Studying Salt-Induced Shifts in Gene Expression Patterns of Glucosinolate Transporters and Glucosinolate Accumulation in Two Contrasting Brassica Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Bioinformatics Analyses
2.3. Biochemical Analyses
2.3.1. Malondialdehyde (MDA) Content
2.3.2. Hydrogen Peroxide (H2O2) Content
2.3.3. Total Free Amino Acids (TFA) Content
2.3.4. Total Soluble Protein (TSP) Content
2.3.5. Phenolic Compounds
2.4. Antioxidant Assays
2.5. RNA Extraction, cDNA Synthesis and qPCR Analysis
2.6. Measurement of Seed Glucosinolates Content
2.7. Statistical Analyses
3. Results
3.1. Bioinformatics Analysis
3.1.1. Identification of GTR Genes in Brassica Complex
3.1.2. Phylogenetic Analysis of GTR Proteins in Brassica Complex
3.1.3. Intron/Exon Composition of GTR Genes
3.1.4. Domain Analysis of GTR Proteins
3.1.5. Motif Analysis of GTR Proteins
3.1.6. Cis-Regulatory Elements Analysis of GTR Genes
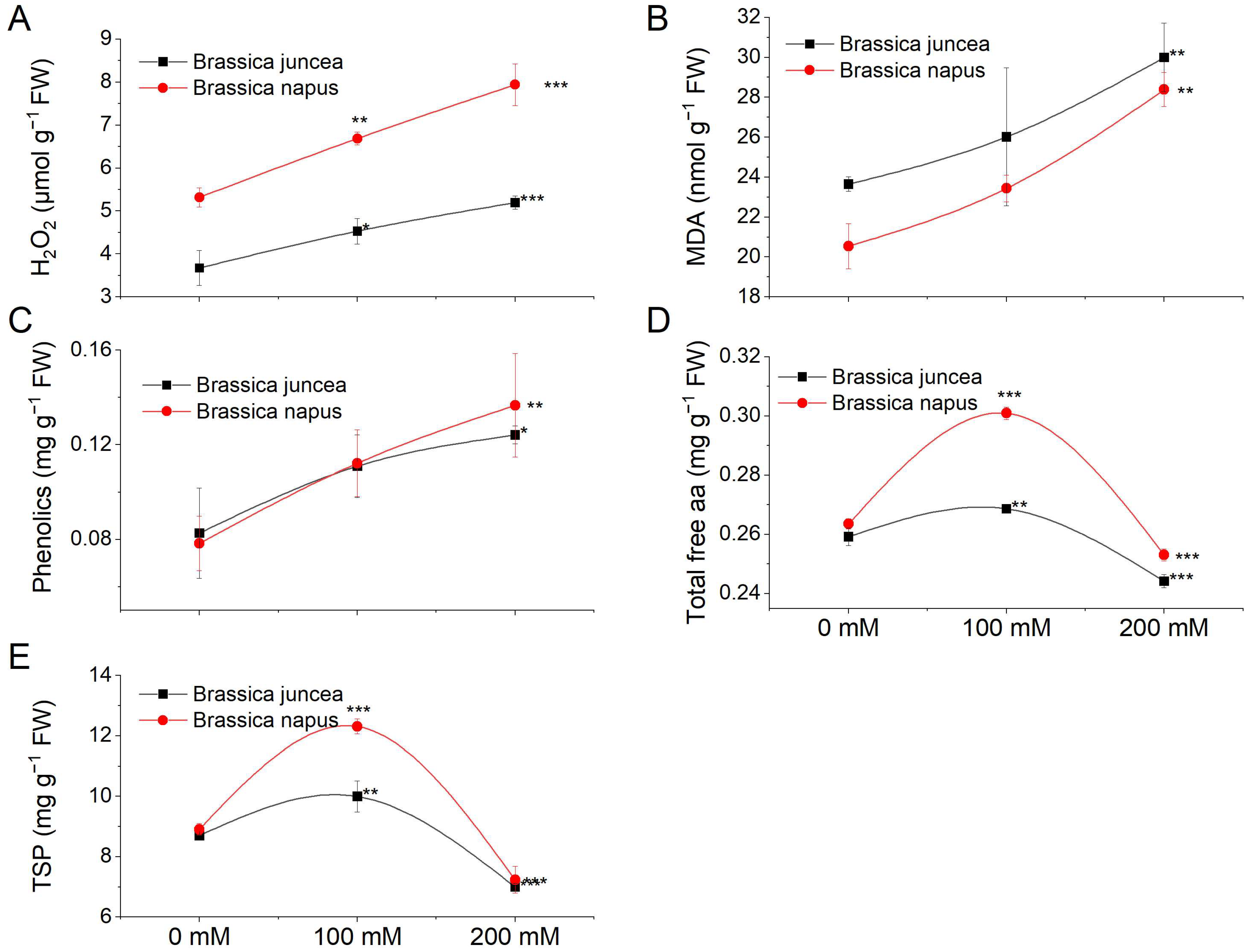
3.2. Effect of NaCl Stress on Different Metabolites
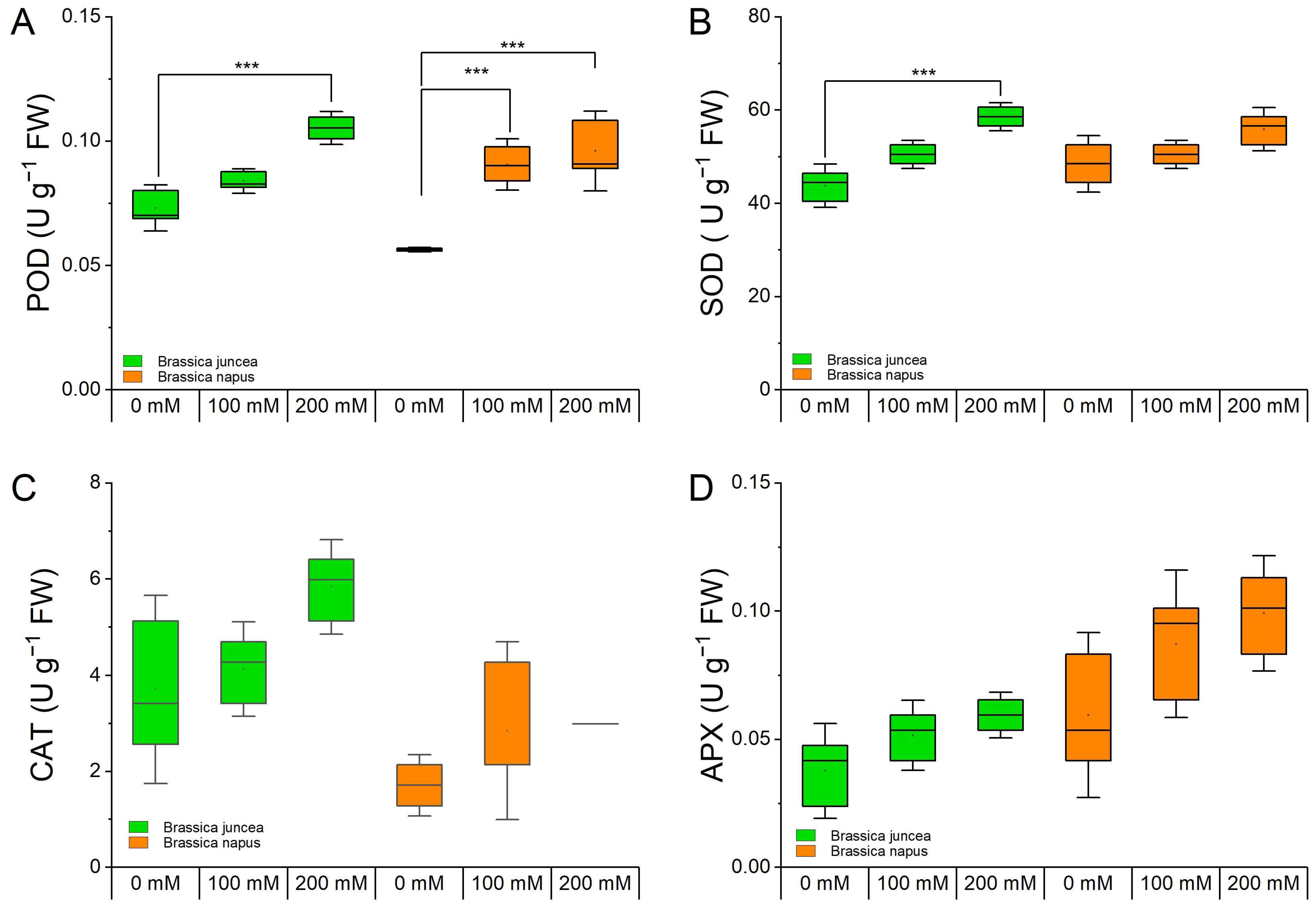
3.3. Effect of NaCl on Enzymatic Antioxidant Activities
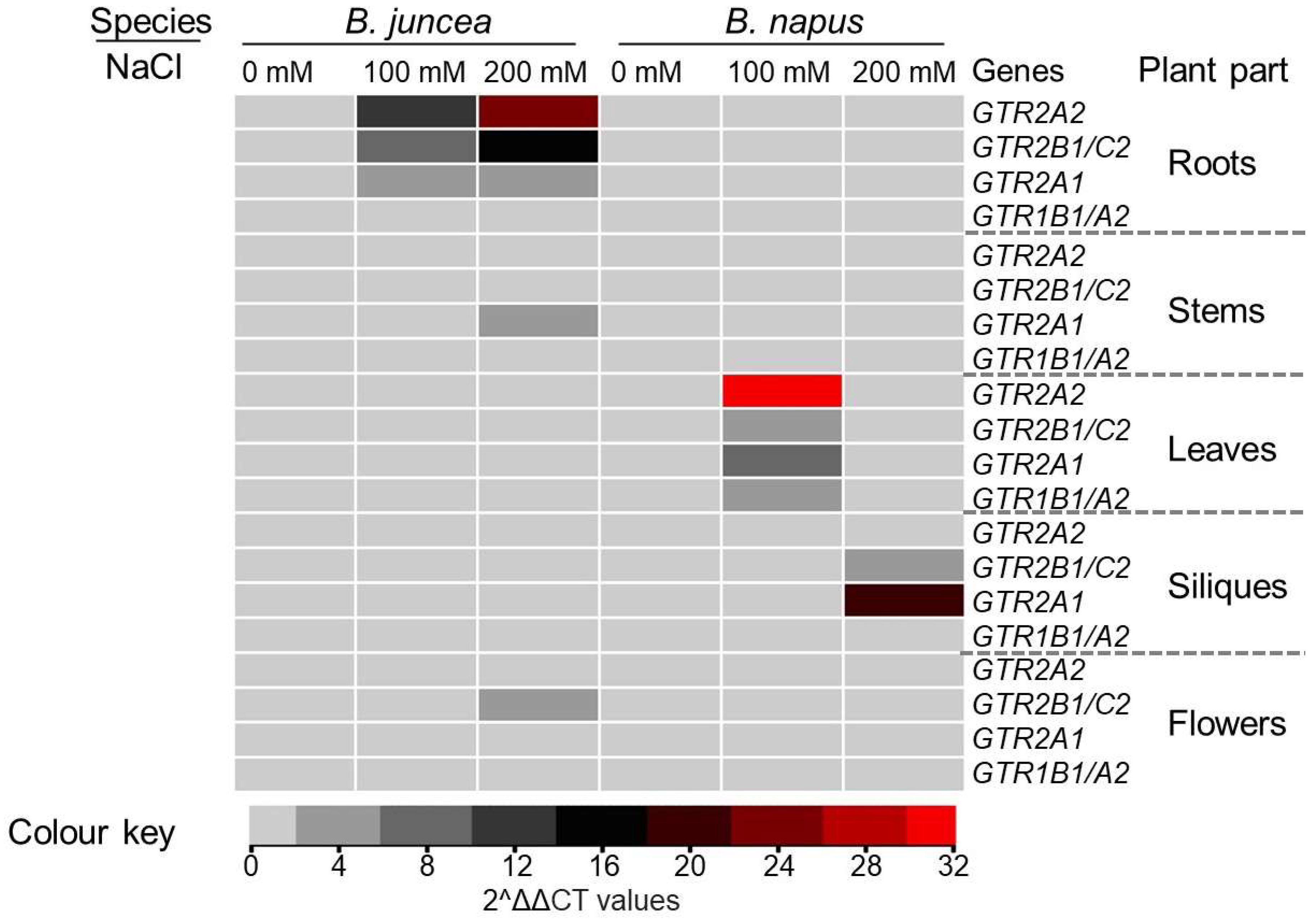
3.4. Effect of NaCl on Selected GTR Genes in Different Tissues
3.5. Determining the Effect of GTR Gene Expression on the Loading of Glucosinolates into the Seed
3.6. Predicting the Effect of NaCl Stress on Glucosinolates Content of B. napus and B. juncea
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cheng, F.; Wu, J.; Wang, X. Genome triplication drove the diversification of Brassica plants. Hortic. Res. 2014, 1, 14024. [Google Scholar] [CrossRef]
- Cheng, F.; Sun, R.; Hou, X.; Zheng, H.; Zhang, F.; Zhang, Y.; Liu, B.; Liang, J.; Zhuang, M.; Liu, Y. Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat. Genet. 2016, 48, 1218–1224. [Google Scholar] [CrossRef]
- Augustine, R.; Arya, G.C.; Nambiar, D.M.; Kumar, R.; Bisht, N.C. Translational genomics in Brassica crops: Challenges, progress, and future prospects. Plant Biotechnol. Rep. 2014, 8, 65–81. [Google Scholar] [CrossRef]
- Nagaharu, U.; Nagaharu, N. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 1935, 7, 389–452. [Google Scholar]
- Zandberg, J.D.; Fernandez, C.T.; Danilevicz, M.F.; Thomas, W.J.W.; Edwards, D.; Batley, J. The Global Assessment of Oilseed Brassica Crop Species Yield, Yield Stability and the Underlying Genetics. Plants 2022, 11, 2740. [Google Scholar] [CrossRef]
- Farooq, N.; Nawaz, M.A.; Mukhtar, Z.; Ali, I.; Hundleby, P.; Ahmad, N. Investigating the in vitro regeneration potential of commercial cultivars of Brassica. Plants 2019, 8, 558. [Google Scholar] [CrossRef] [PubMed]
- Machado, R.M.A.; Serralheiro, R.P. Soil salinity: Effect on vegetable crop growth. Management practices to prevent and mitigate soil salinization. Horticulturae 2017, 3, 30. [Google Scholar] [CrossRef]
- Jamil, M.; Lee, K.B.; Jung, K.Y.; Lee, D.B.; Han, M.S.; Rha, E.S. Salt stress inhibits germination and early seedling growth in cabbage (Brassica oleracea capitata L.). Pak. J. Biol. Sci. 2007, 10, 910–914. [Google Scholar] [CrossRef][Green Version]
- Purty, R.S.; Kumar, G.; Singla-Pareek, S.L.; Pareek, A. Towards salinity tolerance in Brassica: An overview. Physiol. Mol. Biol. Plants 2008, 14, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Ghuge, S.A.; Rai, A.N.; Khandagale, B.G.; Penna, S. Salt-induced stress responses of Brassica (Brassica juncea L.) genotypes. Arch. Agron. Soil Sci. 2011, 57, 127–136. [Google Scholar] [CrossRef]
- Qasim, M.; Ashraf, M.; Ashraf, M.Y.; Rehman, S.U.; Rha, E.S. Salt-induced changes in two canola cultivars differing in salt tolerance. Biol. Plant. 2003, 46, 629–632. [Google Scholar] [CrossRef]
- Pang, Q.; Guo, J.; Chen, S.; Chen, Y.; Zhang, L.; Fei, M.; Jin, S.; Li, M.; Wang, Y.; Yan, X. Effect of salt treatment on the glucosinolate-myrosinase system in Thellungiella salsuginea. Plant Soil 2012, 355, 363–374. [Google Scholar] [CrossRef]
- Ishida, M.; Hara, M.; Fukino, N.; Kakizaki, T.; Morimitsu, Y. Glucosinolate metabolism, functionality and breeding for the improvement of Brassicaceae vegetables. Breed. Sci. 2014, 64, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Fahey, J.W.; Olson, M.E.; Stephenson, K.K.; Wade, K.L.; Chodur, G.M.; Odee, D.; Nouman, W.; Massiah, M.; Alt, J.; Egner, P.A. The diversity of chemoprotective glucosinolates in Moringaceae (Moringa spp.). Sci. Rep. 2018, 8, 7994. [Google Scholar] [CrossRef]
- Annaz, H.; Sane, Y.; Bitchagno, G.T.M.; Ben Bakrim, W.; Drissi, B.; Mahdi, I.; El Bouhssini, M.; Sobeh, M. Caper (Capparis spinosa L.): An updated review on its phytochemistry, nutritional value, traditional uses, and therapeutic potential. Front. Pharmacol. 2022, 13, 878749. [Google Scholar] [CrossRef] [PubMed]
- Jioe, I.P.J.; Lin, H.-L.; Shiesh, C.-C. The investigation of phenylalanine, glucosinolate, benzylisothiocyanate (BITC) and cyanogenic glucoside of papaya fruits (Carica papaya L. cv.‘Tainung No. 2’) under different development stages between seasons and their correlation with bitter taste. Horticulturae 2022, 8, 198. [Google Scholar] [CrossRef]
- Halkier, B.A.; Gershenzon, J. Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 2006, 57, 303–333. [Google Scholar] [CrossRef]
- Hayes, J.D.; Kelleher, M.O.; Eggleston, I.M. The cancer chemopreventive actions of phytochemicals derived from glucosinolates. Eur. J. Nutr 2008, 47, 73–88. [Google Scholar] [CrossRef]
- Dinkova-Kostova, A.T.; Kostov, R.V. Glucosinolates and isothiocyanates in health and disease. Trends Mol. Med. 2012, 18, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Fahey, J.W.; Wehage, S.L.; Holtzclaw, W.D.; Kensler, T.W.; Egner, P.A.; Shapiro, T.A.; Talalay, P. Protection of humans by plant glucosinolates: Efficiency of conversion of glucosinolates to isothiocyanates by the gastrointestinal microflora. Cancer Prev. Res. 2012, 5, 603–611. [Google Scholar] [CrossRef]
- Mithen, R.F.; Dekker, M.; Verkerk, R.; Rabot, S.; Johnson, I.T. The nutritional significance, biosynthesis and bioavailability of glucosinolates in human foods. J. Sci. Food Agric. 2000, 80, 967–984. [Google Scholar] [CrossRef]
- Tanii, H.; Takayasu, T.; Higashi, T.; Leng, S.; Saijoh, K. Allylnitrile: Generation from cruciferous vegetables and behavioral effects on mice of repeated exposure. Food Chem. Toxicol 2004, 42, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Wallig, M.A.; Belyea, R.L.; Tumbleson, M.E. Effect of pelleting on glucosinolate content of Crambe meal. Anim. Feed Sci. Technol. 2002, 99, 205–214. [Google Scholar] [CrossRef]
- Tripathi, M.K.; Mishra, A.S. Glucosinolates in animal nutrition: A review. Anim. Feed Sci. Technol. 2007, 132, 1–27. [Google Scholar] [CrossRef]
- Augustine, R.; Mukhopadhyay, A.; Bisht, N.C. Targeted silencing of BjMYB28 transcription factor gene directs development of low glucosinolate lines in oilseed Brassica juncea. Plant Biotechnol. J. 2013, 11, 855–866. [Google Scholar] [CrossRef] [PubMed]
- Mejicanos, G.; Sanjayan, N.; Kim, I.H.; Nyachoti, C.M. Recent advances in canola meal utilization in swine nutrition. J. Anim. Sci. Technol. 2016, 58, 7. [Google Scholar] [CrossRef] [PubMed]
- Cools, K.; Terry, L.A. The effect of processing on the glucosinolate profile in mustard seed. Food Chem. 2018, 252, 343–348. [Google Scholar] [CrossRef]
- Radojčić Redovniković, I.; Glivetić, T.; Delonga, K.; Vorkapić-Furač, J. Glucosinolates and their potential role in plant. Period. Biol. 2008, 110, 297–309. [Google Scholar]
- Barth, C.; Jander, G. Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J. 2006, 46, 549–562. [Google Scholar] [CrossRef]
- Keum, Y.-S.; Jeong, W.-S.; Kong, A.N.T. Chemoprevention by isothiocyanates and their underlying molecular signaling mechanisms. Mutat. Res.-Fund. Mol. Mech. Mutagen. 2004, 555, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Ellerbrock, B.L.J.; Kim, J.H.; Jander, G. Contribution of glucosinolate transport to Arabidopsis defense responses. Plant Signal. Behav. 2007, 2, 282–283. [Google Scholar] [CrossRef]
- Ahuja, I.; Rohloff, J.; Bones, A.M. Defence mechanisms of Brassicaceae: Implications for plant-insect interactions and potential for integrated pest management. Sustain. Agric. 2011, 2, 623–670. [Google Scholar]
- Touw, A.J.; Verdecia Mogena, A.; Maedicke, A.; Sontowski, R.; Van Dam, N.M.; Tsunoda, T. Both biosynthesis and transport are involved in glucosinolate accumulation during root-herbivory in Brassica rapa. Front. Plant Sci. 2020, 10, 1653. [Google Scholar] [CrossRef]
- Tierens, K.F.M.J.; Thomma, B.P.H.J.; Brouwer, M.; Schmidt, J.; Kistner, K.; Porzel, A.; Mauch-Mani, B.; Cammue, B.P.A.; Broekaert, W.F. Study of the role of antimicrobial glucosinolate-derived isothiocyanates in resistance of Arabidopsis to microbial pathogens. Plant Physiol. 2001, 125, 1688–1699. [Google Scholar] [CrossRef]
- Calmes, B.; N’Guyen, G.; Dumur, J.; Brisach, C.A.; Campion, C.; Iacomi, B.; Pigné, S.; Dias, E.; Macherel, D.; Guillemette, T. Glucosinolate-derived isothiocyanates impact mitochondrial function in fungal cells and elicit an oxidative stress response necessary for growth recovery. Front. Plant Sci. 2015, 6, 414. [Google Scholar] [CrossRef]
- Wittstock, U.; Kurzbach, E.; Herfurth, A.M.; Stauber, E.J. Glucosinolate breakdown. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2016; Volume 80, pp. 125–169. [Google Scholar]
- Abuyusuf, M.; Robin, A.H.K.; Lee, J.-H.; Jung, H.-J.; Kim, H.-T.; Park, J.-I.; Nou, I.-S. Glucosinolate profiling and expression analysis of glucosinolate biosynthesis genes differentiate white mold resistant and susceptible cabbage lines. Int. J. Mol. Sci. 2018, 19, 4037. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Chen, S. Regulation of plant glucosinolate metabolism. Planta 2007, 226, 1343–1352. [Google Scholar] [CrossRef] [PubMed]
- Mann, A.; Kumari, J.; Kumar, R.; Kumar, P.; Pradhan, A.K.; Pental, D.; Bisht, N.C. Targeted editing of multiple homologues of GTR1 and GTR2 genes provides the ideal low-seed, high-leaf glucosinolate oilseed mustard with uncompromised defence and yield. Plant Biotechnol. J. 2023, 21, 2182–2195. [Google Scholar] [CrossRef] [PubMed]
- Nour-Eldin, H.H.; Andersen, T.G.; Burow, M.; Madsen, S.R.; Jørgensen, M.E.; Olsen, C.E.; Dreyer, I.; Hedrich, R.; Geiger, D.; Halkier, B.A. NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 2012, 488, 531–534. [Google Scholar] [CrossRef]
- Léran, S.; Varala, K.; Boyer, J.-C.; Chiurazzi, M.; Crawford, N.; Daniel-Vedele, F.; David, L.; Dickstein, R.; Fernandez, E.; Forde, B. A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 2014, 19, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Nour-Eldin, H.H.; Madsen, S.R.; Engelen, S.; Jørgensen, M.E.; Olsen, C.E.; Andersen, J.S.; Seynnaeve, D.; Verhoye, T.; Fulawka, R.; Denolf, P. Reduction of antinutritional glucosinolates in Brassica oilseeds by mutation of genes encoding transporters. Nat. Biotechnol. 2017, 35, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Nambiar, D.M.; Kumari, J.; Augustine, R.; Kumar, P.; Bajpai, P.K.; Bisht, N.C. GTR1 and GTR2 transporters differentially regulate tissue-specific glucosinolate contents and defence responses in the oilseed crop Brassica juncea. Plant Cell Environ. 2021, 44, 2729–2743. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, T.; Hussain, M.; Mustafa, H.S.B.; Hasan, E.; Aftab, M. Aari canola: Pakistan’s first ever canola quality and short duration mustard (Brassica juncea L.) cultivar resilient to climate change. Int. J. Biol. Pharm. Al. Sci 2017, 6, 777–787. [Google Scholar]
- Sun, B.; Tian, Y.-X.; Chen, Q.; Zhang, Y.; Luo, Y.; Wang, Y.; Li, M.-Y.; Gong, R.-G.; Wang, X.-R.; Zhang, F. Variations in the glucosinolates of the individual edible parts of three stem mustards (Brassica juncea). R. Soc. Open Sci. 2019, 6, 182054. [Google Scholar] [CrossRef]
- Cakmak, I.; Horst, W.J. Effect of aluminium on lipid peroxidation, superoxide dismutase, catalase, and peroxidase activities in root tips of soybean (Glycine max). Physiol. Plant. 1991, 83, 463–468. [Google Scholar] [CrossRef]
- Velikova, V.; Yordanov, I.; Edreva, A. Oxidative stress and some antioxidant systems in acid rain-treated bean plants: Protective role of exogenous polyamines. Plant Sci. 2000, 151, 59–66. [Google Scholar] [CrossRef]
- Hamilton, P.B.; Van Slyke, D.D. Amino acid determination with ninhydrin. J. Biol. Chem. 1943, 150, 231–250. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Aguilar-Garcia, C.; Gavino, G.; Baragaño-Mosqueda, M.; Hevia, P.; Gavino, V.C. Correlation of tocopherol, tocotrienol, γ-oryzanol and total polyphenol content in rice bran with different antioxidant capacity assays. Food Chem. 2007, 102, 1228–1232. [Google Scholar] [CrossRef]
- Beauchamp, C.; Fridovich, I. Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 1971, 44, 276–287. [Google Scholar] [CrossRef]
- Chaoui, A.; Mazhoudi, S.; Ghorbal, M.H.; El Ferjani, E. Cadmium and zinc induction of lipid peroxidation and effects on antioxidant enzyme activities in bean (Phaseolus vulgaris L.). Plant Sci. 1997, 127, 139–147. [Google Scholar] [CrossRef]
- Aebi, H. [13] Catalase in vitro. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1984; Volume 105, pp. 121–126. [Google Scholar]
- Nakano, Y.; Asada, K. Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 1981, 22, 867–880. [Google Scholar]
- Bennett, E.J.; Brignell, C.J.; Carion, P.W.C.; Cook, S.M.; Eastmond, P.J.; Teakle, G.R.; Hammond, J.P.; Love, C.; King, G.J.; Roberts, J.A. Development of a statistical crop model to explain the relationship between seed yield and phenotypic diversity within the Brassica napus genepool. Agronomy 2017, 7, 31. [Google Scholar] [CrossRef]
- Lu, S.; Sturtevant, D.; Aziz, M.; Jin, C.; Li, Q.; Chapman, K.D.; Guo, L. Spatial analysis of lipid metabolites and expressed genes reveals tissue-specific heterogeneity of lipid metabolism in high-and low-oil Brassica napus L. seeds. Plant J. 2018, 94, 915–932. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhu, K.; Li, H.; Han, S.; Meng, Q.; Khan, S.U.; Fan, C.; Xie, K.; Zhou, Y. Precise editing of CLAVATA genes in Brassica napus L. regulates multilocular silique development. Plant Biotechnol. J. 2018, 16, 1322–1335. [Google Scholar] [CrossRef] [PubMed]
- Cartea, M.E.; Velasco, P. Glucosinolates in Brassica foods: Bioavailability in food and significance for human health. Phytochem. Rev. 2008, 7, 213–229. [Google Scholar] [CrossRef]
- Bell, L.; Oloyede, O.O.; Lignou, S.; Wagstaff, C.; Methven, L. Taste and flavor perceptions of glucosinolates, isothiocyanates, and related compounds. Mol. Nutr. Food Res. 2018, 62, 1700990. [Google Scholar] [CrossRef] [PubMed]
- Andersen, T.G.; Nour-Eldin, H.H.; Fuller, V.L.; Olsen, C.E.; Burow, M.; Halkier, B.A. Integration of biosynthesis and long-distance transport establish organ-specific glucosinolate profiles in vegetative Arabidopsis. Plant Cell 2013, 25, 3133–3145. [Google Scholar] [CrossRef] [PubMed]
- O'Brien, D. Canola: Good protein source for dairy cattle. Agric. Res. 2016, 64, 1. [Google Scholar]
- Lysak, M.A.; Koch, M.A. Phylogeny, genome, and karyotype evolution of crucifers (Brassicaceae). In Genetics and Genomics of the Brassicaceae; Springer: New York, NY, USA, 2011; pp. 1–31. [Google Scholar]
- Arias, T.; Beilstein, M.A.; Tang, M.; McKain, M.R.; Pires, J.C. Diversification times among Brassica (Brassicaceae) crops suggest hybrid formation after 20 million years of divergence. Am. J. Bot. 2014, 101, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Lukens, L.N.; Quijada, P.A.; Udall, J.; Pires, J.C.; Schranz, M.E.; Osborn, T.C. Genome redundancy and plasticity within ancient and recent Brassica crop species. Biol. J. Linn. Soc. 2004, 82, 665–674. [Google Scholar] [CrossRef]
- He, J.; He, X.; Chang, P.; Jiang, H.; Gong, D.; Sun, Q. Genome-wide identification and characterization of TCP family genes in Brassica juncea var. tumida. PeerJ 2020, 8, e9130. [Google Scholar] [CrossRef]
- Ahmad, N.; Fatima, S.; Mehmood, M.A.; Zaman, Q.U.; Atif, R.M.; Zhou, W.; Rahman, M.-U.; Gill, R.A. Targeted genome editing in polyploids: Lessons from Brassica. Front. Plant Sci. 2023, 14, 1152468. [Google Scholar] [CrossRef] [PubMed]
- Areej, A.; Nawaz, H.; Aslam, I.; Danial, M.; Qayyum, Z.; Rasool, U.A.; Asif, J.; Khalid, A.; Serfraz, S.; Saleem, F. Investigation of NLR genes reveals divergent evolution on NLRome in diploid and polyploid species in genus trifolium. Genes 2023, 14, 867. [Google Scholar] [CrossRef]
- Verma, D.; Lakhanpal, N.; Singh, K. Genome-wide identification and characterization of abiotic-stress responsive SOD (superoxide dismutase) gene family in Brassica juncea and B. rapa. BMC Genom. 2019, 20, 227. [Google Scholar] [CrossRef]
- Lohani, N.; Babaei, S.; Singh, M.B.; Bhalla, P.L. Genome-wide in silico identification and comparative analysis of Dof gene family in Brassica napus. Plants 2021, 10, 709. [Google Scholar] [CrossRef]
- Wang, P.; Zhao, Z.; Zhang, Z.; Cai, Z.; Liao, J.; Tan, Q.; Xiang, M.; Chang, L.; Xu, D.; Tian, Q. Genome-wide identification and analysis of NPR family genes in Brassica juncea var. tumida. Gene 2021, 769, 145210. [Google Scholar] [CrossRef] [PubMed]
- Afridi, M.; Ahmad, K.; Malik, S.S.; Rehman, N.; Yasin, M.; Khan, S.M.; Hussain, A.; Khan, M.R. Genome-wide identification, phylogeny, and expression profiling analysis of shattering genes in rapeseed and mustard plants. J. Genet. Eng. Biotechnol. 2022, 20, 124. [Google Scholar]
- He, J.; Gu, L.; Tan, Q.; Wang, Y.; Hui, F.; He, X.; Chang, P.; Gong, D.; Sun, Q. Genome-wide analysis and identification of the PEBP genes of Brassica juncea var. Tumida. BMC Genom. 2022, 23, 535. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Qi, W.; Bai, J.; Li, H.; Fang, Y.; Xu, J.; Xu, Y.; Zeng, X.; Pu, Y.; Wang, W. Genome-Wide Identification and Analysis of the Ascorbate Peroxidase (APX) Gene Family of Winter Rapeseed (Brassica rapa L.) under Abiotic Stress. Front. Genet. 2022, 12, 753624. [Google Scholar] [CrossRef]
- Tyagi, S.; Sharma, S.; Taneja, M.; Kumar, R.; Sembi, J.K.; Upadhyay, S.K. Superoxide dismutases in bread wheat (Triticum aestivum L.): Comprehensive characterization and expression analysis during development and, biotic and abiotic stresses. Agri. Gene 2017, 6, 1–13. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, X.; Deng, F.; Yuan, R.; Shen, F. Genome-wide characterization and expression analyses of superoxide dismutase (SOD) genes in Gossypium hirsutum. BMC Genom. 2017, 18, 376. [Google Scholar] [CrossRef]
- Cai, Z.; Chen, Y.; Liao, J.; Wang, D. Genome-wide identification and expression analysis of jasmonate ZIM domain gene family in tuber mustard (Brassica juncea var. tumida). PLoS ONE 2020, 15, e0234738. [Google Scholar] [CrossRef] [PubMed]
- Hölzl, G.; Rezaeva, B.R.; Kumlehn, J.; Dörmann, P. Ablation of glucosinolate accumulation in the oil crop Camelina sativa by targeted mutagenesis of genes encoding the transporters GTR1 and GTR2 and regulators of biosynthesis MYB28 and MYB29. Plant Biotechnol. J. 2023, 21, 189–201. [Google Scholar] [CrossRef]
- Newstead, S. Molecular insights into proton coupled peptide transport in the PTR family of oligopeptide transporters. Biochim. Biophys. Acta 2015, 1850, 488–499. [Google Scholar] [CrossRef]
- Yan, N. Structural biology of the major facilitator superfamily transporters. Annu. Rev. Biophys. 2015, 44, 257–283. [Google Scholar] [CrossRef]
- Jørgensen, M.E.; Xu, D.; Crocoll, C.; Ernst, H.A.; Ramírez, D.; Motawia, M.S.; Olsen, C.E.; Mirza, O.; Nour-Eldin, H.H.; Halkier, B.A. Origin and evolution of transporter substrate specificity within the NPF family. Elife 2017, 6, e19466. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic-and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef]
- Schmitz, R.J.; Grotewold, E.; Stam, M. Cis-regulatory sequences in plants: Their importance, discovery, and future challenges. Plant Cell 2022, 34, 718–741. [Google Scholar] [CrossRef]
- Desikan, R.; A.-H.-Mackerness, S.; Hancock, J.T.; Neill, S.J. Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 2001, 127, 159–172. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, M.D.; Petersen, B.L.; Glawischnig, E.; Jensen, A.B.; Andreasson, E.; Halkier, B.A. Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiol. 2003, 131, 298–308. [Google Scholar] [CrossRef]
- Miao, H.; Wang, J.; Cai, C.; Chang, J.; Zhao, Y.; Wang, Q. Accumulation of glucosinolates in broccoli. In Glucosinolates; Springer: Cham, Switzerland, 2017; pp. 133–162. [Google Scholar]
- Fiorillo, A.; Manai, M.; Visconti, S.; Camoni, L. The Salt Tolerance–Related Protein (STRP) Is a Positive Regulator of the Response to Salt Stress in Arabidopsis thaliana. Plants 2023, 12, 1704. [Google Scholar] [CrossRef]
- Narayani, M.; Srivastava, S. Elicitation: A stimulation of stress in in vitro plant cell/tissue cultures for enhancement of secondary metabolite production. Phytochem. Rev. 2017, 16, 1227–1252. [Google Scholar] [CrossRef]
- Rahman, A.; Albadrani, G.M.; Waraich, E.A.; Awan, T.H.; Yavaş, İ.; Hussain, S. Plant Secondary Metabolites and Abiotic Stress Tolerance: Overview and Implications. In Plant Abiotic Stress Responses and Tolerance Mechanisms; IntechOpen: London, UK, 2023. [Google Scholar]
- Chen, Y.-z.; Pang, Q.-Y.; He, Y.; Zhu, N.; Branstrom, I.; Yan, X.-F.; Chen, S. Proteomics and metabolomics of Arabidopsis responses to perturbation of glucosinolate biosynthesis. Mol. Plant 2012, 5, 1138–1150. [Google Scholar] [CrossRef]
- Farooq, N.; Khan, M.O.; Ahmed, M.Z.; Fatima, S.; Nawaz, M.A.; Abideen, Z.; Nielsen, B.L.; Ahmad, N. Salt-induced modulation of ion transport and psii photoprotection determine the salinity tolerance of amphidiploid Brassicas. Plants 2023, 12, 2590. [Google Scholar] [CrossRef]
- Jiang, D.; Lei, J.; Cao, B.; Wu, S.; Chen, G.; Chen, C. Molecular cloning and characterization of three glucosinolate transporter (GTR) genes from Chinese kale. Genes 2019, 10, 202. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, Z.; Chen, J.; Chen, B.; Tang, W.; Chen, X.; Lai, Z.; Guo, R. Comparative transcriptomic analyses of glucosinolate metabolic genes during the formation of Chinese kale seeds. BMC Plant Biol. 2021, 21, 394. [Google Scholar] [CrossRef]
- Li, M.; Xie, F.; Li, Y.; Gong, L.; Luo, Y.; Zhang, Y.; Chen, Q.; Wang, Y.; Lin, Y.; Zhang, Y. Genome-wide analysis of the heat shock transcription factor gene family in Brassica juncea: Structure, evolution, and expression profiles. DNA Cell Biol. 2020, 39, 1990–2004. [Google Scholar] [CrossRef] [PubMed]
- Arbona, V.; Manzi, M.; de Ollas, C.; Gómez-Cadenas, A. Metabolomics as a tool to investigate abiotic stress tolerance in plants. Int. J. Mol. Sci. 2013, 14, 4885–4911. [Google Scholar] [CrossRef] [PubMed]
- Petretto, G.L.; Urgeghe, P.P.; Massa, D.; Melito, S. Effect of salinity (NaCl) on plant growth, nutrient content, and glucosinolate hydrolysis products trends in rocket genotypes. Plant Physiol. Biochem. 2019, 141, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Maina, S.; Ryu, D.H.; Cho, J.Y.; Jung, D.S.; Park, J.-E.; Nho, C.W.; Bakari, G.; Misinzo, G.; Jung, J.H.; Yang, S.-H. Exposure to salinity and light spectra regulates glucosinolates, phenolics, and antioxidant capacity of Brassica carinata L. microgreens. Antioxidants 2021, 10, 1183. [Google Scholar] [CrossRef] [PubMed]
- Sarikamiş, G.; Cakir, A. Influence of salinity on aliphatic and indole glucosinolates in broccoli (Brassica oleracea var. italica). Appl. Ecol. Environ. Res. 2017, 15, 1781. [Google Scholar] [CrossRef]
- Velasco, P.; Cartea, M.E.; González, C.; Vilar, M.; Ordás, A. Factors affecting the glucosinolate content of kale (Brassica oleracea acephala group). J. Agric. Food Chem. 2007, 55, 955–962. [Google Scholar] [CrossRef] [PubMed]
- Zaghdoud, C.; Alcaraz-Lopez, C.; Mota-Cadenas, C.; Martínez-Ballesta, M.d.C.; Moreno, D.A.; Ferchichi, A.; Carvajal, M. Differential responses of two broccoli (Brassica oleracea L. var Italica) cultivars to salinity and nutritional quality improvement. Sci. World J. 2012, 2012, 291435. [Google Scholar] [CrossRef]
- Gyawali, S.; Parkin, I.A.P.; Steppuhn, H.; Buchwaldt, M.; Adhikari, B.; Wood, R.; Wall, K.; Buchwaldt, L.; Singh, M.; Bekkaoui, D. Seedling, early vegetative, and adult plant growth of oilseed rapes (Brassica napus L.) under saline stress. Can. J. Plant Sci. 2019, 99, 927–941. [Google Scholar] [CrossRef]




| Similarity (%) | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AtGTR1 | AtGTR2 | BnaGTR1A1 | BnaGTR1A2 | BnaGTR1A3 | BnaGTR1C1 | BnaGTR1C2 | BnaGTR1C3 | BnaGTR2A1 | BnaGTR2A2 | BnaGTR2A3 | BnaGTR2C1 | BnaGTR2C2 | BnaGTR2C3 | BjuGTR1A1 | BjuGTR1A2 | BjuGTR1A3 | BjuGTR1B1 | BjuGTR1B2 | BjuGTR1B3 | BjuGTR2A1 | BjuGTR2A2 | BjuGTR2A3 | BjuGTR2B1 | BjuGTR2B2 | BjuGTR2B3 | ||
| Divergence (%) | AtGTR1 | 81.4 | 86.4 | 85.1 | 80.7 | 86.3 | 85.1 | 80.4 | 81.2 | 78.2 | 77.9 | 81.1 | 81.2 | 77.7 | 86.4 | 85.3 | 83.1 | 86.3 | 85.6 | 83.2 | 81.2 | 81.2 | 77.9 | 81.2 | 80.9 | 78.5 | |
| AtGTR2 | 18.2 | 79.2 | 79.7 | 77.0 | 79.0 | 79.8 | 77.0 | 92.5 | 92.2 | 85.5 | 92.3 | 92.2 | 85.4 | 79.2 | 80.3 | 77.1 | 78.5 | 79.7 | 77.9 | 92.5 | 92.2 | 85.0 | 92.7 | 92.2 | 86.1 | ||
| BnaGTR1A1 | 12.9 | 19.6 | 89.7 | 88.0 | 99.2 | 89.9 | 87.7 | 80.3 | 79.8 | 77.2 | 80.2 | 79.8 | 78.3 | 99.8 | 90.2 | 88.4 | 96.9 | 89.9 | 88.4 | 80.5 | 79.8 | 77.5 | 80.0 | 80.0 | 78.3 | ||
| BnaGTR1A2 | 14.3 | 18.4 | 7.7 | 85.4 | 89.4 | 98.6 | 85.2 | 81.0 | 79.8 | 77.3 | 80.8 | 79.8 | 76.9 | 89.7 | 99.2 | 85.4 | 89.0 | 96.4 | 87.2 | 81.0 | 79.8 | 77.3 | 80.8 | 79.7 | 77.3 | ||
| BnaGTR1A3 | 17.3 | 21.8 | 10.3 | 12.0 | 87.9 | 85.5 | 97.5 | 77.5 | 77.8 | 74.0 | 77.3 | 77.8 | 74.5 | 88.0 | 85.4 | 99.7 | 88.9 | 85.4 | 94.5 | 77.6 | 77.8 | 75.3 | 77.1 | 77.5 | 74.8 | ||
| BnaGTR1C1 | 13.4 | 20.1 | 0.4 | 8.2 | 10.7 | 89.5 | 87.4 | 80.2 | 79.7 | 76.9 | 80.0 | 79.7 | 77.2 | 99.4 | 89.9 | 88.2 | 96.8 | 89.5 | 88.2 | 80.3 | 79.7 | 77.3 | 79.8 | 79.8 | 78.2 | ||
| BnaGTR1C2 | 14.5 | 18.4 | 7.7 | 1.2 | 12.0 | 8.2 | 85.4 | 81.0 | 79.8 | 77.1 | 80.8 | 79.8 | 76.9 | 89.9 | 98.9 | 85.5 | 89.2 | 98.9 | 87.2 | 81.0 | 79.8 | 77.3 | 80.8 | 79.7 | 77.3 | ||
| BnaGTR1C3 | 17.5 | 22.0 | 10.7 | 12.5 | 2.0 | 11.2 | 12.5 | 77.3 | 77.8 | 73.5 | 77.1 | 77.8 | 74.2 | 87.7 | 85.7 | 97.9 | 88.4 | 85.5 | 94.3 | 77.5 | 77.8 | 74.5 | 77.0 | 77.5 | 74.3 | ||
| BnaGTR2A1 | 18.0 | 6.7 | 17.7 | 17.0 | 21.1 | 18.2 | 17.0 | 21.5 | 94.8 | 88.4 | 99.8 | 94.8 | 88.2 | 80.3 | 81.3 | 77.6 | 79.5 | 81.6 | 78.4 | 99.8 | 94.8 | 86.9 | 99.8 | 94.6 | 89.9 | ||
| BnaGTR2A2 | 18.4 | 7.3 | 18.7 | 19.4 | 21.3 | 19.1 | 19.1 | 21.5 | 4.0 | 87.9 | 94.6 | 89.4 | 81.2 | 81.0 | 80.2 | 78.0 | 79.3 | 79.8 | 77.8 | 94.8 | 99.8 | 87.9 | 94.6 | 97.9 | 86.9 | ||
| BnaGTR2A3 | 23.0 | 13.8 | 22.5 | 21.8 | 25.5 | 22.8 | 21.8 | 26.3 | 11.2 | 11.0 | 88.2 | 87.9 | 97.7 | 77.2 | 77.4 | 74.2 | 76.6 | 77.9 | 74.9 | 88.6 | 87.9 | 96.2 | 88.4 | 88.1 | 94.1 | ||
| BnaGTR2C1 | 18.2 | 6.9 | 18.0 | 17.3 | 21.3 | 18.4 | 17.3 | 21.8 | 0.2 | 4.2 | 11.4 | 94.6 | 88.1 | 80.2 | 81.2 | 77.5 | 79.3 | 81.5 | 78.2 | 99.8 | 94.6 | 86.7 | 97.9 | 94.4 | 89.8 | ||
| BnaGTR2C2 | 18.7 | 7.3 | 18.9 | 19.4 | 21.3 | 19.4 | 19.1 | 21.5 | 4.0 | 0.2 | 11.0 | 4.2 | 87.7 | 79.8 | 80.2 | 78.0 | 79.3 | 81.0 | 78.2 | 94.8 | 89.4 | 86.9 | 94.2 | 97.7 | 89.4 | ||
| BnaGTR2C3 | 22.5 | 13.8 | 22.0 | 22.0 | 24.7 | 22.3 | 22.0 | 25.2 | 11.6 | 11.2 | 2.0 | 11.8 | 11.2 | 77.5 | 77.2 | 74.7 | 76.9 | 78.0 | 75.7 | 88.4 | 87.7 | 95.8 | 88.1 | 87.9 | 94.6 | ||
| BjuGTR1A1 | 12.9 | 19.6 | 0.0 | 7.7 | 10.3 | 0.4 | 7.7 | 10.7 | 17.7 | 18.7 | 22.5 | 18.0 | 18.9 | 22.0 | 90.2 | 88.4 | 97.1 | 89.9 | 88.4 | 80.5 | 79.8 | 77.5 | 80.0 | 80.0 | 78.3 | ||
| BjuGTR1A2 | 14.1 | 18.0 | 7.5 | 0.6 | 11.8 | 7.9 | 1.0 | 12.3 | 16.8 | 18.9 | 21.3 | 17.0 | 18.9 | 21.5 | 7.5 | 85.4 | 89.5 | 96.7 | 87.7 | 81.3 | 80.2 | 77.7 | 81.2 | 80.0 | 77.7 | ||
| BjuGTR1A3 | 17.0 | 21.5 | 10.1 | 11.8 | 0.2 | 10.5 | 11.8 | 1.8 | 20.8 | 21.1 | 25.2 | 21.1 | 21.1 | 24.5 | 10.1 | 11.6 | 89.2 | 85.4 | 94.8 | 77.8 | 78.0 | 75.3 | 77.3 | 77.6 | 74.9 | ||
| BjuGTR1B1 | 12.9 | 19.6 | 2.4 | 8.2 | 9.4 | 2.8 | 8.2 | 9.9 | 17.7 | 18.2 | 22.5 | 18.0 | 18.4 | 22.0 | 2.4 | 7.9 | 9.2 | 89.4 | 89.2 | 79.7 | 79.3 | 77.3 | 79.2 | 79.5 | 77.5 | ||
| BjuGTR1B2 | 14.1 | 19.4 | 8.2 | 3.2 | 12.5 | 8.6 | 3.4 | 12.5 | 16.6 | 18.0 | 21.1 | 16.8 | 18.0 | 20.8 | 8.2 | 3.2 | 12.3 | 8.4 | 87.4 | 81.6 | 81.0 | 77.9 | 81.6 | 81.0 | 78.3 | ||
| BjuGTR1B3 | 16.1 | 20.1 | 9.6 | 10.7 | 3.8 | 10.1 | 11.0 | 4.2 | 19.4 | 20.3 | 24.2 | 19.6 | 20.3 | 23.2 | 9.6 | 10.5 | 3.6 | 8.6 | 11.0 | 78.5 | 78.2 | 76.0 | 78.2 | 78.0 | 75.7 | ||
| BjuGTR2A1 | 18.0 | 6.7 | 17.7 | 17.0 | 21.1 | 18.2 | 17.0 | 21.5 | 0.0 | 4.0 | 11.2 | 0.2 | 4.0 | 11.6 | 17.7 | 16.8 | 20.8 | 17.7 | 16.6 | 19.4 | 94.8 | 86.9 | 98.0 | 94.6 | 90.1 | ||
| BjuGTR2A2 | 18.4 | 7.3 | 18.7 | 19.4 | 21.3 | 19.1 | 19.1 | 21.5 | 4.0 | 0.0 | 11.0 | 4.2 | 0.2 | 11.2 | 18.7 | 18.9 | 21.1 | 18.2 | 18.0 | 20.3 | 4.0 | 86.9 | 94.6 | 97.9 | 89.4 | ||
| BjuGTR2A3 | 24.7 | 16.3 | 25.0 | 24.2 | 27.3 | 25.2 | 24.2 | 28.3 | 14.1 | 13.8 | 4.0 | 14.3 | 13.8 | 4.4 | 25.0 | 23.7 | 27.3 | 25.0 | 23.5 | 26.3 | 14.1 | 13.8 | 87.3 | 87.3 | 91.7 | ||
| BjuGTR2B1 | 18.0 | 6.5 | 18.4 | 17.0 | 21.5 | 18.9 | 17.0 | 22.0 | 1.6 | 4.2 | 11.0 | 1.8 | 4.2 | 11.4 | 18.4 | 16.8 | 21.3 | 18.4 | 16.6 | 19.6 | 1.6 | 4.2 | 13.6 | 94.4 | 89.6 | ||
| BjuGTR2B2 | 18.4 | 7.5 | 18.2 | 19.1 | 21.1 | 18.7 | 18.9 | 21.3 | 4.8 | 1.2 | 11.0 | 5.0 | 1.4 | 11.2 | 18.2 | 18.7 | 20.8 | 17.7 | 17.5 | 19.8 | 4.8 | 1.2 | 13.6 | 5.0 | 89.4 | ||
| BjuGTR2B3 | 22.5 | 12.7 | 21.8 | 22.5 | 25.2 | 22.3 | 22.5 | 25.7 | 9.4 | 9.0 | 6.1 | 9.6 | 9.0 | 5.8 | 21.8 | 22.0 | 25.0 | 22.0 | 21.3 | 24.0 | 9.4 | 9.0 | 8.8 | 9.4 | 9.4 | ||
| Gene | Coefficients | Estimate | Standard Error | t-Value | Pr (>|t|) | Significance | Cumulative % Variance Explained |
|---|---|---|---|---|---|---|---|
| (Intercept) | 137.31 | 5.67 | 24.2 | 1.3 × 10−14 | *** | - | |
| GTR2A21 | Stem | 21.46 | 7.84 | 2.73 | 0.0153 | * | 52.57 |
| Root | 0.93 | 0.53 | 1.73 | 0.1027 | — | 60.52 | |
| GTR2B1/C22 | Flower | 14.73 | 3.69 | 3.98 | 0.0010 | ** | 49.83 |
| GTR2A13 | Flower | 19.01 | 8.50 | 2.23 | 0.0409 | * | 45.33 |
| Root | 9.54 | 4.30 | 2.21 | 0.0425 | * | 58.82 | |
| GTR1A2/B14 | Root | 2.22 | 1.00 | 2.22 | 0.0417 | * | 47.17 |
| Stem | 16.59 | 7.96 | 2.08 | 0.0548 | — | 59.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fatima, S.; Khan, M.O.; Iqbal, N.; Iqbal, M.M.; Qamar, H.; Imtiaz, M.; Hundleby, P.; Wei, Z.; Ahmad, N. Studying Salt-Induced Shifts in Gene Expression Patterns of Glucosinolate Transporters and Glucosinolate Accumulation in Two Contrasting Brassica Species. Metabolites 2024, 14, 179. https://doi.org/10.3390/metabo14040179
Fatima S, Khan MO, Iqbal N, Iqbal MM, Qamar H, Imtiaz M, Hundleby P, Wei Z, Ahmad N. Studying Salt-Induced Shifts in Gene Expression Patterns of Glucosinolate Transporters and Glucosinolate Accumulation in Two Contrasting Brassica Species. Metabolites. 2024; 14(4):179. https://doi.org/10.3390/metabo14040179
Chicago/Turabian StyleFatima, Samia, Muhammad Omar Khan, Nadia Iqbal, Muhammad Mudassar Iqbal, Huma Qamar, Muhammad Imtiaz, Penny Hundleby, Zhengyi Wei, and Niaz Ahmad. 2024. "Studying Salt-Induced Shifts in Gene Expression Patterns of Glucosinolate Transporters and Glucosinolate Accumulation in Two Contrasting Brassica Species" Metabolites 14, no. 4: 179. https://doi.org/10.3390/metabo14040179
APA StyleFatima, S., Khan, M. O., Iqbal, N., Iqbal, M. M., Qamar, H., Imtiaz, M., Hundleby, P., Wei, Z., & Ahmad, N. (2024). Studying Salt-Induced Shifts in Gene Expression Patterns of Glucosinolate Transporters and Glucosinolate Accumulation in Two Contrasting Brassica Species. Metabolites, 14(4), 179. https://doi.org/10.3390/metabo14040179






