Evaluation of Metabolomics as Diagnostic Targets in Oral Squamous Cell Carcinoma: A Systematic Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Research Question
2.2. Search Strategy
2.3. Study Inclusion and Exclusion Criteria
2.3.1. Inclusion Criteria
- P (Participants): Adult patients (>18 years of age) from any geographic location, any age or gender.
- E (Exposure): Patients with confirmed diagnosis of OSCC.
- C (Comparison): Difference in concentration of metabolites between OSCC and predetermined controls.
- O (Outcomes): Dysregulation of metabolite concentrations between the predetermined study groups, which are reported as either mean ± standard deviation, fold change concentration or log fold change concentration.
- S (Study Design): Human-based observational studies (case-control, cohort, or cross-sectional) published since inception of OSCC metabolomics, i.e., January 2007 and April 2023 that used a metabolomic technique to quantify metabolite concentration.
2.3.2. Exclusion Criteria
- Patients diagnosed with neoplasms other than OSCC either in the past or currently.
- Patients suffering from any reported chronic systemic illness or on medication for the same.
- Patients with oral lesions due to associated dermatological diseases, infections, localised trauma, recurrent aphthous ulcers, and systemic conditions.
- Targeted metabolomic experiments that are used to validate and translate already identified metabolites from hypothesis generating studies.
- Components other than metabolites as biomarkers such as genetic and protein.
- Animal or cell-based studies.
- Non-observational study designs such as case reports, conference proceedings, letters to editor, reviews, and meta-analysis.
- Metabolites quantified other than in concentration such as field of appearance, retention time, m/z ratio, etc.
- Studies published before January 2007 or after April 2023.
2.4. Study Selection
2.5. Data Extraction and Outcomes
2.6. Data Synthesis
2.7. Risk of Bias Assessment
2.8. Statistical Analysis
3. Results
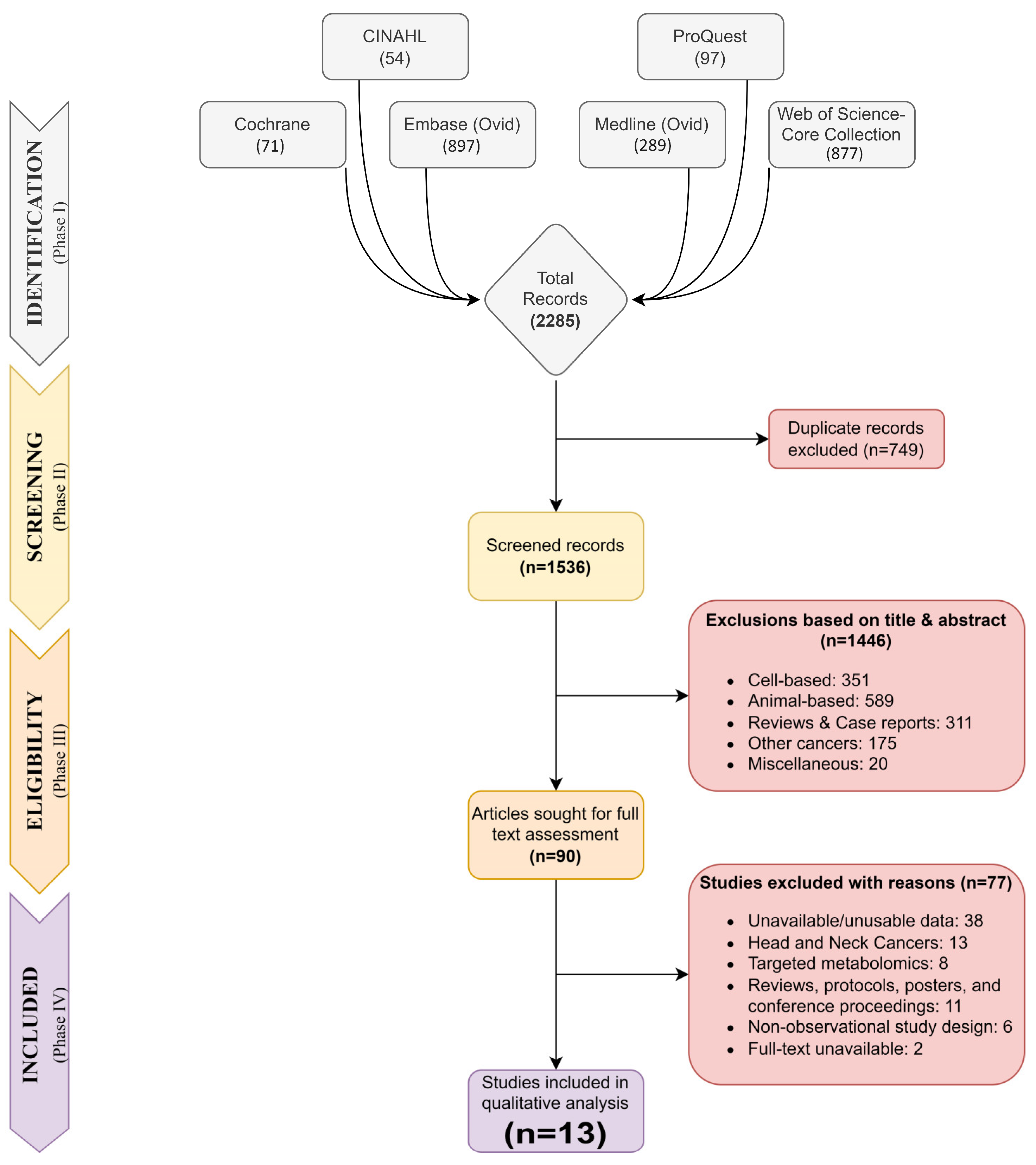
3.1. Search Strategy and Study Selection
3.2. Study Characteristics
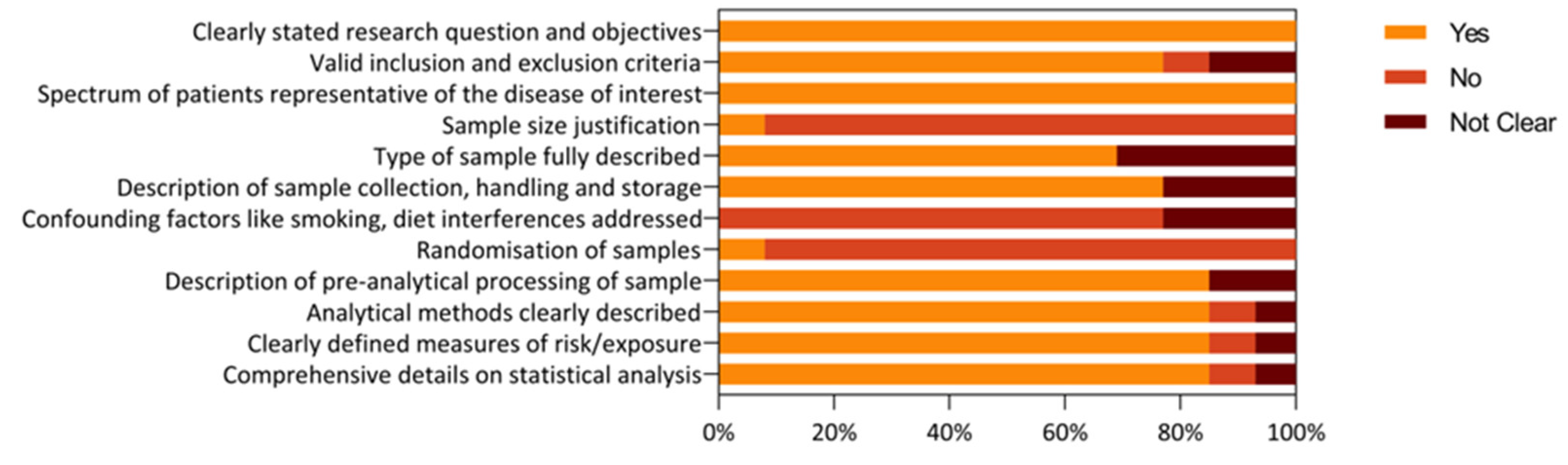
3.3. Risk of Bias and Quality Assessment
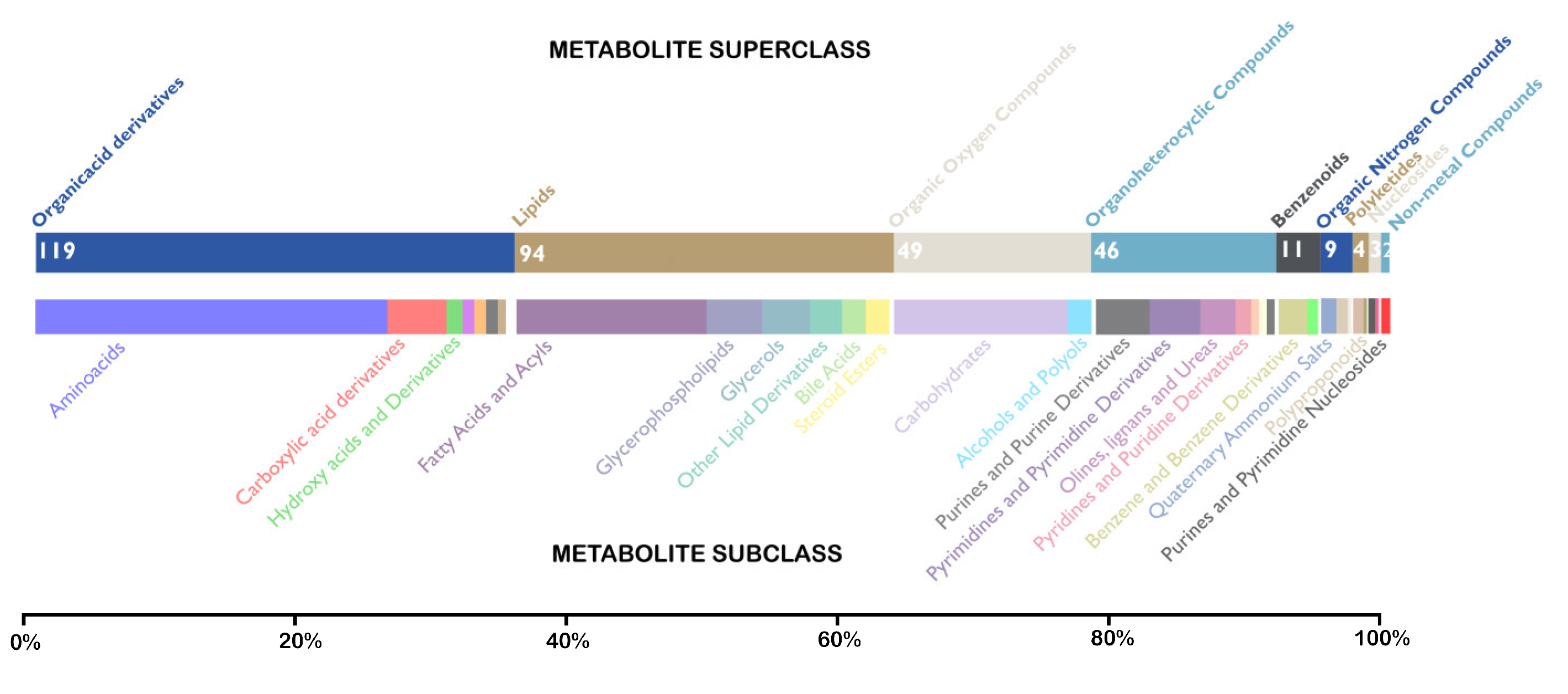
3.4. Identification of Unique Metabolites
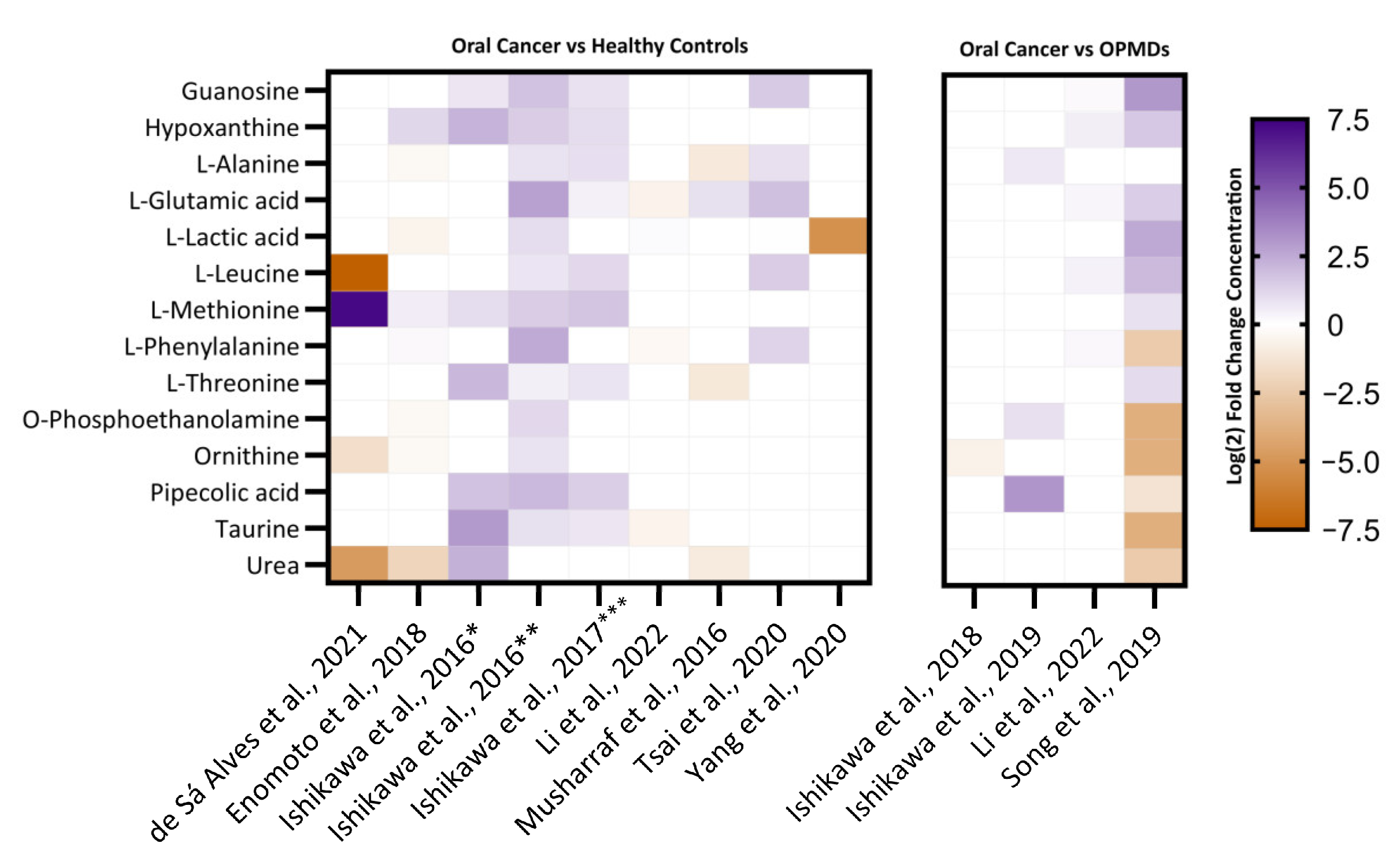
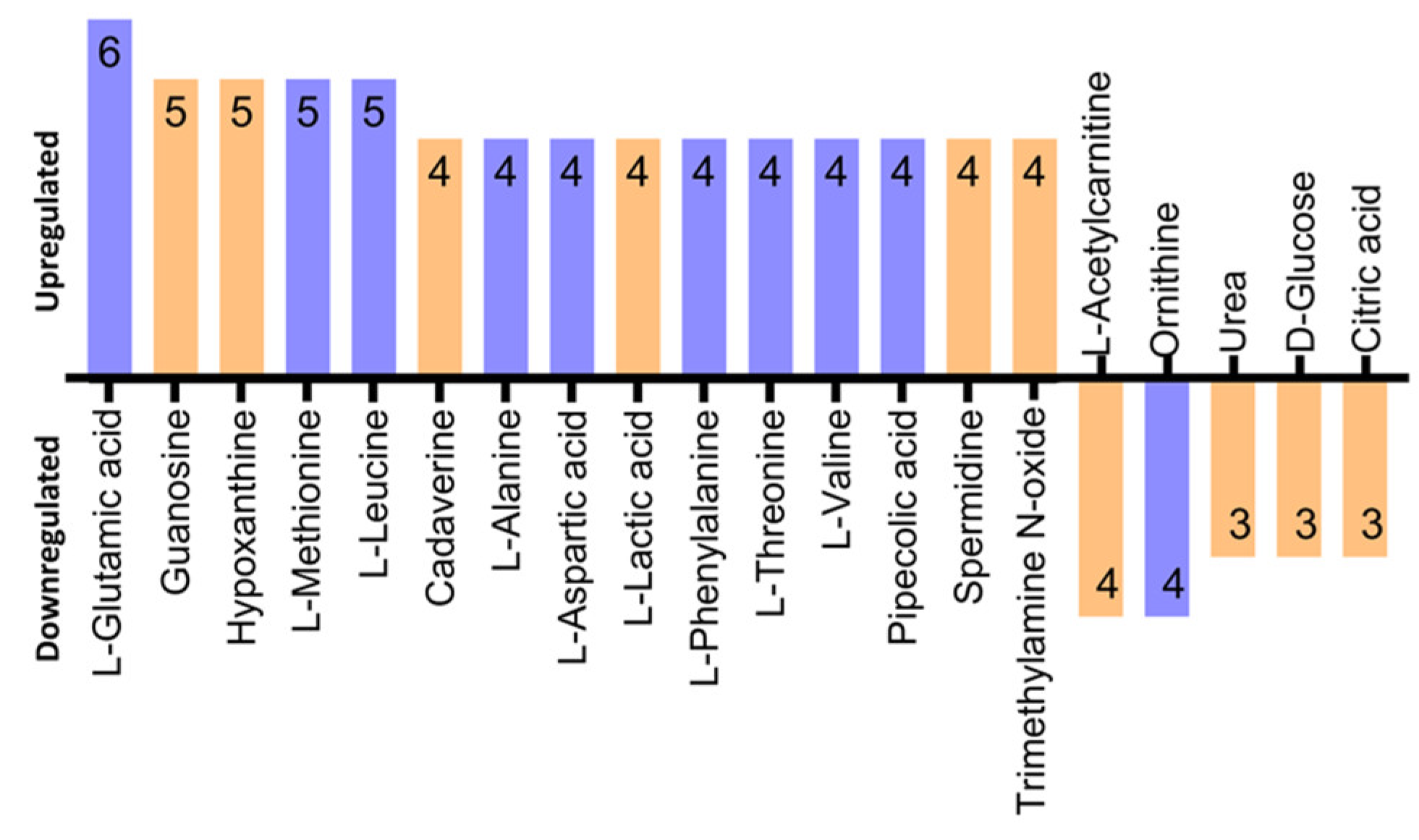
3.5. Identification of Differentially Regulated Metabolites
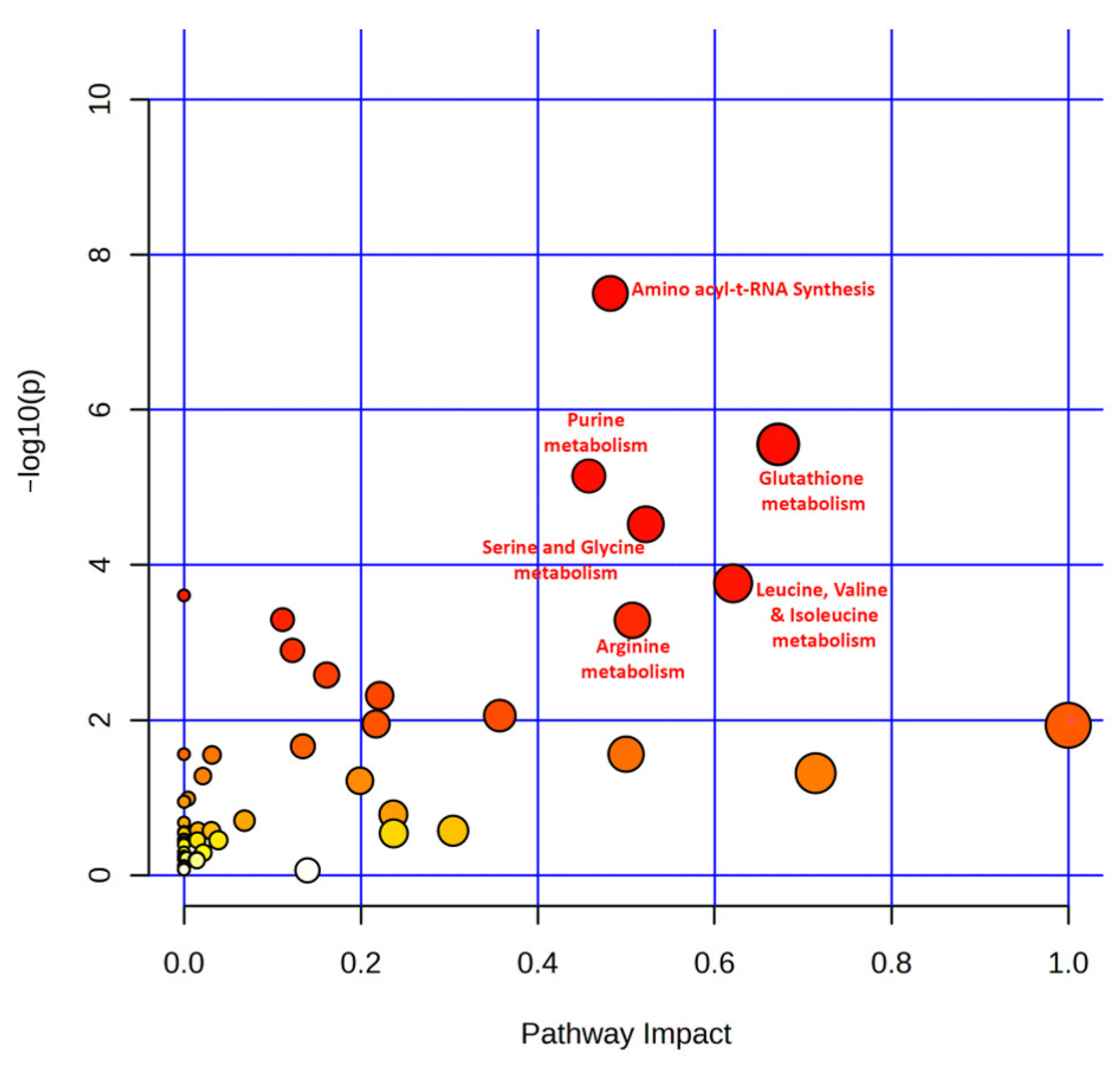
3.6. Pathway Analysis of Top Featured Metabolites
4. Discussion
4.1. Identification of Metabolite Candidate Biomarkers
4.2. Identification of Significantly Enriched Pathways
4.3. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer Statistics for the Year 2020: An Overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Aguirre-Urizar, J.M.; Lafuente-Ibáñez de Mendoza, I.; Warnakulasuriya, S. Malignant Transformation of Oral Leukoplakia: Systematic Review and Meta-Analysis of the Last 5 Years. Oral Dis. 2021, 27, 1881–1895. [Google Scholar] [CrossRef] [PubMed]
- Chaitanya, N.; Allam, N.; Gandhi Babu, D.; Waghray, S.; Badam, R.; Lavanya, R. Systematic Meta-Analysis on Association of Human Papilloma Virus and Oral Cancer. J. Cancer Res. Ther. 2016, 12, 969. [Google Scholar] [CrossRef]
- Pelucchi, C.; Gallus, S.; Garavello, W.; Bosetti, C.; La Vecchia, C. Cancer Risk Associated with Alcohol and Tobacco Use: Focus on Upper Aero-Digestive Tract and Liver. Alcohol Res. Health 2006, 29, 193–198. [Google Scholar] [PubMed]
- Warnakulasuriya, S. Causes of Oral Cancer—An Appraisal of Controversies. Br. Dent. J. 2009, 207, 471–475. [Google Scholar] [CrossRef] [PubMed]
- Speight, P.M.; Epstein, J.; Kujan, O.; Lingen, M.W.; Nagao, T.; Ranganathan, K.; Vargas, P. Screening for Oral Cancer—A Perspective from the Global Oral Cancer Forum. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2017, 123, 680–687. [Google Scholar] [CrossRef]
- Thavarool, S.B.; Muttath, G.; Nayanar, S.; Duraisamy, K.; Bhat, P.; Shringarpure, K.; Nayak, P.; Tripathy, J.P.; Thaddeus, A.; Philip, S.; et al. Improved Survival among Oral Cancer Patients: Findings from a Retrospective Study at a Tertiary Care Cancer Centre in Rural Kerala, India. World J. Surg. Oncol. 2019, 17, 15. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Lum, J.J.; Hatzivassiliou, G.; Thompson, C.B. The Biology of Cancer: Metabolic Reprogramming Fuels Cell Growth and Proliferation. Cell Metab. 2008, 7, 11–20. [Google Scholar] [CrossRef]
- Wei, J.; Xie, G.; Zhou, Z.; Shi, P.; Qiu, Y.; Zheng, X.; Chen, T.; Su, M.; Zhao, A.; Jia, W. Salivary Metabolite Signatures of Oral Cancer and Leukoplakia. Int. J. Cancer 2011, 129, 2207–2217. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.; Wang, S.; Li, Z.; Hu, X.; Yang, X.; Song, Y.; Jing, Y.; Hu, Q.; Ni, Y. Identification of Metabolism-Associated Biomarkers for Early and Precise Diagnosis of Oral Squamous Cell Carcinoma. Biomolecules 2022, 12, 400. [Google Scholar] [CrossRef]
- Rai, V.; Mukherjee, R.; Ghosh, A.K.; Routray, A.; Chakraborty, C. “Omics” in Oral Cancer: New Approaches for Biomarker Discovery. Arch. Oral Biol. 2018, 87, 15–34. [Google Scholar] [CrossRef]
- Xiao, Y.; Bi, M.; Guo, H.; Li, M. Multi-Omics Approaches for Biomarker Discovery in Early Ovarian Cancer Diagnosis. EBioMedicine 2022, 79, 104001. [Google Scholar] [CrossRef]
- Ning, L.; Huixin, H. Topic Evolution Analysis for Omics Data Integration in Cancers. Front. Cell Dev. Biol. 2021, 9, 338. [Google Scholar] [CrossRef] [PubMed]
- Goodacre, R.; Vaidyanathan, S.; Dunn, W.B.; Harrigan, G.G.; Kell, D.B. Metabolomics by Numbers: Acquiring and Understanding Global Metabolite Data. Trends Biotechnol. 2004, 22, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Madama, D.; Martins, R.; Pires, A.S.; Botelho, M.F.; Alves, M.G.; Abrantes, A.M.; Cordeiro, C.R. Metabolomic Profiling in Lung Cancer: A Systematic Review. Metabolites 2021, 11, 630. [Google Scholar] [CrossRef]
- Wang, Q.; Gao, P.; Wang, X.; Duan, Y. The Early Diagnosis and Monitoring of Squamous Cell Carcinoma via Saliva Metabolomics. Sci. Rep. 2014, 4, 6802. [Google Scholar] [CrossRef]
- Chen, X.; Yu, D. Metabolomics Study of Oral Cancers. Metabolomics 2019, 15, 22. [Google Scholar] [CrossRef]
- Panneerselvam, K.; Ishikawa, S.; Krishnan, R.; Sugimoto, M. Salivary Metabolomics for Oral Cancer Detection: A Narrative Review. Metabolites 2022, 12, 436. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Song, X.; Yang, X.; Han, W.; Fu, Y.; Wang, S.; Zhang, X.; Sun, G.; Lu, Y.; Wang, Z.; et al. Big Cohort Metabolomic Profiling of Serum for Oral Squamous Cell Carcinoma Screening and Diagnosis. Nat. Sci. 2022, 2, e20210071. [Google Scholar] [CrossRef]
- Lohavanichbutr, P.; Zhang, Y.; Wang, P.; Gu, H.; Gowda, G.A.N.; Djukovic, D.; Buas, M.F.; Raftery, D.; Chen, C. Salivary Metabolite Profiling Distinguishes Patients with Oral Cavity Squamous Cell Carcinoma from Normal Controls. PLoS ONE 2018, 13, e0204249. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Yang, X.; Narayanan, R.; Shankar, V.; Ethiraj, S.; Wang, X. Oral Squamous Cell Carcinoma Diagnosed from Saliva Metabolic Profiling. Proc. Natl. Acad. Sci. USA 2020, 117, 16167–16173. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, L.; Li, N.; Jia, Q.; Wang, X.; Zuo, L.; Long, J.; Xue, P.; Sun, Z.; Zhao, H. Metabolomics Based Plasma Biomarkers for Diagnosis of Oral Squamous Cell Carcinoma and Oral Erosive Lichen Planus. J. Cancer 2022, 13, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n91. [Google Scholar] [CrossRef]
- Amir-Behghadami, M.; Janati, A. Population, Intervention, Comparison, Outcomes and Study (PICOS) Design as a Framework to Formulate Eligibility Criteria in Systematic Reviews. Emerg. Med. J. 2020, 37, 387. [Google Scholar] [CrossRef] [PubMed]
- Mendeley Reference Manager. Mendeley. Available online: https://www.mendeley.com/reference-management/reference-manager (accessed on 20 February 2023).
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan-a Web and Mobile App for Systematic Reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; De Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the Gap between Raw Spectra and Functional Insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.C.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 Update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef]
- Fahy, E.; Sud, M.; Cotter, D.; Subramaniam, S. LIPID MAPS Online Tools for Lipid Research. Nucleic Acids Res. 2007, 35, W606–W612. [Google Scholar] [CrossRef] [PubMed]
- Study Quality Assessment Tools. NHLBI, NIH. Available online: https://www.nhlbi.nih.gov/health-topics/study-quality-assessment-tools (accessed on 3 August 2022).
- Lumbreras, B.; Porta, M.; Márquez, S.; Pollán, M.; Parker, L.A.; Hernández-Aguado, I. QUADOMICS: An Adaptation of the Quality Assessment of Diagnostic Accuracy Assessment (QUADAS) for the Evaluation of the Methodological Quality of Studies on the Diagnostic Accuracy of ‘-Omics’-Based Technologies. Clin. Biochem. 2008, 41, 1316–1325. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, S.; Sugimoto, M.; Kitabatake, K.; Sugano, A.; Nakamura, M.; Kaneko, M. Identification of Salivary Metabolomic Biomarkers for Oral Cancer Screening. Sci. Rep. 2016, 6, 31520. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, S.; Sugimoto, M.; Kitabatake, K.; Tu, M.; Sugano, A.; Yamamori, I. Effect of Timing of Collection of Salivary Metabolomic Biomarkers on Oral Cancer Detection. Amino Acids 2017, 49, 761–770. [Google Scholar] [CrossRef]
- de Sá Alves, M.; de Sá Rodrigues, N.; Bandeira, C.M.; Chagas JF, S.; Pascoal MB, N.; Nepomuceno, G.L.J.T.; da Silva Martinho, H.; Alves, M.G.O.; Mendes, M.A.; Dias, M.; et al. Identification of Possible Salivary Metabolic Biomarkers and Altered Metabolic Pathways in South American Patients Diagnosed with Oral Squamous Cell Carcinoma. Metabolites 2021, 11, 650. [Google Scholar] [CrossRef]
- Enomoto, Y.; Kimoto, A.; Suzuki, H.; Nishiumi, S.; Yoshida, M.; Komori, T. Exploring a Novel Screening Method for Patients with Oral Squamous Cell Carcinoma: A Plasma Metabolomics Analysis. Kobe J. Med. Sci. 2018, 64, E26. [Google Scholar]
- Ishikawa, S.; Wong, D.T.W.; Sugimoto, M.; Gleber-Netto, F.O.; Li, F.; Tu, M.; Zhang, Y.; Akin, D.; Iino, M. Identification of Salivary Metabolites for Oral Squamous Cell Carcinoma and Oral Epithelial Dysplasia Screening from Persistent Suspicious Oral Mucosal Lesions. Clin. Oral Investig. 2019, 23, 3557–3563. [Google Scholar] [CrossRef]
- Ishikawa, S.; Sugimoto, M.; Edamatsu, K.; Sugano, A.; Kitabatake, K.; Iino, M. Discrimination of Oral Squamous Cell Carcinoma from Oral Lichen Planus by Salivary Metabolomics. Oral Dis. 2020, 26, 35–42. [Google Scholar] [CrossRef]
- Sridharan, G.; Ramani, P.; Patankar, S.; Vijayaraghavan, R. Evaluation of Salivary Metabolomics in Oral Leukoplakia and Oral Squamous Cell Carcinoma. J. Oral Pathol. Med. 2019, 48, 299–306. [Google Scholar] [CrossRef]
- Musharraf, S.G.; Shahid, N.; Naqvi, S.M.A.; Saleem, M.; Siddiqui, A.J.; Ali, A. Metabolite Profiling of Preneoplastic and Neoplastic Lesions of Oral Cavity Tissue Samples Revealed a Biomarker Pattern. Sci. Rep. 2016, 6, 38985. [Google Scholar] [CrossRef]
- Tsai, C.K.; Lin, C.Y.; Kang, C.J.; Liao, C.T.; Wang, W.L.; Chiang, M.H. Nuclear Magnetic Resonance Metabolomics Biomarkers for Identifying High Risk Patients with Extranodal Extension in Oral Squamous Cell Carcinoma. J. Clin. Med. 2020, 9, 951. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.-H.; Jing, Y.; Wang, S.; Ding, F.; Zhang, X.-X.; Chen, S.; Zhang, L.; Hu, Q.-G.; Ni, Y.-H. Integrated Non-Targeted and Targeted Metabolomics Uncovers Amino Acid Markers of Oral Squamous Cell Carcinoma. Front. Oncol. 2020, 10, 426. [Google Scholar] [CrossRef]
- Lee, C.H.; Ko, A.M.S.; Warnakulasuriya, S.; Yin, B.L.; Sunarjo; Zain, R.B.; Ibrahim, S.O.; Liu, Z.W.; Li, W.H.; Zhang, S.S.; et al. Intercountry Prevalences and Practices of Betel-Quid Use in South, Southeast and Eastern Asia Regions and Associated Oral Preneoplastic Disorders: An International Collaborative Study by Asian Betel-Quid Consortium of South and East Asia. Int. J. Cancer 2011, 129, 1741–1751. [Google Scholar] [CrossRef] [PubMed]
- Spielmann, N.; Wong, D. Saliva: Diagnostics and Therapeutic Perspectives. Oral Dis. 2011, 17, 345–354. [Google Scholar] [CrossRef]
- Sugimoto, M. Salivary Metabolomics for Cancer Detection. Expert Rev. Proteom. 2020, 17, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Rubio, A.; Garland, G.D.; Sfakianos, A.; Harvey, R.F.; Willis, A.E. Aberrant Protein Synthesis and Cancer Development: The Role of Canonical Eukaryotic Initiation, Elongation and Termination Factors in Tumorigenesis. Semin. Cancer Biol. 2022, 86, 151–165. [Google Scholar] [CrossRef]
- White-Gilbertson, S.; Kurtz, D.T.; Voelkel-Johnson, C. The Role of Protein Synthesis in Cell Cycling and Cancer. Mol. Oncol. 2009, 3, 402. [Google Scholar] [CrossRef]
- Le, A.; Lane, A.N.; Hamaker, M.; Bose, S.; Gouw, A.; Barbi, J.; Tsukamoto, T.; Rojas, C.J.; Slusher, B.S.; Zhang, H.; et al. Glucose-Independent Glutamine Metabolism via TCA Cycling for Proliferation and Survival in B-Cells. Cell Metab. 2012, 15, 110. [Google Scholar] [CrossRef]
- Fendt, S.M.; Bell, E.L.; Keibler, M.A.; Olenchock, B.A.; Mayers, J.R.; Wasylenko, T.M.; Vokes, N.I.; Guarente, L.; Heiden, M.G.V.; Stephanopoulos, G. Reductive Glutamine Metabolism Is a Function of the α-Ketoglutarate to Citrate Ratio in Cells. Nat. Commun. 2013, 4, 2236. [Google Scholar] [CrossRef]
- Cetindis, M.; Biegner, T.; Munz, A.; Teriete, P.; Reinert, S.; Grimm, M. Glutaminolysis and Carcinogenesis of Oral Squamous Cell Carcinoma. Eur. Arch. Oto Rhino Laryngol. 2016, 273, 495–503. [Google Scholar] [CrossRef]
- Kamarajan, P.; Rajendiran, T.M.; Kinchen, J.; Bermúdez, M.; Danciu, T.; Kapila, Y.L. Head and Neck Squamous Cell Carcinoma Metabolism Draws on Glutaminolysis, and Stemness Is Specifically Regulated by Glutaminolysis via Aldehyde Dehydrogenase. J. Proteome Res. 2017, 16, 1315–1326. [Google Scholar] [CrossRef]
- Ogawa, T.; Washio, J.; Takahashi, T.; Echigo, S.; Takahashi, N. Glucose and Glutamine Metabolism in Oral Squamous Cell Carcinoma: Insight from a Quantitative Metabolomic Approach. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2014, 118, 218–225. [Google Scholar] [CrossRef]
- Wang, T.; Cai, B.; Ding, M.; Su, Z.; Liu, Y.; Shen, L. C-Myc Overexpression Promotes Oral Cancer Cell Proliferation and Migration by Enhancing Glutaminase and Glutamine Synthetase Activity. Am. J. Med. Sci. 2019, 358, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, R.M.; Stern, P.H.; Coalson, D.W.; Douglas Wallace, C.; Erbe, R.W. Altered Methionine Metabolism in Cancer Cells. Methods Mol. Biol. 2019, 1866, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, S.M.; Gao, X.; Dai, Z.; Locasale, J.W. Methionine Metabolism in Health and Cancer: A Nexus of Diet and Precision Medicine. Nat. Rev. Cancer 2019, 19, 625–637. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, A.; Nakanishi, M. Navigating the DNA Methylation Landscape of Cancer. Trends Genet. 2021, 37, 1012–1027. [Google Scholar] [CrossRef]
- Ma, J.; Zhong, M.; Xiong, Y.; Gao, Z.; Wu, Z.; Liu, Y.; Hong, X. Emerging Roles of Nucleotide Metabolism in Cancer Development: Progress and Prospect. Aging 2021, 13, 13349. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Yao, Y.; Scott, A.J.; Wilder-Romans, K.; Dresser, J.J.; Werner, C.K.; Sun, H.; Pratt, D.; Sajjakulnukit, P.; Zhao, S.G.; et al. Purine Metabolism Regulates DNA Repair and Therapy Resistance in Glioblastoma. Nat. Commun. 2020, 11, 3811. [Google Scholar] [CrossRef]
- Camici, M.; Garcia-Gil, M.; Pesi, R.; Allegrini, S.; Tozzi, M.G. Purine-Metabolising Enzymes and Apoptosis in Cancer. Cancers 2019, 11, 1354. [Google Scholar] [CrossRef]
- Potter, M.; Newport, E.; Morten, K.J. The Warburg Effect: 80 Years On. Biochem. Soc. Trans. 2016, 44, 1499. [Google Scholar] [CrossRef]
- Ananieva, E.A.; Wilkinson, A.C. Branched-Chain Amino Acid Metabolism in Cancer. Curr. Opin. Clin. Nutr. Metab. Care 2018, 21, 64. [Google Scholar] [CrossRef] [PubMed]
- Yue, M.; Jiang, J.; Gao, P.; Liu, H.; Qing, G. Oncogenic MYC Activates a Feedforward Regulatory Loop Promoting Essential Amino Acid Metabolism and Tumorigenesis. Cell Rep. 2017, 21, 3819–3832. [Google Scholar] [CrossRef]
- Zhou, Z.; Sun, B.; Nie, A.; Yu, D.; Bian, M. Roles of Aminoacyl-TRNA Synthetases in Cancer. Front. Cell Dev. Biol. 2020, 8, 599765. [Google Scholar] [CrossRef]
- Sangha, A.K.; Kantidakis, T. The Aminoacyl-TRNA Synthetase and TRNA Expression Levels Are Deregulated in Cancer and Correlate Independently with Patient Survival. Curr. Issues Mol. Biol. 2022, 44, 3001–3019. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Guo, R.; Li, Y.; Kang, G.; Wu, Y.; Cheng, J.; Jia, J.; Wang, W.; Li, Z.; Wang, A.; et al. Contribution of Upregulated Aminoacyl-tRNA Biosynthesis to Metabolic Dysregulation in Gastric Cancer. J. Gastroenterol. Hepatol. 2021, 36, 3113. [Google Scholar] [CrossRef]
- He, Y.; Gong, J.; Wang, Y.; Qin, Z.; Jiang, Y.; Ma, H.; Jin, G.; Chen, J.; Hu, Z.; Guan, X.; et al. Potentially Functional Polymorphisms in Aminoacyl-TRNA Synthetases Genes Are Associated with Breast Cancer Risk in a Chinese Population. Mol. Carcinog. 2015, 54, 577–583. [Google Scholar] [CrossRef]
- Wakasugi, K.; Schimmel, P. Highly Differentiated Motifs Responsible for Two Cytokine Activities of a Split Human TRNA Synthetase. J. Biol. Chem. 1999, 274, 23155–23159. [Google Scholar] [CrossRef] [PubMed]
- Albaugh, V.L.; Pinzon-Guzman, C.; Barbul, A. Arginine Metabolism and Cancer. Cancers 2021, 13, 3541. [Google Scholar] [CrossRef]
- Wheatley, D.N.; Campbell, E. Arginine Catabolism, Liver E Xtracts and Cancer. Pathol. Oncol. Res. 2002, 8, 18–25. [Google Scholar] [CrossRef]
- Selvi, I.; Basar, H.; Baydilli, N.; Murat, K.; Kaymaz, O. The Importance of Plasma Arginine Level and Its Downstream Metabolites in Diagnosing Prostate Cancer. Int. Urol. Nephrol. 2019, 51, 1975–1983. [Google Scholar] [CrossRef]
- Hu, L.; Gao, Y.; Cao, Y.; Zhang, Y.; Xu, M.; Wang, Y.; Jing, Y.; Guo, S.; Jing, F.; Hu, X.; et al. Identification of Arginine and Its “Downstream” Molecules as Potential Markers of Breast Cancer. IUBMB Life 2016, 68, 817–822. [Google Scholar] [CrossRef] [PubMed]
- Manna, S.K.; Tanaka, N.; Krausz, K.W.; Haznadar, M.; Xue, X.; Matsubara, T.; Bowman, E.D.; Fearon, E.R.; Harris, C.C.; Shah, Y.M.; et al. Biomarkers of Coordinate Metabolic Reprogramming in Colorectal Tumors in Mice and Humans. Gastroenterology 2014, 146, 1313–1324. [Google Scholar] [CrossRef]
- Townsend, D.E.; Kaenjak, A.; Jayaswal, R.K.; Wilkinson, B.J. Proline Is Biosynthesized from Arginine in Staphylococcus Aureus. Microbiology 1996, 142, 1491–1497. [Google Scholar] [CrossRef][Green Version]
- Patil, M.D.; Bhaumik, J.; Babykutty, S.; Banerjee, U.C.; Fukumura, D. Arginine Dependence of Tumor Cells: Targeting a Chink in Cancer’s Armor. Oncogene 2016, 35, 4957. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Liu, X.; Cheng, C.; Yu, W.; Yi, P. Metabolism of Amino Acids in Cancer. Front. Cell Dev. Biol. 2021, 8, 1628. [Google Scholar] [CrossRef] [PubMed]
- Kamphorst, J.J.; Nofal, M.; Commisso, C.; Hackett, S.R.; Lu, W.; Grabocka, E.; Vander Heiden, M.G.; Miller, G.; Drebin, J.A.; Bar-Sagi, D.; et al. Human Pancreatic Cancer Tumors Are Nutrient Poor and Tumor Cells Actively Scavenge Extracellular Protein. Cancer Res. 2015, 75, 544–553. [Google Scholar] [CrossRef]
- Mayers, J.R.; Torrence, M.E.; Danai, L.V.; Papagiannakopoulos, T.; Davidson, S.M.; Bauer, M.R.; Lau, A.N.; Ji, B.W.; Dixit, P.D.; Hosios, A.M.; et al. Tissue-of-Origin Dictates Branched-Chain Amino Acid Metabolism in Mutant Kras-Driven Cancers. Science 2016, 353, 1161. [Google Scholar] [CrossRef]
- Maddocks, O.D.K.; Labuschagne, C.F.; Adams, P.D.; Vousden, K.H. Serine Metabolism Supports the Methionine Cycle and DNA/RNA Methylation through De Novo ATP Synthesis in Cancer Cells. Mol. Cell 2016, 61, 210–221. [Google Scholar] [CrossRef]
- Jain, M.; Nilsson, R.; Sharma, S.; Madhusudhan, N.; Kitami, T.; Souza, A.L.; Kafri, R.; Kirschner, M.W.; Clish, C.B.; Mootha, V.K. Metabolite Profiling Identifies a Key Role for Glycine in Rapid Cancer Cell Proliferation. Science 2012, 336, 1040–1044. [Google Scholar] [CrossRef]
- Maddocks, O.D.K.; Berkers, C.R.; Mason, S.M.; Zheng, L.; Blyth, K.; Gottlieb, E.; Vousden, K.H. Serine Starvation Induces Stress and P53-Dependent Metabolic Remodelling in Cancer Cells. Nature 2013, 493, 542–546. [Google Scholar] [CrossRef]
- Chen, M.; Meng, L. The Double Faced Role of Xanthine Oxidoreductase in Cancer. Acta Pharmacol. Sin. 2021, 43, 1623–1632. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, L.; Sandhu, J.K.; Harper, M.E.; Cuperlovic-culf, M. Role of Glutathione in Cancer: From Mechanisms to Therapies. Biomolecules 2020, 10, 1429. [Google Scholar] [CrossRef] [PubMed]
- Carretero, J.; Obrador, E.; Anasagasti, M.J.; Martin, J.J.; Vidal-Vanaclocha, F.; Estrela, J.M. Growth-Associated Changes in Glutathione Content Correlate with Liver Metastatic Activity of B16 Melanoma Cells. Clin. Exp. Metastasis 1999, 17, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.Z.; Chen, C.; Zeng, Z.; Yang, H.; Oh, J.; Chen, L.; Lu, S.C. Mechanism and Significance of Increased Glutathione Level in Human Hepatocellular Carcinoma and Liver Regeneration. FASEB J. 2001, 15, 19–21. [Google Scholar] [CrossRef] [PubMed]
- Bansal, A.; Celeste Simon, M. Glutathione Metabolism in Cancer Progression and Treatment Resistance. J. Cell Biol. 2018, 217, 2291. [Google Scholar] [CrossRef]
| Author/Year | Country | Cases/Controls | Age (Range) | Sex (Male/Female) | Cancer Staging (I/II/III/IV) | Inclusion/Exclusion Criteria for Cases | Sample Type | Sample Storage | Sample Preparation (Yes/No) | Analytical Methods |
|---|---|---|---|---|---|---|---|---|---|---|
| De Sa Alves., (2021) [37] | Brazil | 27 OSCC/41 Healthy controls | OSCC: 57 ± 13.87 Healthy Controls: 57.34 ± 11.66 [mean ± SD] (28–88) | OSCC:19/8 Healthy controls: 21/20 | 4/4/6/13 | IC: Patients aged 18 & above with confirmed diagnosis of OSCC. EC: Patients diagnosed with other cancers/have undergone prior treatment with surgery, chemo/radio therapy | Saliva | −80 °C | Yes | GC-MS |
| Enomoto et al., (2018) [38] | Japan | 48 OSCC/29 other oral diseases | OSCC: 66.3 Other oral diseases: 60.3 [mean] | OSCC: 25/23 Other oral diseases: 15/14 | 9/10/11/18 | IC: Confirmed OSCC EC: History of malignant tumour, metabolic disease, or endocrine disease | Serum | −80 °C | Yes | GC-MS |
| Ishikawa et al., (2016) [35] | Japan | 24 OC/44 Healthy controls | OC: 72 (23–94) Healthy controls: 68 (21–90) [median] | OC: 14/10 Healthy controls: 16/28 | 5/6/8/5 | Not reported | Saliva, Tissue | −80 °C | Yes | CE-TOFMS |
| Ishikawa et al., (2017) [36] | Japan | 22 OSCC/44 Healthy controls | OSCC: 72 (23–94) Healthy controls: 68 (21–90) [median] | OSCC: 12/10 Healthy controls: 16/28 | 3/6/8/5 | IC: None of the OSCC patients received prior chemo/radio treatment. EC: History of malignancies or autoimmune disorders | Saliva | −80 °C | Yes | CE-TOFMS |
| Ishikawa et al., (2018) [39] | Japan | 6 OSCC, 10 OED/32 PSOML | OSCC: 63.5 (49–83) OED: 69 (57–81) PSOML: 62.5 (21–86) [median] | OSCC: 6/0 OED: 6/4 PSOML: 21/11 | NR | IC: Pathologically confirmed OSCC, OED and OELP. EC: Prior chemo/radio therapy | Saliva | −80 °C | Yes | CE-TOFMS |
| Ishikawa et al., (2019) [40] | Japan | 34 OSCC/26 OLP | OSCC: 70.5 (29–87) OLP: 67.5 (34–98) [median] | OSCC: 20/14 OLP: 5/21 | 14/9/2/9 | IC: Pathologically confirmed OSCC, OLP. EC: Prior chemo/radio therapy | Saliva | −80 °C | Yes | CE-TOFMS |
| Li et al., (2022) [23] | China | 72 OSCC,75 OELP/47 Healthy controls | OSCC: 66 ± 12 OELP: 61 ± 7 Healthy controls: 65 ± 9 [mean ± SD] | OSCC: 35/37 OELP: 38/37 Healthy controls: 23/24 | 17/21/19/14 Unknown: 1 | IC: Pathologically confirmed OSCC, OELP confirmed as per WHO diagnostic criteria of lichen planus. EC: No released/refractory OSCC/OELP, free from chronic systemic diseases | Serum | −80 °C | Yes | UHPLC-Q-Orbitrap |
| Song et al., (2019) [22] | China | 125 OSCC, 124 PML/124 Healthy controls | OSCC: 35–65 PML: 35–65 Healthy controls: 30–60 | OSCC: 65/60 PML: 64/60 Healthy controls: 64/60 | 29/40/23/33 | IC: Histologically confirmed OSCC, PML. EC: Prior chemo/radio therapy | Saliva | −80 °C | Yes | CPSI-MS coupled with ML |
| Sridharan et al., (2019) [41] | India | 22 OSCC, 21 OLK/18 Healthy controls | OSCC: 43 OLK: 48 Healthy controls: 32 [median] | OSCC: 81.9%/18.1% OLK: 90.5%/9.5% Healthy controls: 66.7%/33.3% | NR | IC: OSCC: clinical and histopathological confirmed OSCC; OLK: clinically diagnosed OLK. EC: history of systemic illness and medications; history of therapy for OLK and OSCC and with recurrent oral lesions. | Saliva | −80 °C | Yes | UPLC-QTOFMS |
| Syed et al., (2016) [42] | Pakistan | 21 OSCC, 15 OSF/15 Healthy controls | NR | NR | NR | IC: Clinically confirmed OSCC and OSF. EC: Prior therapy and in either remission or relapse stage. | Tissue | −80 °C | Yes | GC-MS |
| Tsai et al., (2020) [43] | Taiwan | 110 OSCC (37 normal tissue, 36 tumour tissue, 44 plasma, 98 urine) | 52.4 (28–79) [median] | NR | NR | IC: Oral cavity cancers EC: Any other tumours including oropharyngeal cancers | Tumour tissue, plasma & urine | −80 °C | Yes | NMR |
| Yang et al., (2020) [44] | China | 8 OSCC/8 Healthy controls | NR | NR | NR | Not Reported | Tumour tissue | −80 °C | Yes | GC-MS |
| Yang et al., (2021) [20] | China | 578 OSCC/241 Healthy controls | NR | NR | NR | IC: Histopathological confirmed OSCC. EC: Not reported. | Serum | −80 °C | Yes | CPSI-MS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alapati, S.; Fortuna, G.; Ramage, G.; Delaney, C. Evaluation of Metabolomics as Diagnostic Targets in Oral Squamous Cell Carcinoma: A Systematic Review. Metabolites 2023, 13, 890. https://doi.org/10.3390/metabo13080890
Alapati S, Fortuna G, Ramage G, Delaney C. Evaluation of Metabolomics as Diagnostic Targets in Oral Squamous Cell Carcinoma: A Systematic Review. Metabolites. 2023; 13(8):890. https://doi.org/10.3390/metabo13080890
Chicago/Turabian StyleAlapati, Susanth, Giulio Fortuna, Gordon Ramage, and Christopher Delaney. 2023. "Evaluation of Metabolomics as Diagnostic Targets in Oral Squamous Cell Carcinoma: A Systematic Review" Metabolites 13, no. 8: 890. https://doi.org/10.3390/metabo13080890
APA StyleAlapati, S., Fortuna, G., Ramage, G., & Delaney, C. (2023). Evaluation of Metabolomics as Diagnostic Targets in Oral Squamous Cell Carcinoma: A Systematic Review. Metabolites, 13(8), 890. https://doi.org/10.3390/metabo13080890






