Mitochondrial Abundance and Function Differ Across Muscle Within Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sampling
2.2. Mitochondrial DNA Copy Number
2.3. SDS-Page and Western Blotting
2.4. Mitochondrial Isolation
2.5. Mitochondrial Respiration
2.6. Statistical Analysis
3. Results
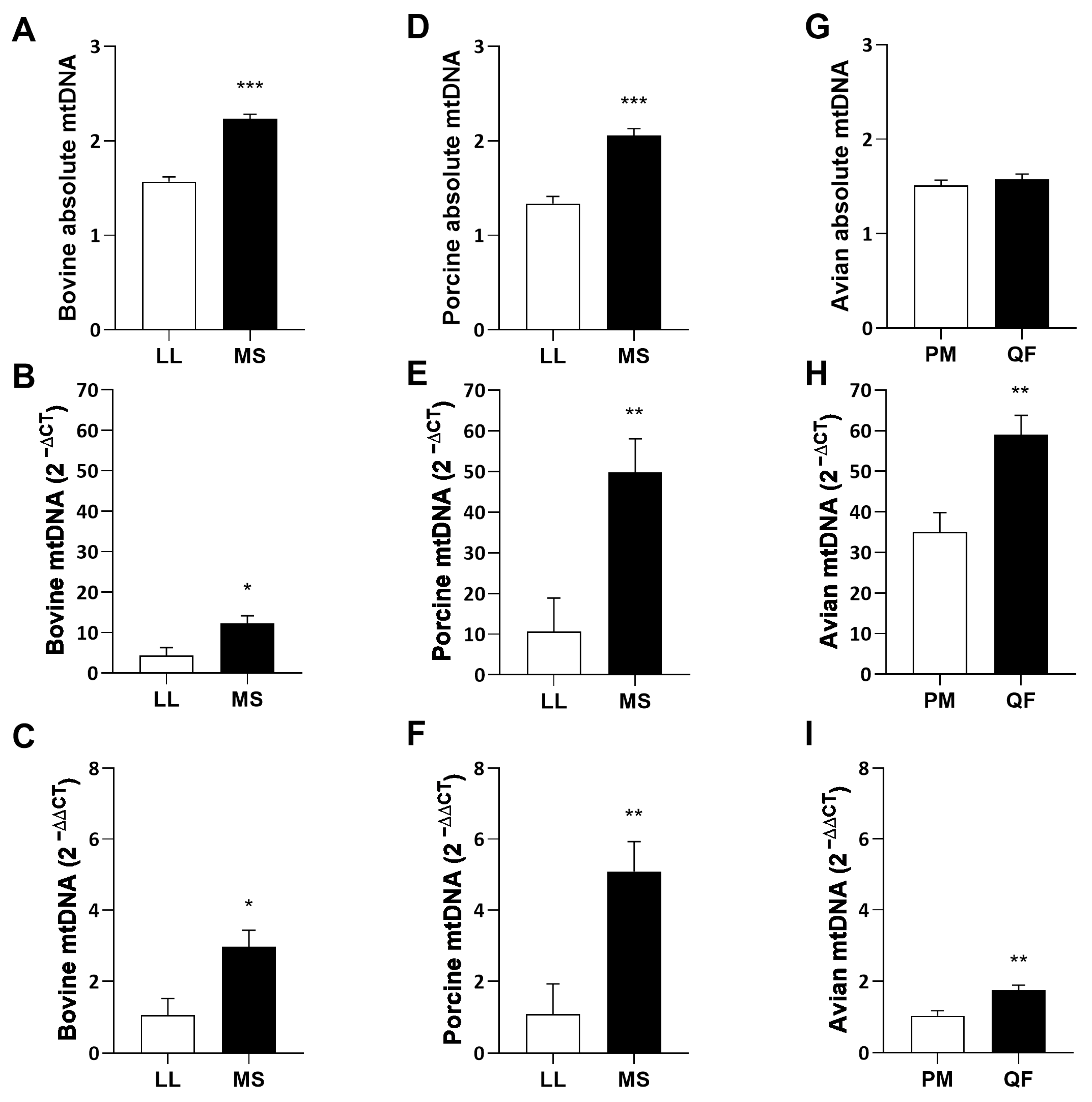
3.1. Mitochondrial DNA Copy Number
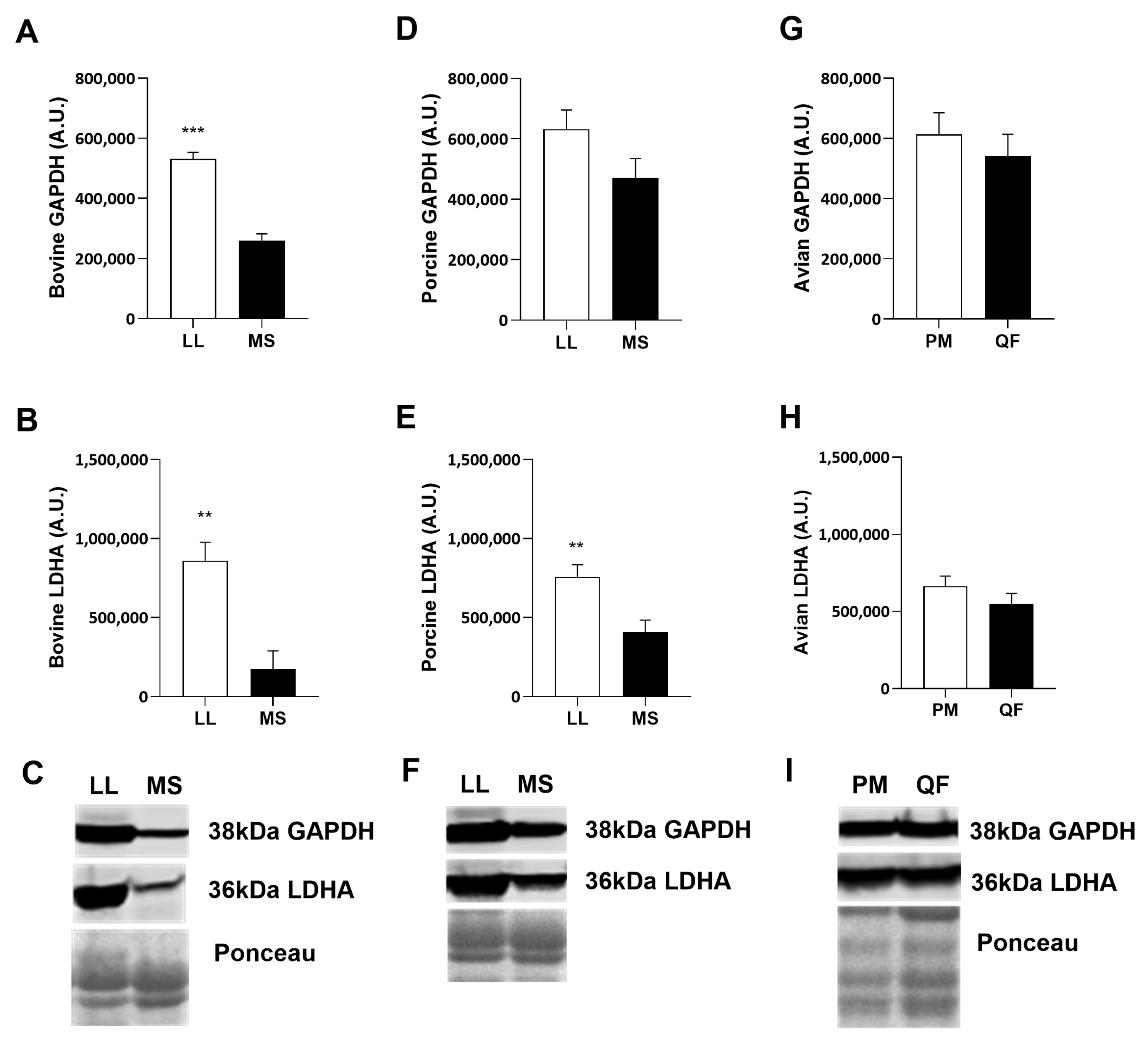
3.2. Muscle Protein Abundance
3.3. Abundance of Proteins in Mitochondrial Fraction
3.4. Mitochondrial Respiration
4. Discussion
4.1. Mitochondrial DNA Copy Number
4.2. Muscle Protein Abundance
4.3. Abundance of Proteins in Mitochondrial Fraction
4.4. Mitochondrial Respiration
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Capper, J.L. The environmental impact of beef production in the United States: 1977 compared with 2007. J. Anim. Sci. 2011, 89, 4249–4261. [Google Scholar] [CrossRef] [PubMed]
- Terry, S.A.; Basarab, J.A.; Guan, L.L.; McAllister, T.A. Strategies to improve the efficiency of beef cattle production. Can. J. Anim. Sci. 2021, 101, 1–19. [Google Scholar] [CrossRef]
- Wu, F.; Vierck, K.R.; DeRouchey, J.M.; O’Quinn, T.G.; Tokach, M.D.; Goodband, R.D.; Dritz, S.S.; Woodworth, J.C. A review of heavy weight market pigs: Status of knowledge and future needs assessment. Transl. Anim. Sci. 2017, 1, 1–15. [Google Scholar] [CrossRef]
- Zuidhof, M.J.; Schneider, B.L.; Carney, V.L.; Korver, D.R.; Robinson, F.E. Growth, efficiency, and yield of commercial broilers from 1957, 1978, and 2005. Poult. Sci. 2014, 93, 2970–2982. [Google Scholar] [CrossRef]
- Drouillard, J.S. Current situation and future trends for beef production in the United States of America—A review. Asian-Australas. J. Anim. Sci. 2018, 31, 1007–1016. [Google Scholar] [CrossRef] [PubMed]
- Wicks, J.; Beline, M.; Gomez, J.F.M.; Luzardo, S.; Silva, S.L.; Gerrard, D. Muscle Energy Metabolism, Growth, and Meat Quality in Beef Cattle. Agriculture 2019, 9, 195. [Google Scholar] [CrossRef]
- Schiaffino, S.; Reggiani, C. Molecular diversity of myofibrillar proteins: Gene regulation and functional significance. Physiol. Rev. 1996, 76, 371–423. [Google Scholar] [CrossRef]
- Schiaffino, S.; Reggiani, C. Fiber Types in Mammalian Skeletal Muscles. Physiol. Rev. 2011, 91, 1447–1531. [Google Scholar] [CrossRef]
- Arany, Z.; Lebrasseur, N.; Morris, C.; Smith, E.; Yang, W.; Ma, Y.; Chin, S.; Spiegelman, B.M. The Transcriptional Coactivator PGC-1β Drives the Formation of Oxidative Type IIX Fibers in Skeletal Muscle. Cell Metab. 2007, 5, 35–46. [Google Scholar] [CrossRef]
- Bottje, W.G.; Carstens, G.E. Variation in metabolism: Biological efficiency of energy production and utilization that affects feed efficiency. In Feed Efficiency in the Beef Industry; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2012; pp. 251–273. [Google Scholar]
- Mishra, P.; Varuzhanyan, G.; Pham, A.H.; Chan, D.C. Mitochondrial Dynamics is a Distinguishing Feature of Skeletal Muscle Fiber Types and Regulates Organellar Compartmentalization. Cell Metab. 2015, 22, 1033–1044. [Google Scholar] [CrossRef]
- Glancy, B.; Balaban, R.S. Protein composition and function of red and white skeletal muscle mitochondria. Am. J. Physiol.-Cell Physiol. 2011, 300, C1280–C1290. [Google Scholar] [CrossRef] [PubMed]
- Apaoblaza, A.; Gerrard, S.D.; Matarneh, S.K.; Wicks, J.C.; Kirkpatrick, L.; England, E.M.; Scheffler, T.L.; Duckett, S.K.; Shi, H.; Silva, S.L.; et al. Muscle from grass- and grain-fed cattle differs energetically. Meat Sci. 2020, 161, 107996. [Google Scholar] [CrossRef] [PubMed]
- Morales Gómez, J.F.; Antonelo, D.S.; Beline, M.; Pavan, B.; Bambil, D.B.; Fantinato-Neto, P.; Saran-Netto, A.; Leme, P.R.; Goulart, R.S.; Gerrard, D.E.; et al. Feeding strategies impact animal growth and beef color and tenderness. Meat Sci. 2022, 183, 108599. [Google Scholar] [CrossRef]
- Antonelo, D.S.; Gómez, J.F.M.; Silva, S.L.; Beline, M.; Zhang, X.; Wang, Y.; Pavan, B.; Koulicoff, L.A.; Rosa, A.F.; Goulart, R.S.; et al. Proteome basis for the biological variations in color and tenderness of longissimus thoracis muscle from beef cattle differing in growth rate and feeding regime. Food Res. Int. 2022, 153, 110947. [Google Scholar] [CrossRef] [PubMed]
- Rehfeldt, C.; Henning, M.; Fiedler, I. Consequences of pig domestication for skeletal muscle growth and cellularity. Livest. Sci. 2008, 116, 30–41. [Google Scholar] [CrossRef]
- Reverter, A.; Okimoto, R.; Sapp, R.; Bottje, W.G.; Hawken, R.; Hudson, N.J. Chicken muscle mitochondrial content appears co-ordinately regulated and is associated with performance phenotypes. Biol. Open 2016, 6, 50–58. [Google Scholar] [CrossRef]
- Zeng, C.; Shi, H.; Kirkpatrick, L.T.; Ricome, A.; Park, S.; Scheffler, J.M.; Hannon, K.M.; Grant, A.L.; Gerrard, D.E. Driving an Oxidative Phenotype Protects Myh4 Null Mice From Myofiber Loss During Postnatal Growth. Front. Physiol. 2022, 12, 785151. [Google Scholar] [CrossRef]
- López-Andreo, M.; Lugo, L.; Garrido-Pertierra, A.; Prieto, M.I.; Puyet, A. Identification and quantitation of species in complex DNA mixtures by real-time polymerase chain reaction. Anal. Biochem. 2005, 339, 73–82. [Google Scholar] [CrossRef]
- Laker, R.C.; Drake, J.C.; Wilson, R.J.; Lira, V.A.; Lewellen, B.M.; Ryall, K.A.; Fisher, C.C.; Zhang, M.; Saucerman, J.J.; Goodyear, L.J.; et al. Ampk phosphorylation of Ulk1 is required for targeting of mitochondria to lysosomes in exercise-induced mitophagy. Nat. Commun. 2017, 8, 548. [Google Scholar] [CrossRef]
- Scheffler, T.L.; Matarneh, S.K.; England, E.M.; Gerrard, D.E. Mitochondria influence postmortem metabolism and pH in an in vitro model. Meat Sci. 2015, 110, 118–125. [Google Scholar] [CrossRef]
- Boutagy, N.E.; Pyne, E.; Rogers, G.W.; Ali, M.; Hulver, M.W.; Frisard, M.I. Isolation of Mitochondria from Minimal Quantities of Mouse Skeletal Muscle for High Throughput Microplate Respiratory Measurements. J. Vis. Exp. 2015, 105, e53217. [Google Scholar] [CrossRef]
- Herbst, A.; Widjaja, K.; Nguy, B.; Lushaj, E.B.; Moore, T.M.; Hevener, A.L.; McKenzie, D.; Aiken, J.M.; Wanagat, J. Digital PCR Quantitation of Muscle Mitochondrial DNA: Age, Fiber Type, and Mutation-Induced Changes. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2017, 72, 1327–1333. [Google Scholar] [CrossRef] [PubMed]
- Cole, L.W. The Evolution of Per-cell Organelle Number. Front. Cell Dev. Biol. 2016, 4, 85. [Google Scholar] [CrossRef]
- Hoeks, J.; Hesselink, M.; Schrauwen, P. Mitochondrial Respiration. In Encyclopedia of Exercise Medicine in Health and Disease; Mooren, F.C., Ed.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 587–590. [Google Scholar]
- Groot, G.S.P.; Kroon, A.M. Mitochondrial DNA from various organisms does not contain internally methylated cytosine in -CCGG- sequences. Biochim. Biophys. Acta (BBA)—Nucleic Acids Protein Synth. 1979, 564, 355–357. [Google Scholar] [CrossRef]
- Malik, A.N.; Shahni, R.; Iqbal, M.M. Increased peripheral blood mitochondrial DNA in type 2 diabetic patients with nephropathy. Diabetes Res. Clin. Pract. 2009, 86, e22–e24. [Google Scholar] [CrossRef]
- Malik, A.N.; Czajka, A. Is mitochondrial DNA content a potential biomarker of mitochondrial dysfunction? Mitochondrion 2013, 13, 481–492. [Google Scholar] [CrossRef] [PubMed]
- Ajaz, S.; Czajka, A.; Malik, A. Accurate Measurement of Circulating Mitochondrial DNA Content from Human Blood Samples Using Real-Time Quantitative PCR. In Mitochondrial Medicine: Volume I, Probing Mitochondrial Function; Weissig, V., Edeas, M., Eds.; Springer: New York, NY, USA, 2015; pp. 117–131. [Google Scholar]
- Hock, M.B.; Kralli, A. Transcriptional Control of Mitochondrial Biogenesis and Function. Annu. Rev. Physiol. 2009, 71, 177–203. [Google Scholar] [CrossRef]
- Williams, R.S. Mitochondrial gene expression in mammalian striated muscle. Evidence that variation in gene dosage is the major regulatory event. J. Biol. Chem. 1986, 261, 12390–12394. [Google Scholar] [CrossRef]
- Veltri, K.L.; Espiritu, M.; Singh, G. Distinct genomic copy number in mitochondria of different mammalian organs. J. Cell. Physiol. 1990, 143, 160–164. [Google Scholar] [CrossRef]
- Song, S.; Ahn, C.H.; Kim, G.D. Muscle Fiber Typing in Bovine and Porcine Skeletal Muscles Using Immunofluorescence with Monoclonal Antibodies Specific to Myosin Heavy Chain Isoforms. Food Sci. Anim. Resour. 2020, 40, 132–144. [Google Scholar] [CrossRef]
- Tanabe, R.I.; Susumu, M.; Koichi, C. Sequencing of the 2a, 2x, and slow isoforms of the bovine myosin heavy chain and the different expression among muscles. Mamm. Genome 1998, 9, 1056–1058. [Google Scholar] [CrossRef] [PubMed]
- Wegner, J.; Albrecht, E.; Fiedler, I.; Teuscher, F.; Papstein, H.-J.; Ender, K. Growth- and breed-related changes of muscle fiber characteristics in cattle1. J. Anim. Sci. 2000, 78, 1485–1496. [Google Scholar] [CrossRef] [PubMed]
- Hammond, E.L.; Sayer, D.; Nolan, D.; Walker, U.A.; Ronde, A.; Montaner, J.S.; Cote, H.C.; Gahan, M.E.; Cherry, C.L.; Wesselingh, S.L.; et al. Assessment of precision and concordance of quantitative mitochondrial DNA assays: A collaborative international quality assurance study. J. Clin. Virol. 2003, 27, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Klont, R.E.; Brocks, L.; Eikelenboom, G. Muscle fibre type and meat quality. Meat Sci. 1998, 49, S219–S229. [Google Scholar] [CrossRef]
- England, E.M.; Matarneh, S.K.; Oliver, E.M.; Apaoblaza, A.; Scheffler, T.L.; Shi, H.; Gerrard, D.E. Excess glycogen does not resolve high ultimate pH of oxidative muscle. Meat Sci. 2016, 114, 95–102. [Google Scholar] [CrossRef]
- Ono, Y.; Iwamoto, H.; Takahara, H. The Relationship Between Muscle Growth and the Growth of Different Fiber Types in the Chicken. Poult. Sci. 1993, 72, 568–576. [Google Scholar] [CrossRef]
- Noskov, S.Y.; Rostovtseva, T.K.; Chamberlin, A.C.; Teijido, O.; Jiang, W.; Bezrukov, S.M. Current state of theoretical and experimental studies of the voltage-dependent anion channel (VDAC). Biochim. Biophys. Acta (BBA) Biomembr. 2016, 1858, 1778–1790. [Google Scholar] [CrossRef]
- Schuh, R.A.; Jackson, K.C.; Khairallah, R.J.; Ward, C.W.; Spangenburg, E.E. Measuring mitochondrial respiration in intact single muscle fibers. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 302, R712–R719. [Google Scholar] [CrossRef]
- Picard, M.; Taivassalo, T.; Gouspillou, G.; Hepple, R.T. Mitochondria: Isolation, structure and function. J. Physiol. 2011, 589, 4413–4421. [Google Scholar] [CrossRef]
- Lanza, I.R.; Nair, K.S. Functional assessment of isolated mitochondria in vitro. Methods Enzymol. 2009, 457, 349–372. [Google Scholar] [CrossRef]
- Gnaiger, E. Capacity of oxidative phosphorylation in human skeletal muscle: New perspectives of mitochondrial physiology. Int. J. Biochem. Cell Biol. 2009, 41, 1837–1845. [Google Scholar] [CrossRef] [PubMed]
- Horan, M.P.; Pichaud, N.; Ballard, J.W.O. Review: Quantifying Mitochondrial Dysfunction in Complex Diseases of Aging. J. Gerontol. Ser. A 2012, 67, 1022–1035. [Google Scholar] [CrossRef] [PubMed]
- Leverve, X.M.; Fontaine, E. Role of Substrates in the Regulation of Mitochondrial Function In Situ. Int. Union Biochem. Mol. Biol. 2001, 52, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Kerner, J.; Hoppel, C. Fatty acid import into mitochondria. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2000, 1486, 1–17. [Google Scholar] [CrossRef]
- Maekawa, S.; Takada, S.; Furihata, T.; Fukushima, A.; Yokota, T.; Kinugawa, S. Mitochondrial respiration of complex II is not lower than that of complex I in mouse skeletal muscle. Biochem. Biophys. Rep. 2020, 21, 100717. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yen, C.-N.; Bodmer, J.S.; Wicks, J.C.; Zumbaugh, M.D.; Persia, M.E.; Shi, T.H.; Gerrard, D.E. Mitochondrial Abundance and Function Differ Across Muscle Within Species. Metabolites 2024, 14, 553. https://doi.org/10.3390/metabo14100553
Yen C-N, Bodmer JS, Wicks JC, Zumbaugh MD, Persia ME, Shi TH, Gerrard DE. Mitochondrial Abundance and Function Differ Across Muscle Within Species. Metabolites. 2024; 14(10):553. https://doi.org/10.3390/metabo14100553
Chicago/Turabian StyleYen, Con-Ning, Jocelyn S. Bodmer, Jordan C. Wicks, Morgan D. Zumbaugh, Michael E. Persia, Tim H. Shi, and David E. Gerrard. 2024. "Mitochondrial Abundance and Function Differ Across Muscle Within Species" Metabolites 14, no. 10: 553. https://doi.org/10.3390/metabo14100553
APA StyleYen, C.-N., Bodmer, J. S., Wicks, J. C., Zumbaugh, M. D., Persia, M. E., Shi, T. H., & Gerrard, D. E. (2024). Mitochondrial Abundance and Function Differ Across Muscle Within Species. Metabolites, 14(10), 553. https://doi.org/10.3390/metabo14100553






