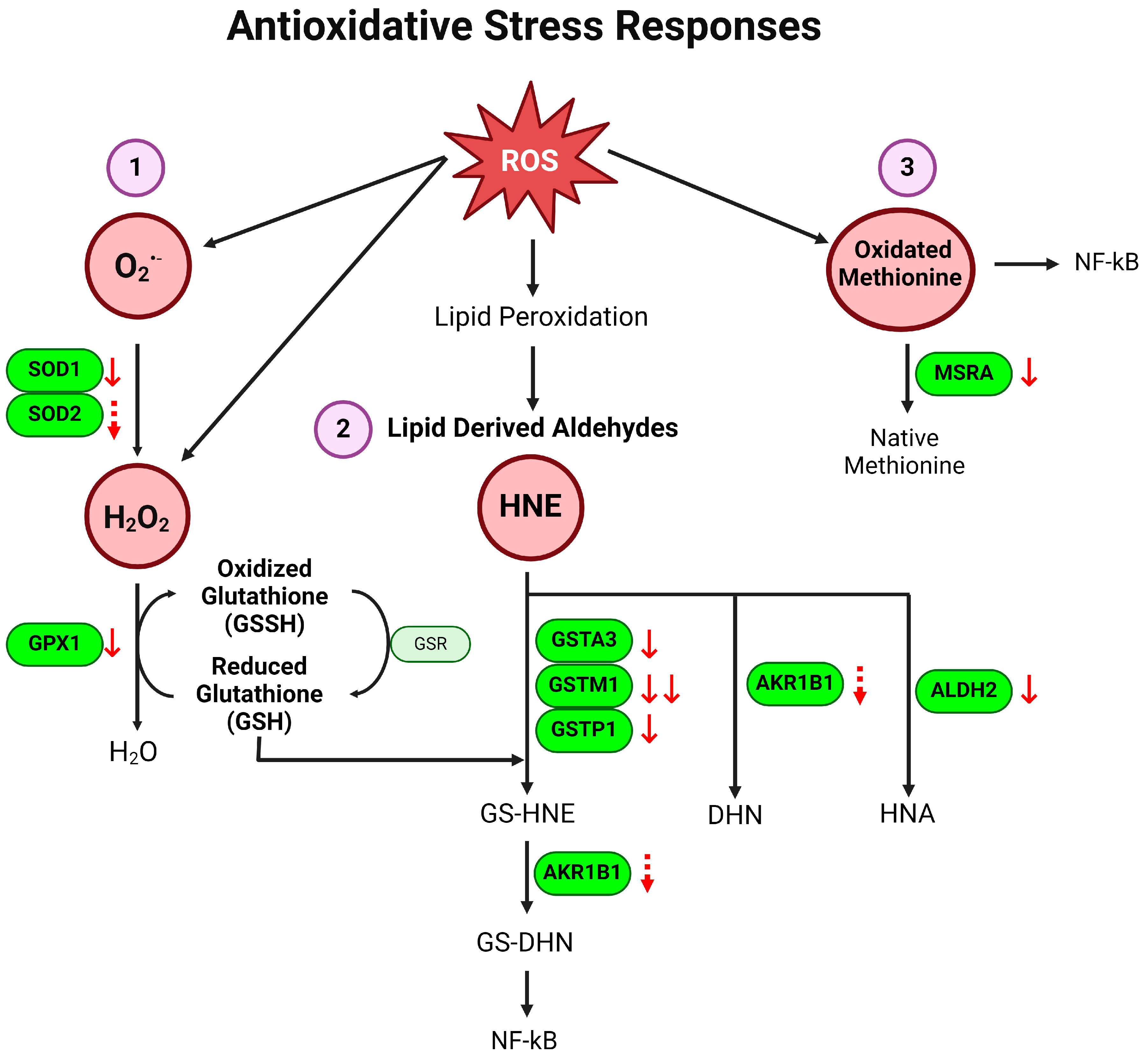
3.1. Antioxidative Defense Pathway Is Downregulated in Colon of Colitis
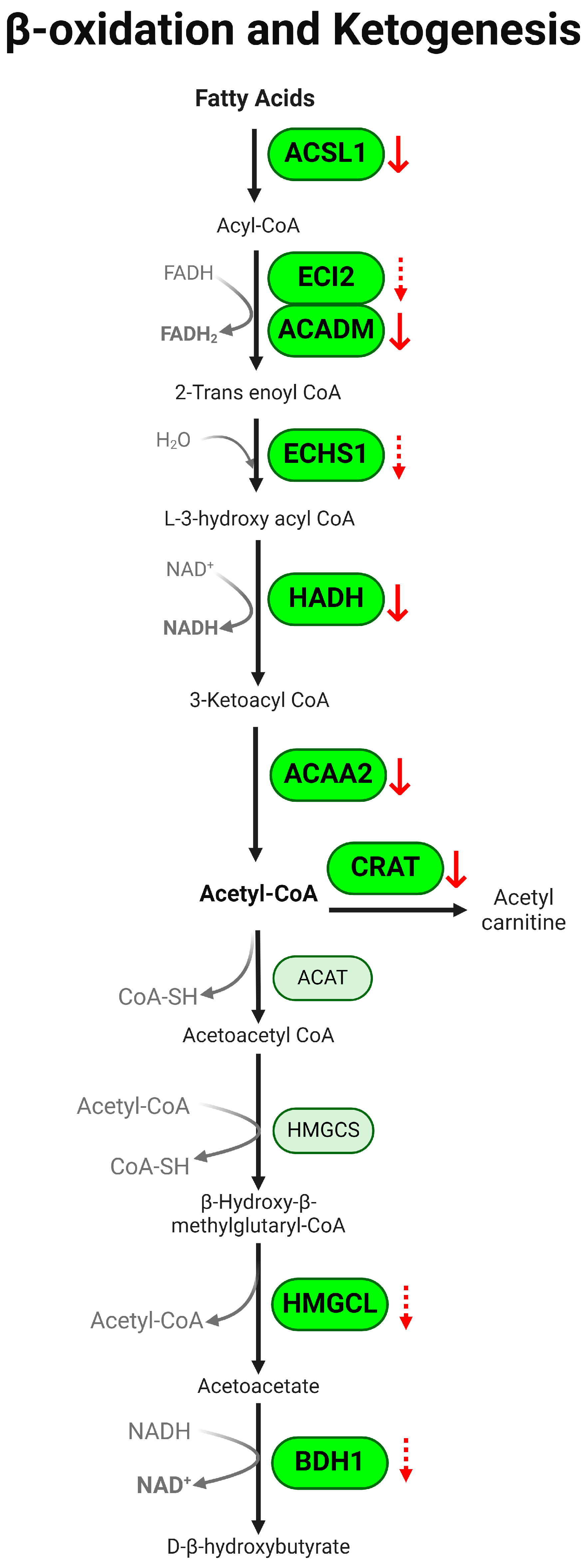
3.2. β-Oxidation Pathway Is Downregulated in Colon of Colitis
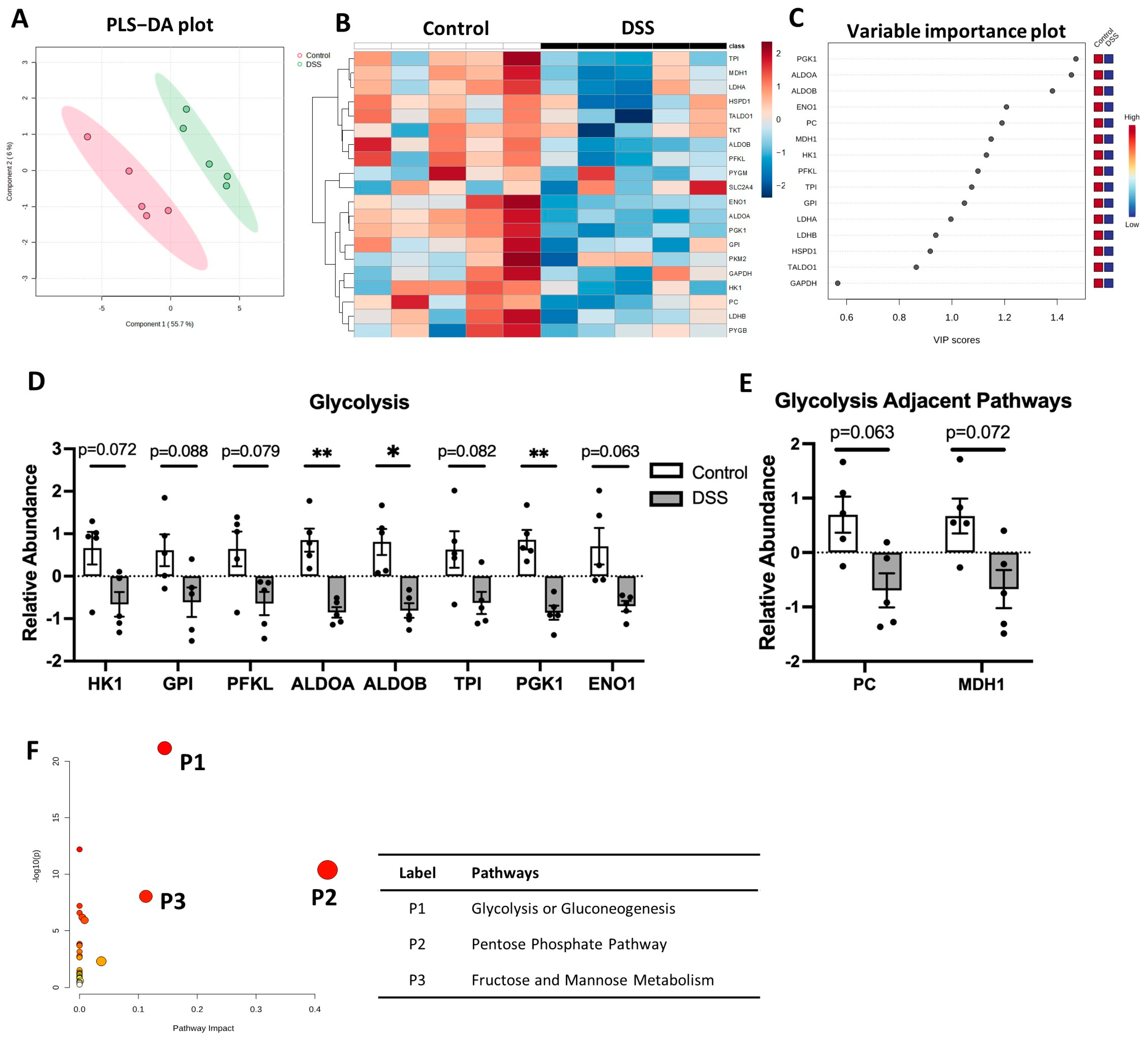
3.3. Glycolysis Pathway Is Downregulated in Colon of Colitis
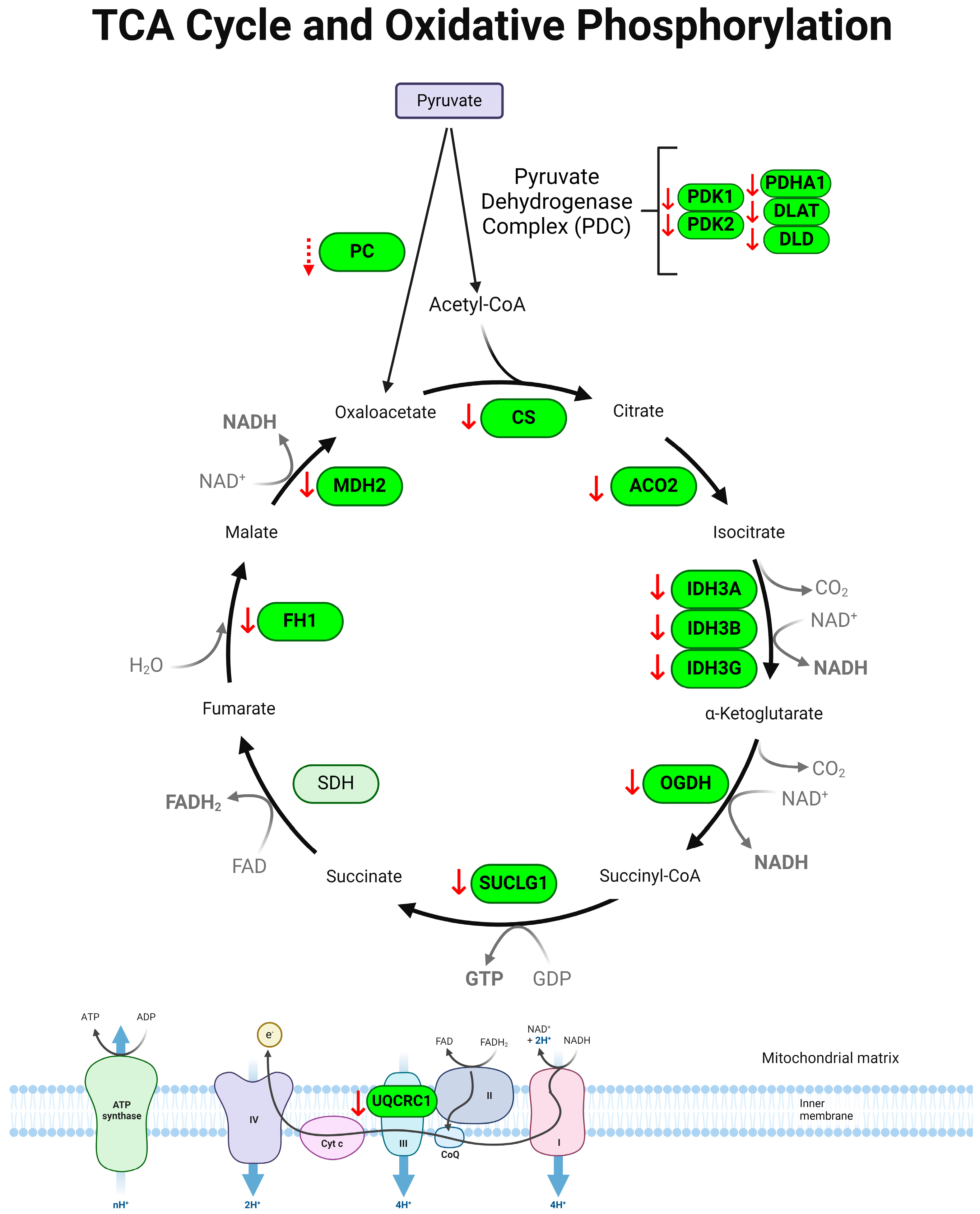
3.4. Tricarboxylic Acid (TCA) Cycle Is Downregulated in Colon of Colitis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bernstein, C.N.; Eliakim, A.; Fedail, S.; Fried, M.; Gearry, R.; Goh, K.L.; Hamid, S.; Khan, A.G.; Khalif, I.; Ng, S.C.; et al. World Gastroenterology Organisation Global Guidelines Inflammatory Bowel Disease: Update August 2015. J. Clin. Gastroenterol. 2016, 50, 803–818. [Google Scholar] [CrossRef] [PubMed]
- Flynn, S.; Eisenstein, S. Inflammatory Bowel Disease Presentation and Diagnosis. Surg. Clin. N. Am. 2019, 99, 1051–1062. [Google Scholar] [CrossRef] [PubMed]
- Nobrega, V.G.; Silva, I.N.N.; Brito, B.S.; Silva, J.; Silva, M.; Santana, G.O. The Onset of Clinical Manifestations in Inflammatory Bowel Disease Patients. Arq. Gastroenterol. 2018, 55, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Kwon, J.E.; Cho, M.L. Immunological pathogenesis of inflammatory bowel disease. Intest. Res. 2018, 16, 26–42. [Google Scholar] [CrossRef] [PubMed]
- Fakhoury, M.; Negrulj, R.; Mooranian, A.; Al-Salami, H. Inflammatory bowel disease: Clinical aspects and treatments. J. Inflamm. Res. 2014, 7, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Jung, S.A.; Lee, K.M.; Park, S.J.; Kim, T.O.; Choi, C.H.; Kim, H.G.; Moon, W.; Moon, C.M.; Song, H.K.; et al. Impact of inflammatory bowel disease on daily life: An online survey by the Korean Association for the Study of Intestinal Diseases. Intest. Res. 2017, 15, 338–344. [Google Scholar] [CrossRef][Green Version]
- Lonnfors, S.; Vermeire, S.; Greco, M.; Hommes, D.; Bell, C.; Avedano, L. IBD and health-related quality of life—Discovering the true impact. J. Crohn’s Colitis 2014, 8, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Mitropoulou, M.A.; Fradelos, E.C.; Lee, K.Y.; Malli, F.; Tsaras, K.; Christodoulou, N.G.; Papathanasiou, I.V. Quality of Life in Patients With Inflammatory Bowel Disease: Importance of Psychological Symptoms. Cureus 2022, 14, e28502. [Google Scholar] [CrossRef] [PubMed]
- Axelrad, J.E.; Lichtiger, S.; Yajnik, V. Inflammatory bowel disease and cancer: The role of inflammation, immunosuppression, and cancer treatment. World J. Gastroenterol. 2016, 22, 4794–4801. [Google Scholar] [CrossRef]
- Samadder, N.J.; Valentine, J.F.; Guthery, S.; Singh, H.; Bernstein, C.N.; Leighton, J.A.; Wan, Y.; Wong, J.; Boucher, K.; Pappas, L.; et al. Family History Associates With Increased Risk of Colorectal Cancer in Patients With Inflammatory Bowel Diseases. Clin. Gastroenterol. Hepatol. 2019, 17, 1807–1813.e1. [Google Scholar] [CrossRef]
- Xu, F.; Dahlhamer, J.M.; Zammitti, E.P.; Wheaton, A.G.; Croft, J.B. Health-Risk Behaviors and Chronic Conditions Among Adults with Inflammatory Bowel Disease—United States, 2015 and 2016. MMWR Morb. Mortal. Wkly. Rep. 2018, 67, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Roman, A.L.; Munoz, F. Comorbidity in inflammatory bowel disease. World J. Gastroenterol. 2011, 17, 2723–2733. [Google Scholar] [CrossRef] [PubMed]
- Scoville, E.A.; Allaman, M.M.; Brown, C.T.; Motley, A.K.; Horst, S.N.; Williams, C.S.; Koyama, T.; Zhao, Z.; Adams, D.W.; Beaulieu, D.B.; et al. Alterations in Lipid, Amino Acid, and Energy Metabolism Distinguish Crohn’s Disease from Ulcerative Colitis and Control Subjects by Serum Metabolomic Profiling. Metabolomics 2018, 14, 17. [Google Scholar] [CrossRef]
- Aldars-Garcia, L.; Gisbert, J.P.; Chaparro, M. Metabolomics Insights into Inflammatory Bowel Disease: A Comprehensive Review. Pharmaceuticals 2021, 14, 1190. [Google Scholar] [CrossRef] [PubMed]
- Hua, X.; Ungaro, R.C.; Petrick, L.M.; Chan, A.T.; Porter, C.K.; Khalili, H.; Nurses’ Health, S.; Collaborators, P. Inflammatory Bowel Disease Is Associated With Prediagnostic Perturbances in Metabolic Pathways. Gastroenterology 2023, 164, 147–150.e2. [Google Scholar] [CrossRef] [PubMed]
- Ooi, M.; Nishiumi, S.; Yoshie, T.; Shiomi, Y.; Kohashi, M.; Fukunaga, K.; Nakamura, S.; Matsumoto, T.; Hatano, N.; Shinohara, M.; et al. GC/MS-based profiling of amino acids and TCA cycle-related molecules in ulcerative colitis. Inflamm. Res. 2011, 60, 831–840. [Google Scholar] [CrossRef] [PubMed]
- Santoru, M.L.; Piras, C.; Murgia, F.; Leoni, V.P.; Spada, M.; Murgia, A.; Liggi, S.; Lai, M.A.; Usai, P.; Caboni, P.; et al. Metabolic Alteration in Plasma and Biopsies From Patients With IBD. Inflamm. Bowel Dis. 2021, 27, 1335–1345. [Google Scholar] [CrossRef] [PubMed]
- Bauset, C.; Gisbert-Ferrandiz, L.; Cosin-Roger, J. Metabolomics as a Promising Resource Identifying Potential Biomarkers for Inflammatory Bowel Disease. J. Clin. Med. 2021, 10, 622. [Google Scholar] [CrossRef] [PubMed]
- Noh, J.Y.; Wu, C.S.; DeLuca, J.A.A.; Devaraj, S.; Jayaraman, A.; Alaniz, R.C.; Tan, X.D.; Allred, C.D.; Sun, Y. Novel Role of Ghrelin Receptor in Gut Dysbiosis and Experimental Colitis in Aging. Int. J. Mol. Sci. 2022, 23, 2219. [Google Scholar] [CrossRef]
- Kinter, C.S.; Lundie, J.M.; Patel, H.; Rindler, P.M.; Szweda, L.I.; Kinter, M. A quantitative proteomic profile of the Nrf2-mediated antioxidant response of macrophages to oxidized LDL determined by multiplexed selected reaction monitoring. PLoS ONE 2012, 7, e50016. [Google Scholar] [CrossRef]
- Fu, Z.; Lofqvist, C.A.; Liegl, R.; Wang, Z.; Sun, Y.; Gong, Y.; Liu, C.H.; Meng, S.S.; Burnim, S.B.; Arellano, I.; et al. Photoreceptor glucose metabolism determines normal retinal vascular growth. EMBO Mol. Med. 2018, 10, 76–90. [Google Scholar] [CrossRef] [PubMed]
- Chiurchiu, V.; Maccarrone, M. Chronic inflammatory disorders and their redox control: From molecular mechanisms to therapeutic opportunities. Antioxid. Redox Signal 2011, 15, 2605–2641. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Wang, Z.; Zhang, J. Pathomechanisms of Oxidative Stress in Inflammatory Bowel Disease and Potential Antioxidant Therapies. Oxid. Med. Cell Longev. 2017, 2017, 4535194. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Zhang, L.; Ocansey, D.K.W.; Tu, Q.; Mao, F.; Sheng, X. Role of glycolysis in inflammatory bowel disease and its associated colorectal cancer. Front. Endocrinol. 2023, 14, 1242991. [Google Scholar] [CrossRef] [PubMed]
- Soto-Heredero, G.; Gomez de Las Heras, M.M.; Gabande-Rodriguez, E.; Oller, J.; Mittelbrunn, M. Glycolysis—A key player in the inflammatory response. FEBS J. 2020, 287, 3350–3369. [Google Scholar] [CrossRef] [PubMed]
- Arnold, P.K.; Finley, L.W.S. Regulation and function of the mammalian tricarboxylic acid cycle. J. Biol. Chem. 2023, 299, 102838. [Google Scholar] [CrossRef]
- Martinez-Reyes, I.; Chandel, N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 2020, 11, 102. [Google Scholar] [CrossRef]
- Patel, M.S.; Nemeria, N.S.; Furey, W.; Jordan, F. The pyruvate dehydrogenase complexes: Structure-based function and regulation. J. Biol. Chem. 2014, 289, 16615–16623. [Google Scholar] [CrossRef]
- Chi, H. Immunometabolism at the intersection of metabolic signaling, cell fate, and systems immunology. Cell Mol. Immunol. 2022, 19, 299–302. [Google Scholar] [CrossRef]
- Haberman, Y.; Karns, R.; Dexheimer, P.J.; Schirmer, M.; Somekh, J.; Jurickova, I.; Braun, T.; Novak, E.; Bauman, L.; Collins, M.H.; et al. Ulcerative colitis mucosal transcriptomes reveal mitochondriopathy and personalized mechanisms underlying disease severity and treatment response. Nat. Commun. 2019, 10, 38. [Google Scholar] [CrossRef]
- Schicho, R.; Shaykhutdinov, R.; Ngo, J.; Nazyrova, A.; Schneider, C.; Panaccione, R.; Kaplan, G.G.; Vogel, H.J.; Storr, M. Quantitative metabolomic profiling of serum, plasma, and urine by (1)H NMR spectroscopy discriminates between patients with inflammatory bowel disease and healthy individuals. J. Proteome Res. 2012, 11, 3344–3357. [Google Scholar] [CrossRef]
- Dawiskiba, T.; Deja, S.; Mulak, A.; Zabek, A.; Jawien, E.; Pawelka, D.; Banasik, M.; Mastalerz-Migas, A.; Balcerzak, W.; Kaliszewski, K.; et al. Serum and urine metabolomic fingerprinting in diagnostics of inflammatory bowel diseases. World J. Gastroenterol. 2014, 20, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Stephens, N.S.; Siffledeen, J.; Su, X.; Murdoch, T.B.; Fedorak, R.N.; Slupsky, C.M. Urinary NMR metabolomic profiles discriminate inflammatory bowel disease from healthy. J. Crohn’s Colitis 2013, 7, e42–e48. [Google Scholar] [CrossRef] [PubMed]
- Yoon, G.; Davidson, L.A.; Goldsby, J.S.; Mullens, D.A.; Ivanov, I.; Donovan, S.M.; Chapkin, R.S. Exfoliated epithelial cell transcriptome reflects both small and large intestinal cell signatures in piglets. Am. J. Physiol. Gastrointest. Liver Physiol. 2021, 321, G41–G51. [Google Scholar] [CrossRef] [PubMed]
- Krzystek-Korpacka, M.; Kempinski, R.; Bromke, M.A.; Neubauer, K. Oxidative Stress Markers in Inflammatory Bowel Diseases: Systematic Review. Diagnostics 2020, 10, 601. [Google Scholar] [CrossRef] [PubMed]
- Ashique, S.; Mishra, N.; Garg, A.; Sibuh, B.Z.; Taneja, P.; Rai, G.; Djearamane, S.; Wong, L.S.; Al-Dayan, N.; Roychoudhury, S.; et al. Recent updates on correlation between reactive oxygen species and synbiotics for effective management of ulcerative colitis. Front. Nutr. 2023, 10, 1126579. [Google Scholar] [CrossRef]
- Bourgonje, A.R.; Feelisch, M.; Faber, K.N.; Pasch, A.; Dijkstra, G.; van Goor, H. Oxidative Stress and Redox-Modulating Therapeutics in Inflammatory Bowel Disease. Trends Mol. Med. 2020, 26, 1034–1046. [Google Scholar] [CrossRef] [PubMed]
- Aviello, G.; Knaus, U.G. ROS in gastrointestinal inflammation: Rescue Or Sabotage? Br. J. Pharmacol. 2017, 174, 1704–1718. [Google Scholar] [CrossRef]
- Fridovich, I. Superoxide anion radical (O2-.), superoxide dismutases, and related matters. J. Biol. Chem. 1997, 272, 18515–18517. [Google Scholar] [CrossRef]
- El-Kheshen, G.; Moeini, M.; Saadat, M. Susceptibility to Ulcerative Colitis and Genetic Polymorphisms of A251G SOD1 and C-262T CAT. J. Med. Biochem. 2016, 35, 333–336. [Google Scholar] [CrossRef]
- Kosaka, T.; Yoshino, J.; Inui, K.; Wakabayashi, T.; Kobayashi, T.; Watanabe, S.; Hayashi, S.; Hirokawa, Y.; Shiraishi, T.; Yamamoto, T.; et al. Involvement of NAD(P)H:quinone oxidoreductase 1 and superoxide dismutase polymorphisms in ulcerative colitis. DNA Cell Biol. 2009, 28, 625–631. [Google Scholar] [CrossRef]
- Kruidenier, L.; Kuiper, I.; van Duijn, W.; Marklund, S.L.; van Hogezand, R.A.; Lamers, C.B.; Verspaget, H.W. Differential mucosal expression of three superoxide dismutase isoforms in inflammatory bowel disease. J. Pathol. 2003, 201, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Naito, Y.; Yoshikawa, T.; Ando, T.; Kishi, A.; Ueda, S.; Oyamada, H.; Kondo, M. Changes in superoxide dismutase activity in the gastric mucosa of peptic ulcer patients. J. Clin. Gastroenterol. 1992, 14 (Suppl. 1), S131–S134. [Google Scholar] [CrossRef] [PubMed]
- Segui, J.; Gironella, M.; Sans, M.; Granell, S.; Gil, F.; Gimeno, M.; Coronel, P.; Pique, J.M.; Panes, J. Superoxide dismutase ameliorates TNBS-induced colitis by reducing oxidative stress, adhesion molecule expression, and leukocyte recruitment into the inflamed intestine. J. Leukoc. Biol. 2004, 76, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Jin, J.; Jeon, S.; Moon, S.H.; Park, M.Y.; Yum, D.Y.; Kim, J.H.; Kang, J.E.; Park, M.H.; Kim, E.J.; et al. SOD1 suppresses pro-inflammatory immune responses by protecting against oxidative stress in colitis. Redox Biol. 2020, 37, 101760. [Google Scholar] [CrossRef] [PubMed]
- Esworthy, R.S.; Aranda, R.; Martin, M.G.; Doroshow, J.H.; Binder, S.W.; Chu, F.F. Mice with combined disruption of Gpx1 and Gpx2 genes have colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2001, 281, G848–G855. [Google Scholar] [CrossRef]
- Hertervig, E.; Nilsson, A.; Seidegard, J. The expression of glutathione transferase mu in patients with inflammatory bowel disease. Scand. J. Gastroenterol. 1994, 29, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.J.; Zhao, B.L.; Qian, Z.; Xu, Y.; Ding, Y.Q. Association of Glutathione S-Transferase M1 null genotype with inflammatory bowel diseases: A systematic review and meta-analysis. Medicine 2019, 98, e17722. [Google Scholar] [CrossRef] [PubMed]
- Broekman, M.M.; Bos, C.; Te Morsche, R.H.; Hoentjen, F.; Roelofs, H.M.; Peters, W.H.; Wanten, G.J.; de Jong, D.J. GST Theta null genotype is associated with an increased risk for ulcerative colitis: A case-control study and meta-analysis of GST Mu and GST Theta polymorphisms in inflammatory bowel disease. J. Hum. Genet. 2014, 59, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Magnusson, M.K.; Brynjolfsson, S.F.; Dige, A.; Uronen-Hansson, H.; Borjesson, L.G.; Bengtsson, J.L.; Gudjonsson, S.; Ohman, L.; Agnholt, J.; Sjovall, H.; et al. Macrophage and dendritic cell subsets in IBD: ALDH+ cells are reduced in colon tissue of patients with ulcerative colitis regardless of inflammation. Mucosal Immunol. 2016, 9, 171–182. [Google Scholar] [CrossRef]
- Jones, G.-R.; Bain, C.C.; Fenton, T.M.; Kelly, A.; Brown, S.L.; Ivens, A.C.; Travis, M.A.; Cook, P.C.; MacDonald, A.S. Dynamics of Colon Monocyte and Macrophage Activation During Colitis. Front. Immunol. 2018, 9, 2764. [Google Scholar] [CrossRef] [PubMed]
- Carpentino, J.E.; Hynes, M.J.; Appelman, H.D.; Zheng, T.; Steindler, D.A.; Scott, E.W.; Huang, E.H. Aldehyde dehydrogenase-expressing colon stem cells contribute to tumorigenesis in the transition from colitis to cancer. Cancer Res. 2009, 69, 8208–8215. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.P.; Kim, K.Y.; Kwak, G.H.; Baek, S.H.; Kim, H.Y. Methionine sulfoxide reductase A protects against lipopolysaccharide-induced septic shock via negative regulation of the proinflammatory responses. Arch. Biochem. Biophys. 2017, 631, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Roediger, W.E. The colonic epithelium in ulcerative colitis: An energy-deficiency disease? Lancet 1980, 2, 712–715. [Google Scholar] [CrossRef] [PubMed]
- De Preter, V.; Arijs, I.; Windey, K.; Vanhove, W.; Vermeire, S.; Schuit, F.; Rutgeerts, P.; Verbeke, K. Impaired butyrate oxidation in ulcerative colitis is due to decreased butyrate uptake and a defect in the oxidation pathway. Inflamm. Bowel Dis. 2012, 18, 1127–1136. [Google Scholar] [CrossRef] [PubMed]
- Heimerl, S.; Moehle, C.; Zahn, A.; Boettcher, A.; Stremmel, W.; Langmann, T.; Schmitz, G. Alterations in intestinal fatty acid metabolism in inflammatory bowel disease. Biochim. Biophys. Acta 2006, 1762, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Coleman, R.A.; Lewin, T.M.; Van Horn, C.G.; Gonzalez-Baro, M.R. Do long-chain acyl-CoA synthetases regulate fatty acid entry into synthetic versus degradative pathways? J. Nutr. 2002, 132, 2123–2126. [Google Scholar] [CrossRef] [PubMed]
- Chapman, M.A.; Grahn, M.F.; Boyle, M.A.; Hutton, M.; Rogers, J.; Williams, N.S. Butyrate oxidation is impaired in the colonic mucosa of sufferers of quiescent ulcerative colitis. Gut 1994, 35, 73–76. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, C.; Jiang, Q.; Yin, Y. Butyrate in Energy Metabolism: There Is Still More to Learn. Trends Endocrinol. Metab. 2021, 32, 159–169. [Google Scholar] [CrossRef]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; Gonzalez, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef]
- Hamer, H.M.; Jonkers, D.; Venema, K.; Vanhoutvin, S.; Troost, F.J.; Brummer, R.J. Review article: The role of butyrate on colonic function. Aliment. Pharmacol. Ther. 2008, 27, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, O.; Creanza, T.M.; Bossa, F.; Palumbo, O.; Maglietta, R.; Ancona, N.; Corritore, G.; Latiano, T.; Martino, G.; Biscaglia, G.; et al. Genome-wide Pathway Analysis Using Gene Expression Data of Colonic Mucosa in Patients with Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2015, 21, 1260–1268. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Plug, L.G.; Patente, T.A.; de Jonge-Muller, E.S.M.; Elmagd, A.A.; van der Meulen-de Jong, A.E.; Everts, B.; Barnhoorn, M.C.; Hawinkels, L. Increased stromal PFKFB3-mediated glycolysis in inflammatory bowel disease contributes to intestinal inflammation. Front. Immunol. 2022, 13, 966067. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Huang, Y.; Gao, Y.; Zhu, S.; Yuan, J. Exploring the glycolytic cross-talk genes between inflammatory bowel disease and colorectal cancer. Funct. Integr. Genom. 2023, 23, 230. [Google Scholar] [CrossRef] [PubMed]
- Cha, H.; Lee, S.; Hwan Kim, S.; Kim, H.; Lee, D.S.; Lee, H.S.; Lee, J.H.; Park, J.W. Increased susceptibility of IDH2-deficient mice to dextran sodium sulfate-induced colitis. Redox Biol. 2017, 13, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.; Julia, A.; Vinaixa, M.; Domenech, E.; Fernandez-Nebro, A.; Canete, J.D.; Ferrandiz, C.; Tornero, J.; Gisbert, J.P.; Nos, P.; et al. Urine metabolome profiling of immune-mediated inflammatory diseases. BMC Med. 2016, 14, 133. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, K.; Kumar, S.; Singh, R.R.; Sharma, U.; Ahuja, V.; Makharia, G.K.; Jagannathan, N.R. Metabolism of the colonic mucosa in patients with inflammatory bowel diseases: An in vitro proton magnetic resonance spectroscopy study. Magn. Reson. Imaging 2009, 27, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Fernie, A.R.; Carrari, F.; Sweetlove, L.J. Respiratory metabolism: Glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 2004, 7, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Yan, P.; Han, T.; Cheng, X.; Li, J. Identification of key genes and biological processes contributing to colitis associated dysplasia in ulcerative colitis. PeerJ 2021, 9, e11321. [Google Scholar] [CrossRef]
- Li, W.; Wubulikasimu, G.; Zhao, X.; Wang, C.; Liu, R.; Wang, L.; Zhu, X.; Chen, Z. UQCRC1 downregulation is correlated with lymph node metastasis and poor prognosis in CRC. Eur. J. Surg. Oncol. 2019, 45, 1005–1010. [Google Scholar] [CrossRef]
- Greuter, T.; Manser, C.; Pittet, V.; Vavricka, S.R.; Biedermann, L.; on behalf of Swiss IBDnet. Gender Differences in Inflammatory Bowel Disease. Digestion 2020, 101, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Rustgi, S.D.; Kayal, M.; Shah, S.C. Sex-based differences in inflammatory bowel diseases: A review. Ther. Adv. Gastroenterol. 2020, 13, 1756284820915043. [Google Scholar] [CrossRef]
- Blumenstein, I.; Herrmann, E.; Filmann, N.; Zosel, C.; Tacke, W.; Bock, H.; Dignass, A.; Hartmann, F.; Zeuzem, S.; Stein, J.; et al. Female patients suffering from inflammatory bowel diseases are treated less frequently with immunosuppressive medication and have a higher disease activity: A subgroup analysis of a large multi-centre, prospective, internet-based study. J. Crohn’s Colitis 2011, 5, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Blumenstein, I.; Sonnenberg, E. Sex- and gender-related differences in inflammatory bowel diseases. Front. Gastroenterol. 2023, 2, 1199687. [Google Scholar] [CrossRef]
- Bábíčková, J.; Tóthová, Ľ.; Lengyelová, E.; Bartoňová, A.; Hodosy, J.; Gardlík, R.; Celec, P. Sex Differences in Experimentally Induced Colitis in Mice: A Role for Estrogens. Inflammation 2015, 38, 1996–2006. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, C.M.; Allred, K.F.; Weeks, B.R.; Chapkin, R.S.; Allred, C.D. Estradiol Has Differential Effects on Acute Colonic Inflammation in the Presence and Absence of Estrogen Receptor beta Expression. Dig. Dis. Sci. 2017, 62, 1977–1984. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Villatoro, E.L.; Allred, C.D. Estrogen receptor actions in colitis. Essays Biochem. 2021, 65, 1003–1013. [Google Scholar] [CrossRef] [PubMed]
- Cader, M.Z.; Kaser, A. Recent advances in inflammatory bowel disease: Mucosal immune cells in intestinal inflammation. Gut 2013, 62, 1653–1664. [Google Scholar] [CrossRef] [PubMed]
- Okumura, R.; Takeda, K. Maintenance of gut homeostasis by the mucosal immune system. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2016, 92, 423–435. [Google Scholar] [CrossRef]
- Noh, J.Y.; Wang, H.; Farnell, Y.; Tan, X.-D.; Wright, G.; Hillhouse, A.; Sun, Y. Inflamm-aging is associated with pro-inflammatory programming of innate immune cells in the colon. Innov. Aging 2022, 6, 443. [Google Scholar] [CrossRef]
- Kratofil, R.M.; Shim, H.B.; Shim, R.; Lee, W.Y.; Labit, E.; Sinha, S.; Keenan, C.M.; Surewaard, B.G.J.; Noh, J.Y.; Sun, Y.; et al. A monocyte-leptin-angiogenesis pathway critical for repair post-infection. Nature 2022, 609, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Kishore, M.; Cheung, K.C.P.; Fu, H.; Bonacina, F.; Wang, G.; Coe, D.; Ward, E.J.; Colamatteo, A.; Jangani, M.; Baragetti, A.; et al. Regulatory T Cell Migration Is Dependent on Glucokinase-Mediated Glycolysis. Immunity 2017, 47, 875–889. [Google Scholar] [CrossRef] [PubMed]
- Noh, J.Y.; Herrera, M.; Patil, B.S.; Tan, X.D.; Wright, G.A.; Sun, Y. The expression and function of growth hormone secretagogue receptor in immune cells: A current perspective. Exp. Biol. Med. 2022, 247, 2184–2191. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Xuan, Y.; Zhang, X.; Liu, Y.; Yang, S.; Yang, K. Immune cell metabolism and metabolic reprogramming. Mol. Biol. Rep. 2022, 49, 9783–9795. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noh, J.Y.; Farhataziz, N.; Kinter, M.T.; Yan, X.; Sun, Y. Colonic Dysregulation of Major Metabolic Pathways in Experimental Ulcerative Colitis. Metabolites 2024, 14, 194. https://doi.org/10.3390/metabo14040194
Noh JY, Farhataziz N, Kinter MT, Yan X, Sun Y. Colonic Dysregulation of Major Metabolic Pathways in Experimental Ulcerative Colitis. Metabolites. 2024; 14(4):194. https://doi.org/10.3390/metabo14040194
Chicago/Turabian StyleNoh, Ji Yeon, Naser Farhataziz, Michael T. Kinter, Xin Yan, and Yuxiang Sun. 2024. "Colonic Dysregulation of Major Metabolic Pathways in Experimental Ulcerative Colitis" Metabolites 14, no. 4: 194. https://doi.org/10.3390/metabo14040194
APA StyleNoh, J. Y., Farhataziz, N., Kinter, M. T., Yan, X., & Sun, Y. (2024). Colonic Dysregulation of Major Metabolic Pathways in Experimental Ulcerative Colitis. Metabolites, 14(4), 194. https://doi.org/10.3390/metabo14040194







