Anti-Alzheimer Potential of a New (+)-Pinitol Glycoside Isolated from Tamarindus indica Pulp: In Vivo and In Silico Evaluations
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Chemicals and Reagents
2.3. Spectral Analyses
2.4. Extraction and Fractionation of Tamarindus Indica Pulp
2.5. Isolation and Purification of Compounds
2.6. Acid Hydrolysis and Sugar Analysis
2.7. Animals and Ethics
2.8. Experimental Design
2.9. T-Maze Test
2.10. Biochemical Analysis
2.11. ELISA Assays
2.12. In Silico Investigation
2.12.1. Prediction of the Potential Targets
2.12.2. Possible Targets of Alzheimer’s Disease
2.12.3. Molecular Docking MD Simulation and Network Construction
2.13. Statistical Analysis
3. Results
3.1. Phytochemical Investigation
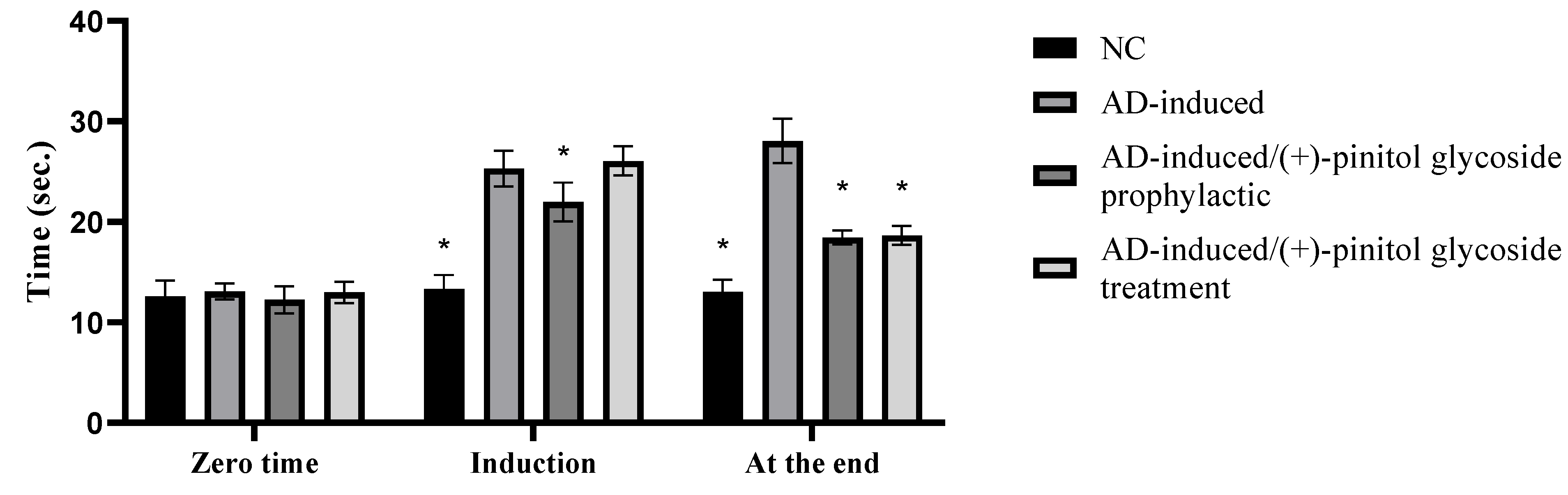
3.2. Behavioral Assessment Using the T-Maze Test
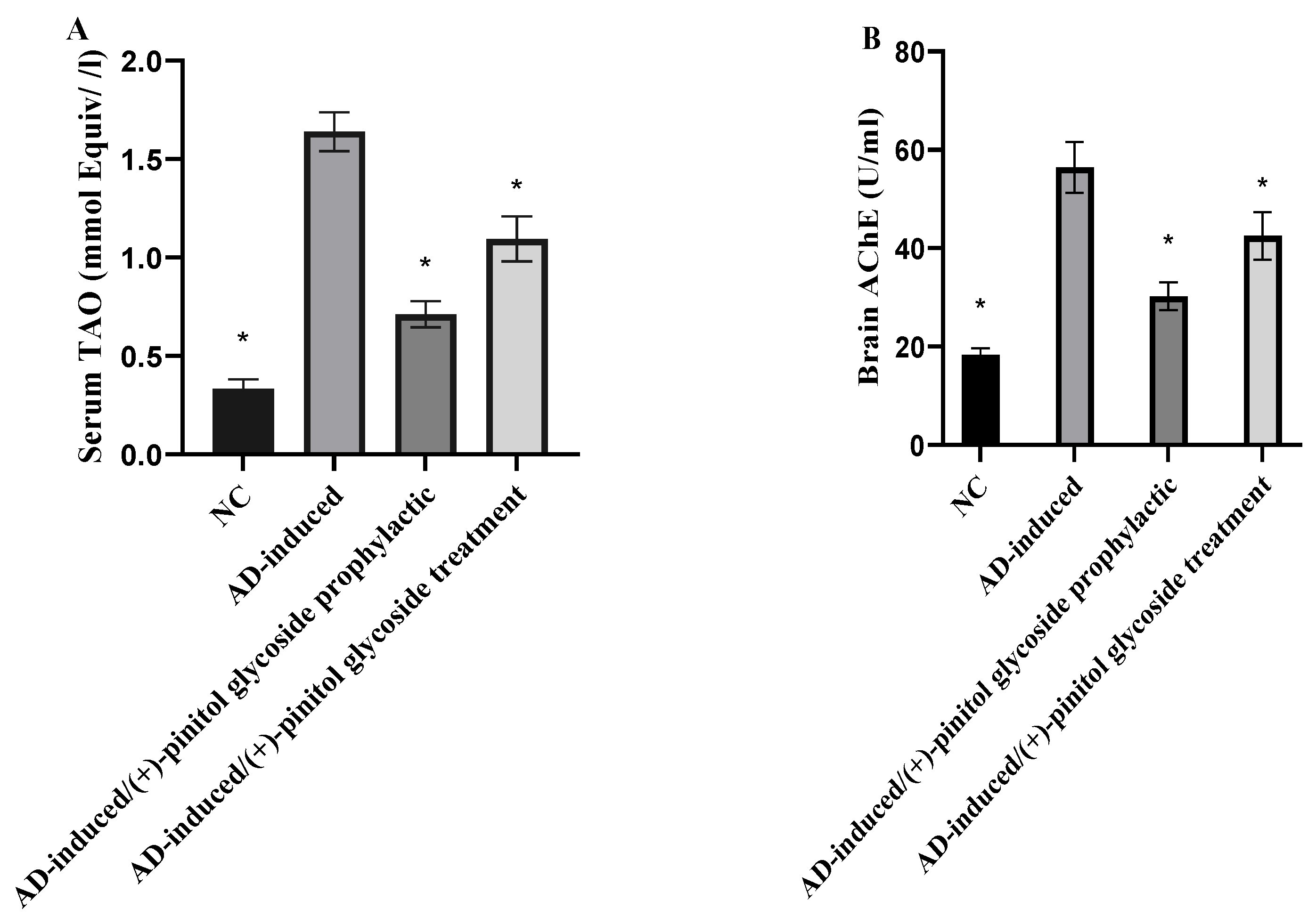
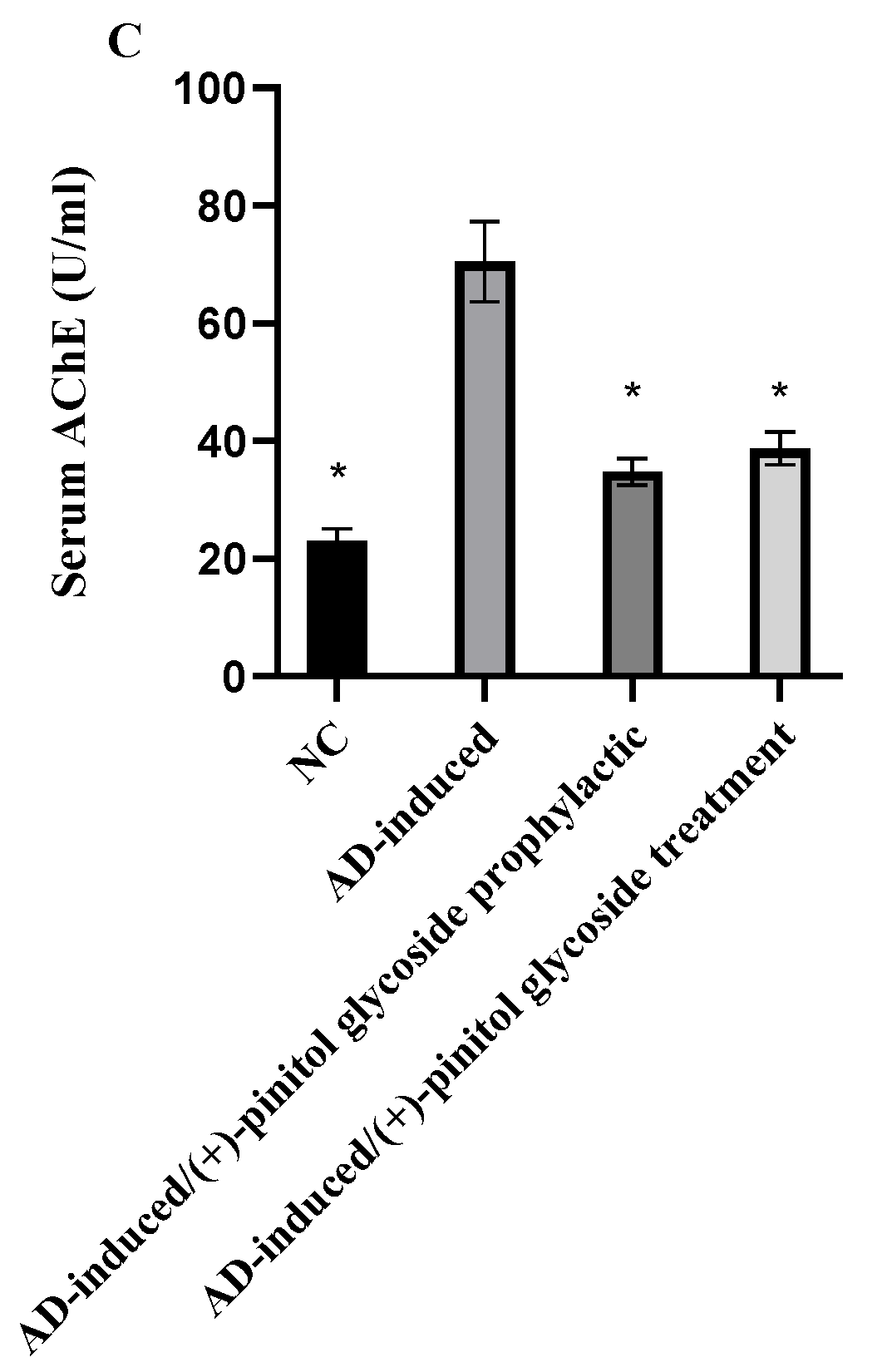
3.3. Biochemical Analysis
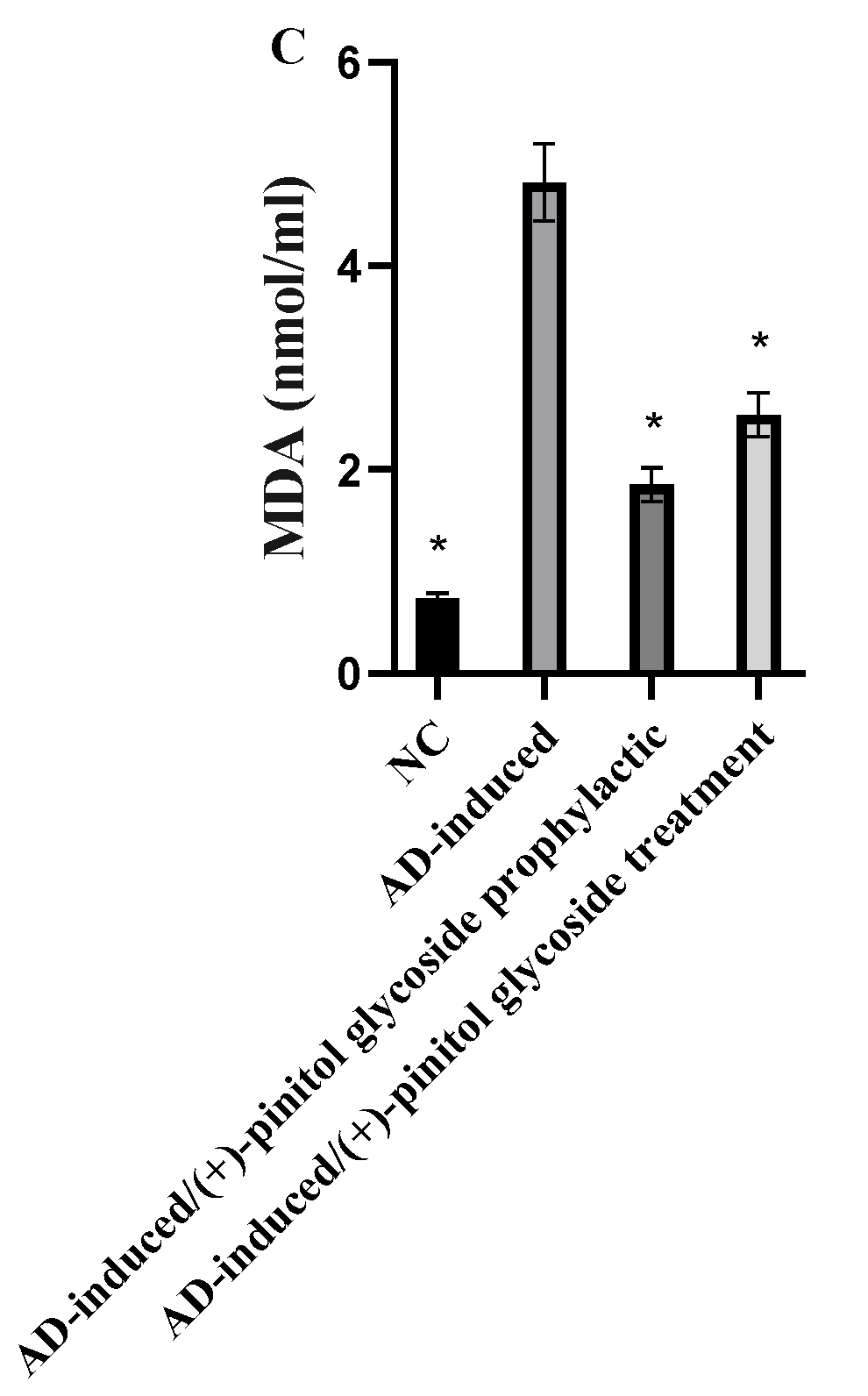
3.4. Evaluation of Oxidative Markers
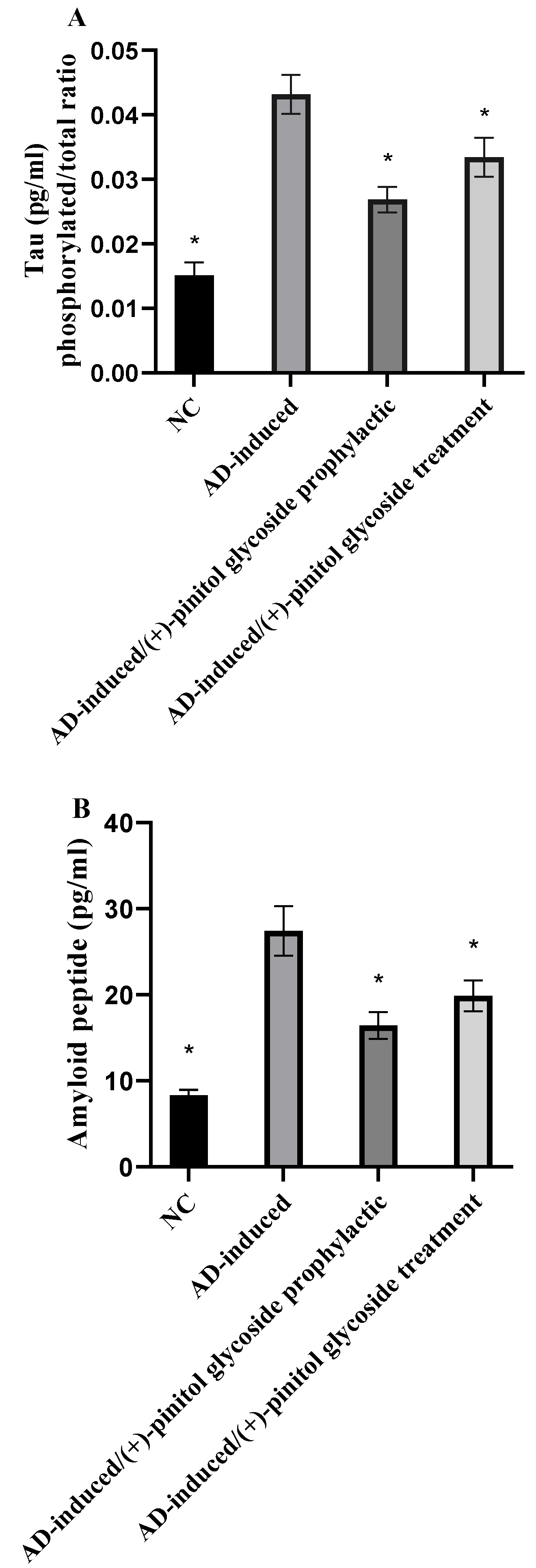
3.5. Evaluation of Tau and Amyloid Peptide
3.6. In Silico-Based Study
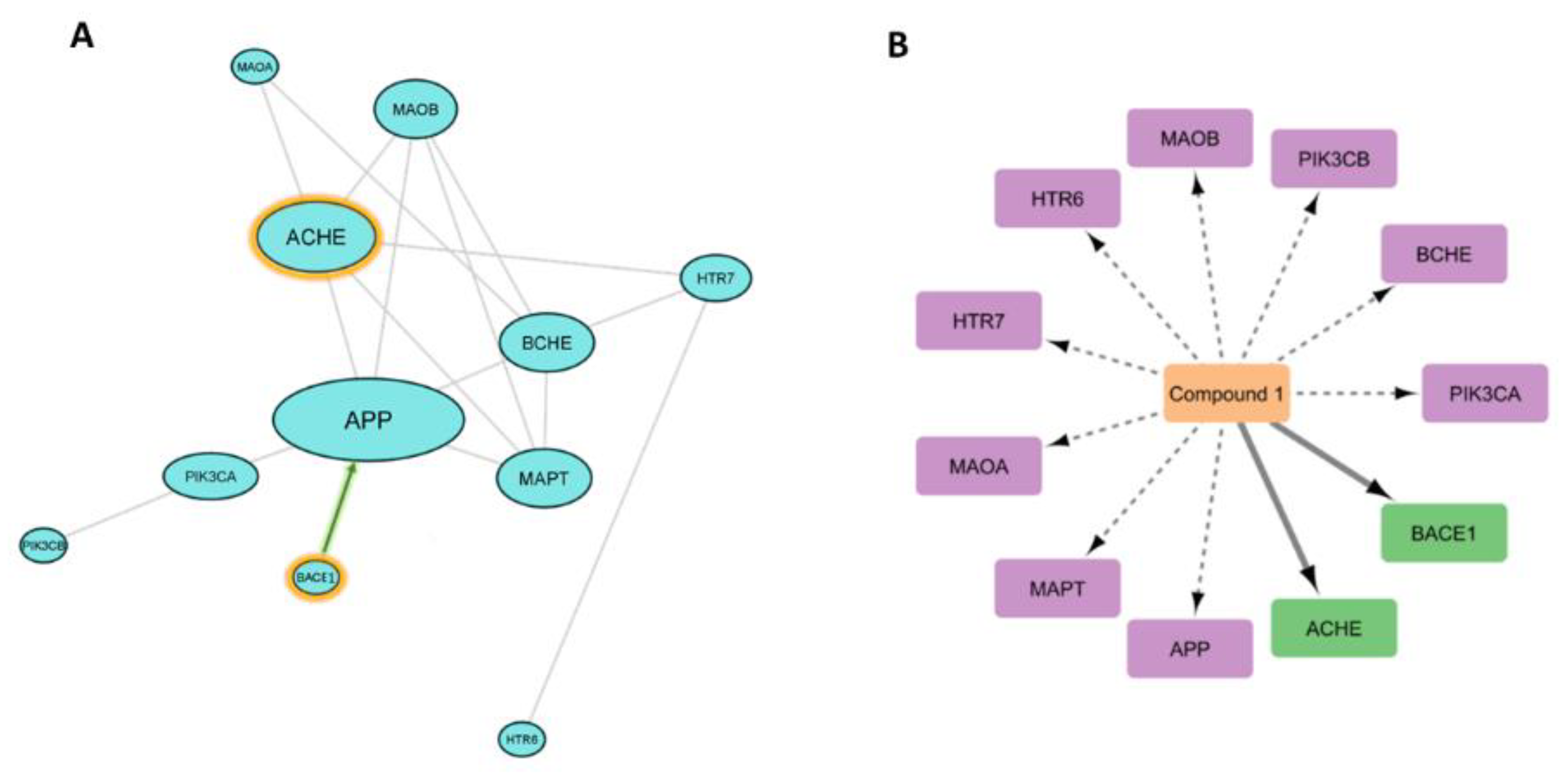
3.6.1. PPI Network of the Predicted Targets and KEGG-Based Enrichment Analysis
3.6.2. Prediction of AD Target Proteins
3.6.3. Analysis of Possible Molecular Mechanisms
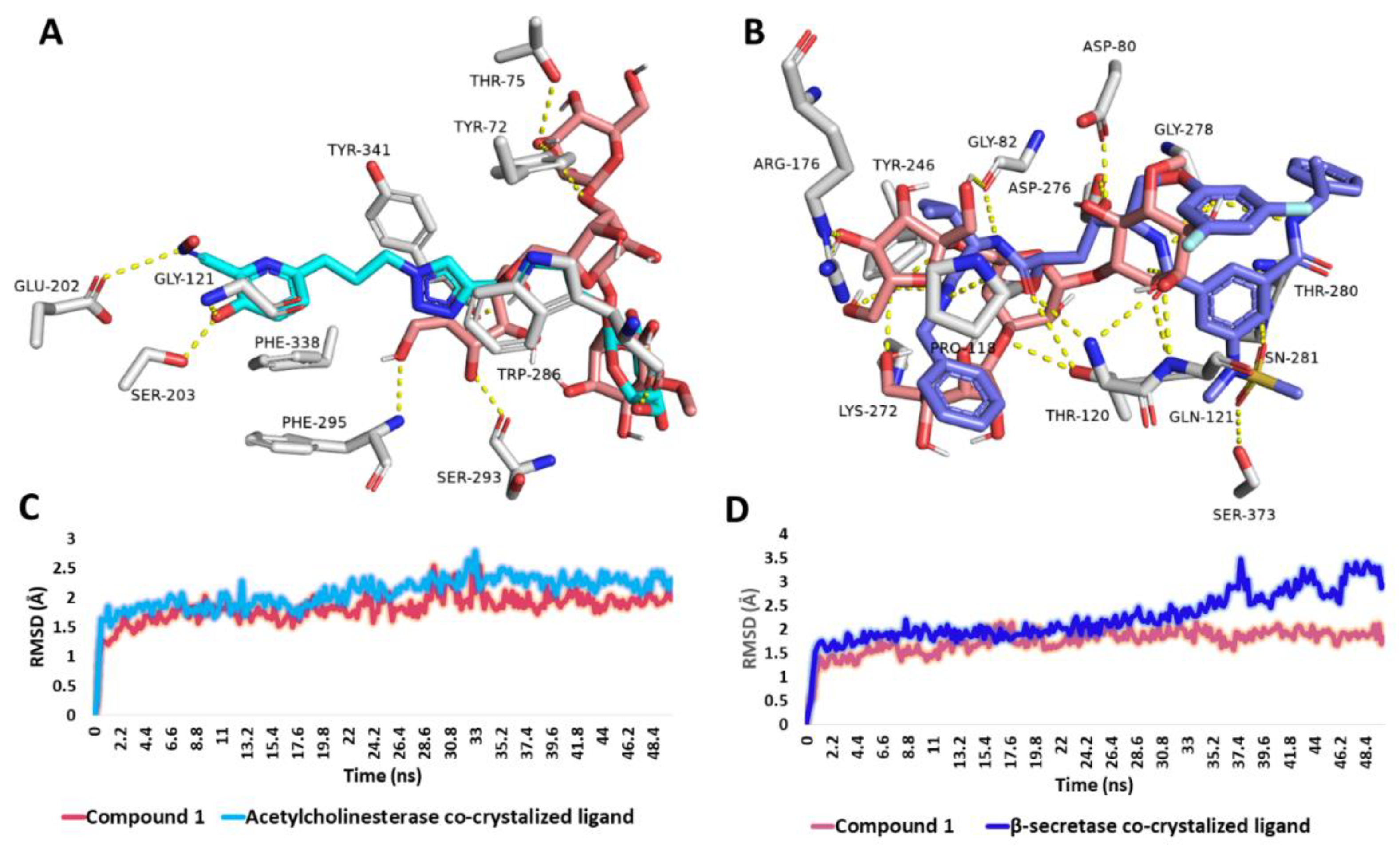
3.6.4. Binding Mode Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Babri, S.; Mohaddes, G.; Feizi, I.; Mohammadnia, A.; Niapour, A.; Alihemmati, A.; Amani, M. Effect of troxerutin on synaptic plasticity of hippocampal dentate gyrus neurons in a β-amyloid model of Alzheimer’s disease: An electrophysiological study. Eur. J. Pharmacol. 2014, 732, 19–25. [Google Scholar] [CrossRef]
- Zheng, T.; Wang, Q.; Bian, F.; Zhao, Y.; Ma, W.; Zhang, Y.; Lu, W.; Lei, P.; Zhang, L.; Hao, X. Salidroside alleviates diabetic neuropathic pain through regulation of the AMPK-NLRP3 inflammasome axis. Toxicol. Appl. Pharmacol. 2021, 416, 115468. [Google Scholar] [CrossRef]
- Raschetti, R.; Albanese, E.; Vanacore, N.; Maggini, M. Cholinesterase inhibitors in mild cognitive impairment: A systematic review of randomised trials. PLoS Med. 2007, 4, e338. [Google Scholar] [CrossRef]
- Kimura, N. Diabetes mellitus induces Alzheimer’s disease pathology: Histopathological evidence from animal models. Int. J. Mol. Sci. 2016, 17, 503. [Google Scholar] [CrossRef] [PubMed]
- Cole, G.M.; Lim, G.P.; Yang, F.; Teter, B.; Begum, A.; Ma, Q.; Harris-White, M.E.; Frautschy, S.A. Prevention of Alzheimer’s disease: Omega-3 fatty acid and phenolic anti-oxidant interventions. Neurobiol. Aging 2005, 26, 133–136. [Google Scholar] [CrossRef] [PubMed]
- Boligon, A.A.; Pereira, R.P.; Feltrin, A.C.; Machado, M.M.; Janovik, V.; Rocha, J.B.T.; Athayde, M.L. Antioxidant activities of flavonol derivatives from the leaves and stem bark of Scutia buxifolia Reiss. Bioresour. Technol. 2009, 100, 6592–6598. [Google Scholar] [CrossRef]
- Elmaidomy, A.H.; Abdelmohsen, U.R.; Alsenani, F.; Aly, H.F.; Shams, S.G.E.; Younis, E.A.; Ahmed, K.A.; Sayed, A.M.; Owis, A.I.; Afifi, N. The anti-Alzheimer potential of Tamarindus indica: An in vivo investigation supported by in vitro and in silico approaches. RSC Adv. 2022, 12, 11769–11785. [Google Scholar] [CrossRef] [PubMed]
- Bui, T.T.; Nguyen, T.H. Natural product for the treatment of Alzheimer’s disease. J. Basic Clin. Physiol. Pharmacol. 2017, 28, 413–423. [Google Scholar] [CrossRef]
- Hoffmann-Ostenhof, O.; Pittner, F. The biosynthesis of myo-inositol and its isomers. Can. J. Chem. 1982, 60, 1863–1871. [Google Scholar] [CrossRef]
- Watkins, O.C.; Yong, H.E.; Sharma, N.; Chan, S.-Y. A review of the role of inositols in conditions of insulin dysregulation and in uncomplicated and pathological pregnancy. Crit. Rev. Food Sci. Nutr. 2022, 62, 1626–1673. [Google Scholar] [CrossRef] [PubMed]
- Kennington, A.S.; Hill, C.R.; Craig, J.; Bogardus, C.; Raz, I.; Ortmeyer, H.K.; Hansen, B.C.; Romero, G.; Larner, J. Low urinary chiro-inositol excretion in non-insulin-dependent diabetes mellitus. N. Engl. J. Med. 1990, 323, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Hallman, M.; Bry, K.; Hoppu, K.; Lappi, M.; Pohjavuori, M. Inositol supplementation in premature infants with respiratory distress syndrome. N. Engl. J. Med. 1992, 326, 1233–1239. [Google Scholar] [CrossRef]
- Asplin, I.; Galasko, G.; Larner, J. chiro-inositol deficiency and insulin resistance: A comparison of the chiro-inositol-and the myo-inositol-containing insulin mediators isolated from urine, hemodialysate, and muscle of control and type II diabetic subjects. Proc. Natl. Acad. Sci. USA 1993, 90, 5924–5928. [Google Scholar] [CrossRef]
- Nestler, J.E.; Jakubowicz, D.J.; Reamer, P.; Gunn, R.D.; Allan, G. Ovulatory and metabolic effects of D-chiro-inositol in the polycystic ovary syndrome. N. Engl. J. Med. 1999, 340, 1314–1320. [Google Scholar] [CrossRef] [PubMed]
- McLaurin, J.; Golomb, R.; Jurewicz, A.; Antel, J.P.; Fraser, P.E. Inositol stereoisomers stabilize an oligomeric aggregate of Alzheimer amyloid β peptide and inhibit Aβ-induced toxicity. J. Biol. Chem. 2000, 275, 18495–18502. [Google Scholar] [CrossRef] [PubMed]
- Mancini, M.; Andreassi, A.; Salvioni, M.; Pelliccione, F.; Mantellassi, G.; Banderali, G. Myoinositol and D-chiro inositol in improving insulin resistance in obese male children: Preliminary data. Int. J. Endocrinol. 2016, 2016, 8720342. [Google Scholar] [CrossRef]
- Showell, M.G.; Mackenzie-Proctor, R.; Jordan, V.; Hodgson, R.; Farquhar, C. Inositol for subfertile women with polycystic ovary syndrome. Cochrane Database Syst. Rev. 2018, 12, 256–266. [Google Scholar] [CrossRef]
- Chhetri, D.R. Myo-inositol and its derivatives: Their emerging role in the treatment of human diseases. Front. Pharmacol. 2019, 10, 1172. [Google Scholar] [CrossRef]
- Abdelrahman, G.H.; Mariod, A.A. Tamarindus indica: Phytochemical Constituents, Bioactive Compounds and Traditional and Medicinal Uses. In Wild Fruits: Composition, Nutritional Value and Products; Springer: Berlin/Heidelberg, Germany, 2019; pp. 229–238. [Google Scholar]
- Kuru, P. Tamarindus indica and its health related effects. Asian Pac. J. Trop. Biomed. 2014, 4, 676–681. [Google Scholar] [CrossRef]
- Jain, R.; Jain, S.; Sharma, A.; Ito, H.; Hatano, T. Isolation of (+)-pinitol and other constituents from the root bark of Tamarindus indica Linn. J. Nat. Med. 2007, 61, 355–356. [Google Scholar] [CrossRef]
- Azab, A. D-Pinitol—Active Natural Product from Carob with Notable Insulin Regulation. Nutrients 2022, 14, 1453. [Google Scholar] [CrossRef]
- Fenili, D.; Weng, Y.-Q.; Aubert, I.; Nitz, M.; McLaurin, J. Sodium/myo-Inositol transporters: Substrate transport requirements and regional brain expression in the TgCRND8 mouse model of amyloid pathology. PLoS ONE 2011, 6, e24032. [Google Scholar] [CrossRef]
- Pitt, J.; Thorner, M.; Brautigan, D.; Larner, J.; Klein, W.L. Protection against the synaptic targeting and toxicity of Alzheimer’s-associated Aβ oligomers by insulin mimetic chiro-inositols. FASEB J. 2013, 27, 199–207. [Google Scholar] [CrossRef]
- Ravindran, R.; Chakrapani, G.; Mitra, K.; Doble, M. Inhibitory activity of traditional plants against Mycobacterium smegmatis and their action on Filamenting temperature sensitive mutant Z (FtsZ)—A cell division protein. PLoS ONE 2020, 15, e0232482. [Google Scholar] [CrossRef] [PubMed]
- Sethi, G.; Ahn, K.S.; Sung, B.; Aggarwal, B.B. Pinitol targets nuclear factor-κB activation pathway leading to inhibition of gene products associated with proliferation, apoptosis, invasion, and angiogenesis. Mol. Cancer Ther. 2008, 7, 1604–1614. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.-C.; Bang, T.-H.; Kang, H.-M.; Park, B.-S.; Kim, I.-R. Anticancer effects of D-pinitol in human oral squamous carcinoma cells. Int. J. Oral Biol. 2020, 45, 152–161. [Google Scholar] [CrossRef]
- Lin, Y.; Wu, Y.; Su, J.; Wang, M.; Wu, X.; Su, Z.; Yi, X.; Wei, L.; Cai, J.; Sun, Z. Therapeutic role of D-pinitol on experimental colitis via activating Nrf2/ARE and PPAR-γ/NF-κB signaling pathways. Food Funct. 2021, 12, 2554–2568. [Google Scholar] [CrossRef]
- Narayanan, C.; Joshi, D.; Mujumdar, A.; Dhekne, V. Pinitol—A new anti-diabetic compound from the leaves of Bougainvillea spectabilis. Curr. Sci. 1987, 56, 139–141. [Google Scholar]
- Srivastava, K.; Dubey, A.; Tiwari, M.; Dubey, A. To evaluate the synergistic effect of Pinitol with Glimepride in diabetic Wistar rats. J. Crit. Rev. 2020, 7, 2058–2062. [Google Scholar]
- Koh, E.S.; Kim, S.; Kim, M.; Hong, Y.A.; Shin, S.J.; Park, C.W.; Chang, Y.S.; Chung, S.; Kim, H.S. D-Pinitol alleviates cyclosporine A-induced renal tubulointerstitial fibrosis via activating Sirt1 and Nrf2 antioxidant pathways. Int. J. Mol. Med. 2018, 41, 1826–1834. [Google Scholar] [CrossRef]
- Singh, R.; Pandey, B.; Tripathi, M.; Pandey, V. Anti-inflammatory effect of (+)-pinitol. Fitoterapia 2001, 72, 168–170. [Google Scholar] [CrossRef]
- Bolliger, H.R.; Brenner, M.; Gänshirt, H.; Mangold, H.K.; Seiler, H.; Stahl, E.; Waldi, D. Thin-Layer Chromatography; a Laboratory Handbook; Springer: Berlin/Heidelberg, Germany, 1965. [Google Scholar]
- Ahmed, W.M.; Ibrahim, M.A.; Helmy, N.A.; ElKashlan, A.M.; Elmaidomy, A.H.; Zaki, A.R. Amelioration of aluminum-induced hepatic and nephrotoxicity by Premna odorata extract is mediated by lowering MMP9 and TGF-β gene alterations in Wistar rat. Environ. Sci. Pollut. Res. 2022, 29, 72827–72838. [Google Scholar] [CrossRef] [PubMed]
- Elmaidomy, A.H.; Zahran, E.M.; Soltane, R.; Alasiri, A.; Saber, H.; Ngwa, C.J.; Pradel, G.; Alsenani, F.; Sayed, A.M.; Abdelmohsen, U.R. New Halogenated Compounds from Halimeda macroloba Seaweed with Potential Inhibitory Activity against Malaria. Molecules 2022, 27, 5617. [Google Scholar] [CrossRef]
- Al-Warhi, T.; Elmaidomy, A.H.; Maher, S.A.; Abu-Baih, D.H.; Selim, S.; Albqmi, M.; Al-Sanea, M.M.; Alnusaire, T.S.; Ghoneim, M.M.; Mostafa, E.M. The Wound-Healing Potential of Olea europaea L. Cv. Arbequina Leaves Extract: An Integrated In Vitro, In Silico, and In Vivo Investigation. Metabolites 2022, 12, 791. [Google Scholar] [CrossRef] [PubMed]
- Bagalagel, A.A.; El-Hawary, S.S.; Alaaeldin, R.; Elmaidomy, A.H.; Altemani, F.H.; Waggas, D.S.; Algehainy, N.A.; Saeedi, N.H.; Alsenani, F.; Mokhtar, F.A. The Protective and Therapeutic Anti-Alzheimer Potential of Olea europaea L. cv. Picual: An In Silico and In Vivo Study. Metabolites 2022, 12, 1178. [Google Scholar] [CrossRef] [PubMed]
- Zahran, E.M.; Abdel-Maqsoud, N.M.R.; Tammam, O.Y.; Abdel-Rahman, I.M.; Elrehany, M.A.; Bakhsh, H.T.; Altemani, F.H.; Algehainy, N.A.; Alzubaidi, M.A.; Abdelmohsen, U.R. Scabicidal Potential of Coconut Seed Extract in Rabbits via Downregulating Inflammatory/Immune Cross Talk: A Comprehensive Phytochemical/GC-MS and In Silico Proof. Antibiotics 2022, 12, 43. [Google Scholar] [CrossRef] [PubMed]
- Abbet, C.; Neuburger, M.; Wagner, T.; Quitschau, M.; Hamburger, M.; Potterat, O. Phyteumosides A and B: New saponins with unique triterpenoid aglycons from Phyteuma orbiculare L. Org. Lett. 2011, 13, 1354–1357. [Google Scholar] [CrossRef]
- Chai, X.Y.; Xu, Z.R.; Ren, H.Y.; Shi, H.M.; Lu, Y.N.; Li, F.F.; Tu, P.F. Itosides A–I, new phenolic glycosides from Itoa orientalis. Helv. Chim. Acta 2007, 90, 2176–2185. [Google Scholar] [CrossRef]
- Borai, I.H.; Ezz, M.K.; Rizk, M.Z.; Aly, H.F.; El-Sherbiny, M.; Matloub, A.A.; Fouad, G.I. Therapeutic impact of grape leaves polyphenols on certain biochemical and neurological markers in AlCl3-induced Alzheimer’s disease. Biomed. Pharm. 2017, 93, 837–851. [Google Scholar] [CrossRef]
- Wang, L.; Geng, C.; Jiang, L.; Gong, D.; Liu, D.; Yoshimura, H.; Zhong, L. The anti-atherosclerotic effect of olive leaf extract is related to suppressed inflammatory response in rabbits with experimental atherosclerosis. Eur. J. Nutr. 2008, 47, 235–243. [Google Scholar] [CrossRef]
- Deacon, R.M.; Rawlins, J.N.P. T-maze alternation in the rodent. Nat. Protoc. 2006, 1, 7–12. [Google Scholar] [CrossRef]
- Brown, T. ChemDraw. Sci. Teach. 2014, 81, 67. [Google Scholar]
- Wang, Y.; Bryant, S.H.; Cheng, T.; Wang, J.; Gindulyte, A.; Shoemaker, B.A.; Thiessen, P.A.; He, S.; Zhang, J. Pubchem bioassay: 2017 update. Nucleic Acids Res. 2017, 45, D955–D963. [Google Scholar] [CrossRef]
- Rebhan, M.; Chalifa-Caspi, V.; Prilusky, J.; Lancet, D. GeneCards: A novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics 1998, 14, 656–664. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, S.; Li, F.; Zhou, Y.; Zhang, Y.; Wang, Z.; Zhang, R.; Zhu, J.; Ren, Y.; Tan, Y. Therapeutic target database 2020: Enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020, 48, D1031–D1041. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef] [PubMed]
- Huey, R.; Morris, G.M.; Forli, S. Using AutoDock 4 and AutoDock vina with AutoDockTools: A tutorial. Scripps Res. Inst. Mol. Graph. Lab. 2012, 10550, 1000. [Google Scholar]
- Yuan, S.; Chan, H.S.; Hu, Z. Using PyMOL as a platform for computational drug design. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2017, 7, e1298. [Google Scholar] [CrossRef]
- Bowers, K.J.; Chow, E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D. Scalable algorithms for molecular dynamics simulations on commodity clusters. In Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, FL, USA, 11–17 November 2006; p. 84. [Google Scholar]
- Galande, A.K.; Rohane, S.H. Insilico Molecular docking analysis in Maestro Software. Indian J. 2021, 14, 1–4. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Gapsys, V.; Michielssens, S.; Peters, J.H.; de Groot, B.L.; Leonov, H. Calculation of binding free energies. Mol. Model. Proteins 2015, 173–209. [Google Scholar]
- Mering, C.v.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef]
- Saito, R.; Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.-L.; Lotia, S.; Pico, A.R.; Bader, G.D.; Ideker, T. A travel guide to Cytoscape plugins. Nat. Methods 2012, 9, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhang, L.-M. Chemical structural and chain conformational characterization of some bioactive polysaccharides isolated from natural sources. Carbohydr. Polym. 2009, 76, 349–361. [Google Scholar] [CrossRef]
- Bubb, W.A. NMR spectroscopy in the study of carbohydrates: Characterizing the structural complexity. Concepts Magn. Reson. Part A Educ. J. 2003, 19, 1–19. [Google Scholar] [CrossRef]
- Roslund, M.U.; Tähtinen, P.; Niemitz, M.; Sjöholm, R. Complete assignments of the 1H and 13C chemical shifts and JH, H coupling constants in NMR spectra of D-glucopyranose and all D-glucopyranosyl-D-glucopyranosides. Carbohydr. Res. 2008, 343, 101–112. [Google Scholar] [CrossRef]
- Watson, A.; Hackbusch, S.; Franz, A.H. NMR solution geometry of saccharides containing the 6-O-(α-D-glucopyranosyl)-α/β-D-glucopyranose (isomaltose) or 6-O-(α-D-galactopyranosyl)-α/β-D-glucopyranose (melibiose) core. Carbohydr. Res. 2019, 473, 18–35. [Google Scholar] [CrossRef]
- Elmaidomy, A.H.; Mohyeldin, M.M.; Ibrahim, M.M.; Hassan, H.M.; Amin, E.; Rateb, M.E.; Hetta, M.H.; El Sayed, K.A. Acylated iridoids and rhamnopyranoses from premna odorata (lamiaceae) as novel mesenchymal–epithelial transition factor receptor inhibitors for the control of breast cancer. Phytother. Res. 2017, 31, 1546–1556. [Google Scholar] [CrossRef] [PubMed]
- Seymour, F.R.; Knapp, R.D.; Zweig, J.E.; Bishop, S.H. 13C-nuclear magnetic resonance spectra of compounds containing β-D-fructofuranosyl groups or residues. Carbohydr. Res. 1979, 72, 57–69. [Google Scholar] [CrossRef]
- Safran, M.; Dalah, I.; Alexander, J.; Rosen, N.; Iny Stein, T.; Shmoish, M.; Nativ, N.; Bahir, I.; Doniger, T.; Krug, H. GeneCards Version 3: The human gene integrator. Database 2010, 2010, baq020. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Srivastava, P.; Seth, A.; Tripathi, P.N.; Banerjee, A.G.; Shrivastava, S.K. Comprehensive review of mechanisms of pathogenesis involved in Alzheimer’s disease and potential therapeutic strategies. Prog. Neurobiol. 2019, 174, 53–89. [Google Scholar] [CrossRef]
- Holsinger, R.D.; McLean, C.A.; Beyreuther, K.; Masters, C.L.; Evin, G. Increased expression of the amyloid precursor β-secretase in Alzheimer’s disease. Ann. Neurol. 2002, 51, 783–786. [Google Scholar] [CrossRef]
- Björklund, C.; Oscarson, S.; Benkestock, K.; Borkakoti, N.; Jansson, K.; Lindberg, J.; Vrang, L.; Hallberg, A.; Rosenquist, Å.; Samuelsson, B. Design and synthesis of potent and selective BACE-1 inhibitors. J. Med. Chem. 2010, 53, 1458–1464. [Google Scholar] [CrossRef]
- Bartolucci, C.; Perola, E.; Pilger, C.; Fels, G.; Lamba, D. Three-dimensional structure of a complex of galanthamine (Nivalin®) with acetylcholinesterase from Torpedo californica: Implications for the design of new anti-Alzheimer drugs. Proteins 2001, 42, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Underwood, M.A.; Gaerlan, S.; De Leoz, M.L.A.; Dimapasoc, L.; Kalanetra, K.M.; Lemay, D.G.; German, J.B.; Mills, D.A.; Lebrilla, C.B. Human milk oligosaccharides in premature infants: Absorption, excretion, and influence on the intestinal microbiota. Pediatr. Res. 2015, 78, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Pan, Q.; Zhou, J.; Weng, Y.; Chen, K.; Shi, L.; Zhu, G.; Chen, C.; Li, L.; Geng, M. Pharmacokinetics, distribution, and excretion of sodium oligomannate, a recently approved anti-Alzheimer’s disease drug in China. J. Pharm. Anal. 2022, 12, 145–155. [Google Scholar] [CrossRef]
- Griñán-Ferré, C.; Bellver-Sanchis, A.; Olivares-Martín, M.; Bañuelos-Hortigüela, O.; Pallàs, M. Synergistic neuroprotective effects of a natural product Mixture against AD hallmarks and cognitive decline in Caenorhabditis elegans and an SAMP8 mice model. Nutrients 2021, 13, 2411. [Google Scholar] [CrossRef]
- John, J.; Nampoothiri, M.; Kumar, N.; Mudgal, J.; Nampurath, G.K.; Chamallamudi, M.R. Sesamol, a lipid lowering agent, ameliorates aluminium chloride induced behavioral and biochemical alterations in rats. Pharmacogn. Mag. 2015, 11, 327. [Google Scholar]
- Singh, M.; Kaur, M.; Kukreja, H.; Chugh, R.; Silakari, O.; Singh, D. Acetylcholinesterase inhibitors as Alzheimer therapy: From nerve toxins to neuroprotection. Eur. J. Med. Chem. 2013, 70, 165–188. [Google Scholar] [CrossRef] [PubMed]
- Singla, N.; Dhawan, D. Regulatory role of zinc during aluminium-induced altered carbohydrate metabolism in rat brain. J. Neurosci. Res. 2012, 90, 698–705. [Google Scholar] [CrossRef]
- Mohamd, E.; Ahmed, H.; Estefan, S.; Farrag, A.; Salah, R. Windows into estradiol effects in Alzheimer’s disease therapy. Eur. Rev. Med. Pharm. Sci 2011, 15, 1131–1140. [Google Scholar]
- Aly, H.; Elrigal, N.; Ali, S.; Rizk, M.; Ebrahim, N. Modulatory effects of Casimiroa edulis on aluminium nanoparticles-associated neurotoxicity in a rat model of induced Alzheimer’s disease. J. Mater. Environ. Sci 2018, 9, 1931–1941. [Google Scholar]
- Sumathi, T.; Shobana, C.; Mahalakshmi, V.; Sureka, R.; Subathra, M.; Vishali, A.; Rekha, K. Oxidative Stress in Brains of Male Rats Intoxicated with Aluminium and Neuromodulating Effect of Celastrus Paniculatus Alcoholic Seed Extract. Asian J. Pharm. Clin. Res. 2013, 6, 80–90. [Google Scholar]
- Yokel, R.A.; Allen, D.D.; Ackley, D.C. The distribution of aluminum into and out of the brain. J. Inorg. Biochem. 1999, 76, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Nayak, P. Aluminum: Impacts and disease. Environ. Res. 2002, 89, 101–115. [Google Scholar] [CrossRef]
- Pérez-Grijalba, V.; Arbizu, J.; Romero, J.; Prieto, E.; Pesini, P.; Sarasa, L.; Guillen, F.; Monleón, I.; San-José, I.; Martínez-Lage, P. Plasma Aβ42/40 ratio alone or combined with FDG-PET can accurately predict amyloid-PET positivity: A cross-sectional analysis from the AB255 Study. Alzheimer’s Res. Ther. 2019, 11, 96. [Google Scholar] [CrossRef] [PubMed]
- Medeiros, R.; Baglietto-Vargas, D.; LaFerla, F.M. The role of tau in Alzheimer’s disease and related disorders. CNS Neurosci. Ther. 2011, 17, 514–524. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, K.; Zaidi, T.; Wen, G.; Grundke-Iqbal, I.; Merz, P.; Shaikh, S.; Wisniewski, H.; Alafuzoff, I.; Winblad, B. Defective brain microtubule assembly in Alzheimer’s disease. Lancet 1986, 328, 421–426. [Google Scholar] [CrossRef]
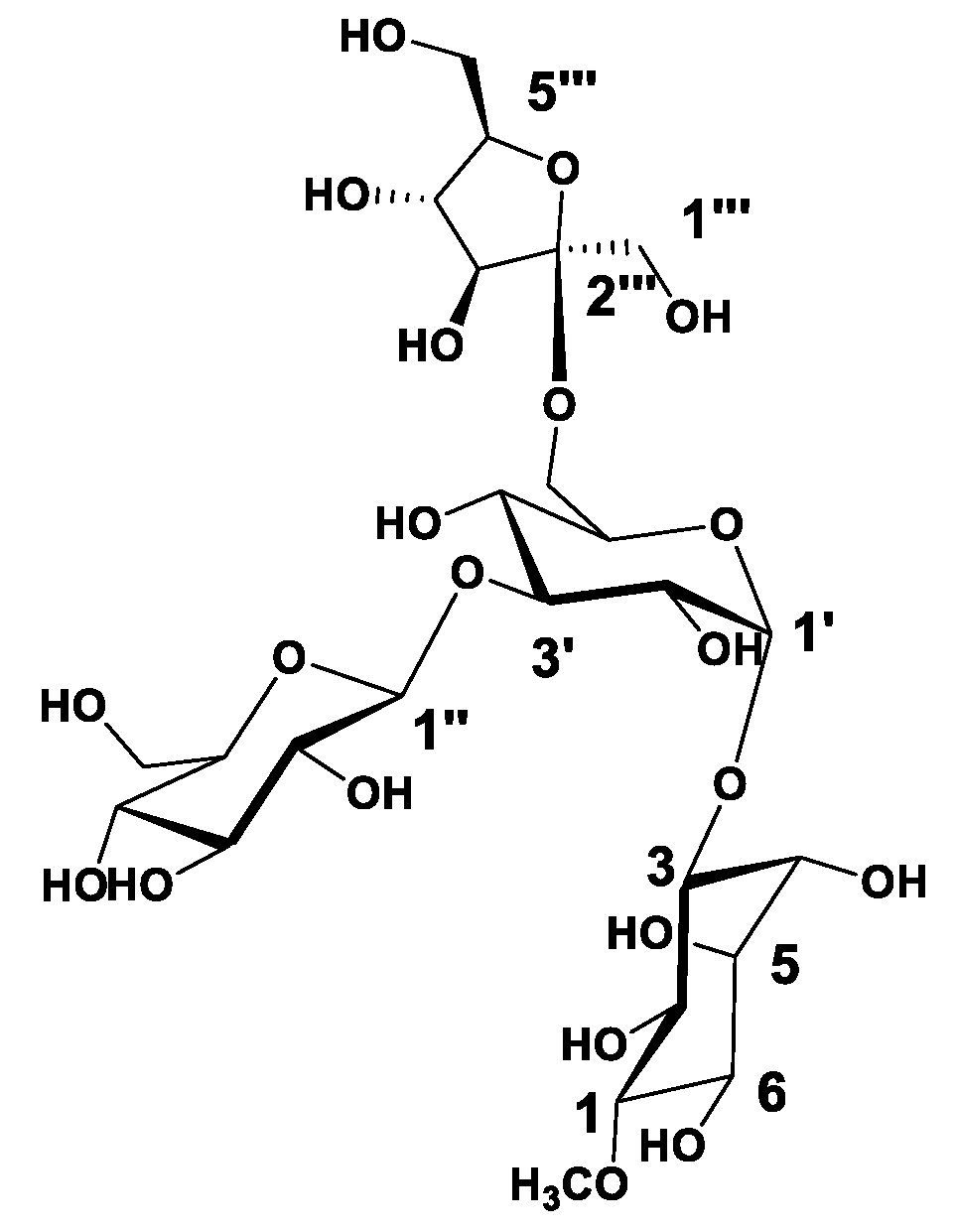
 ) correlations of compound 1.
) correlations of compound 1.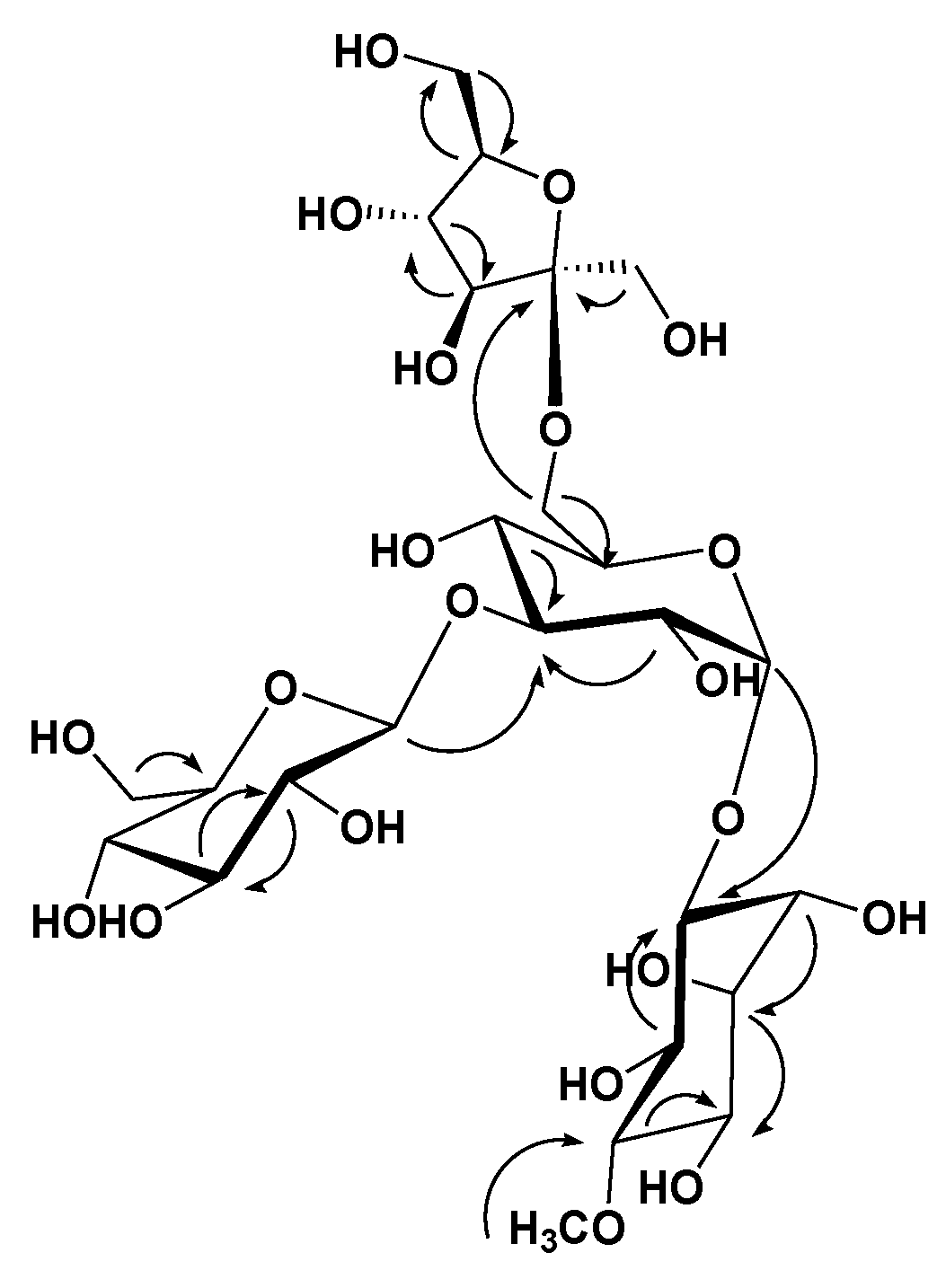


| Position | 1 | |
|---|---|---|
| Moiety | δC | δH (J in Hz) |
| (+)-pinitol | ||
| 1 | 84.4, CH | 3.28, overlapped |
| 2 | 73.0, CH | 3.92, overlapped |
| 3 | 73.3, CH | 4.55, m |
| 4 | 72.2, CH | 3.71, m |
| 5 | 74.5, CH | 3.70, overlapped |
| 6 | 71.5, CH | 3.81, overlapped |
| -OCH3 | 60.6 | 3.63, s |
| α-d-glucopyranosyl | ||
| 1′ | 93.5, CH | 5.14, d (3.5) |
| 2′ | 71.4, CH | 3.81, overlapped |
| 3′ | 77.2, CH | 4.06, overlapped |
| 4′ | 69.1, CH | 3.81, overlapped |
| 5′ | 71.6, CH | 3.81, overlapped |
| 6′ | 64.0, CH2 | 3.62, 4.04, m |
| β-d-glucopyranosyl | ||
| 1″ | 97.7, CH | 4.51, d (8) |
| 2″ | 73.8, CH | 3.62, overlapped |
| 3″ | 76.3, CH | 3.16, t |
| 4″ | 70.8, CH | 3.87, overlapped |
| 5″ | 77.5, CH | 3.35, overlapped |
| 6″ | 62.4, CH2 | 3.81, 4.04, m |
| β-d-fructofuranosyl | ||
| 1‴ | 65.5, CH2 | 3.50, 3.69, m |
| 2‴ | 99.0, qC | |
| 3‴ | 77.6, CH | 3.35, overlapped |
| 4‴ | 71.2, CH | 3.31, m |
| 5‴ | 82.7, CH | 3.78, overlapped |
| 6‴ | 64.3, CH2 | 3.51, 4.04, m |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohamed, E.M.; H. Elmaidomy, A.; Alaaeldin, R.; Alsenani, F.; Altemani, F.H.; Algehainy, N.A.; Alanazi, M.A.; Bagalagel, A.; Althagafi, A.; Elrehany, M.A.; et al. Anti-Alzheimer Potential of a New (+)-Pinitol Glycoside Isolated from Tamarindus indica Pulp: In Vivo and In Silico Evaluations. Metabolites 2023, 13, 732. https://doi.org/10.3390/metabo13060732
Mohamed EM, H. Elmaidomy A, Alaaeldin R, Alsenani F, Altemani FH, Algehainy NA, Alanazi MA, Bagalagel A, Althagafi A, Elrehany MA, et al. Anti-Alzheimer Potential of a New (+)-Pinitol Glycoside Isolated from Tamarindus indica Pulp: In Vivo and In Silico Evaluations. Metabolites. 2023; 13(6):732. https://doi.org/10.3390/metabo13060732
Chicago/Turabian StyleMohamed, Esraa M., Abeer H. Elmaidomy, Rania Alaaeldin, Faisal Alsenani, Faisal H. Altemani, Naseh A. Algehainy, Mohammad A Alanazi, Alaa Bagalagel, Abdulhamid Althagafi, Mahmoud A Elrehany, and et al. 2023. "Anti-Alzheimer Potential of a New (+)-Pinitol Glycoside Isolated from Tamarindus indica Pulp: In Vivo and In Silico Evaluations" Metabolites 13, no. 6: 732. https://doi.org/10.3390/metabo13060732
APA StyleMohamed, E. M., H. Elmaidomy, A., Alaaeldin, R., Alsenani, F., Altemani, F. H., Algehainy, N. A., Alanazi, M. A., Bagalagel, A., Althagafi, A., Elrehany, M. A., & Abdelmohsen, U. R. (2023). Anti-Alzheimer Potential of a New (+)-Pinitol Glycoside Isolated from Tamarindus indica Pulp: In Vivo and In Silico Evaluations. Metabolites, 13(6), 732. https://doi.org/10.3390/metabo13060732







