Urinary Metabolite Profiling to Non-Invasively Monitor the Omega-3 Index: An Exploratory Secondary Analysis of a Randomized Clinical Trial in Young Adults
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants and Study Design
2.2. Supplements
2.3. Erythrocyte Fatty Acid Content
2.4. Untargeted Urine Metabolite Analysis
2.4.1. Chemicals
2.4.2. Preparation of Urine Samples and Quality Controls
2.4.3. Urine Metabolome Analysis
2.4.4. Comprehensive Analysis of Ionic Urinary Metabolites with Quality Control
2.5. Statistical Analysis
2.5.1. Multivariate Analysis
2.5.2. ANOVA Tests
2.5.3. Integrated Data
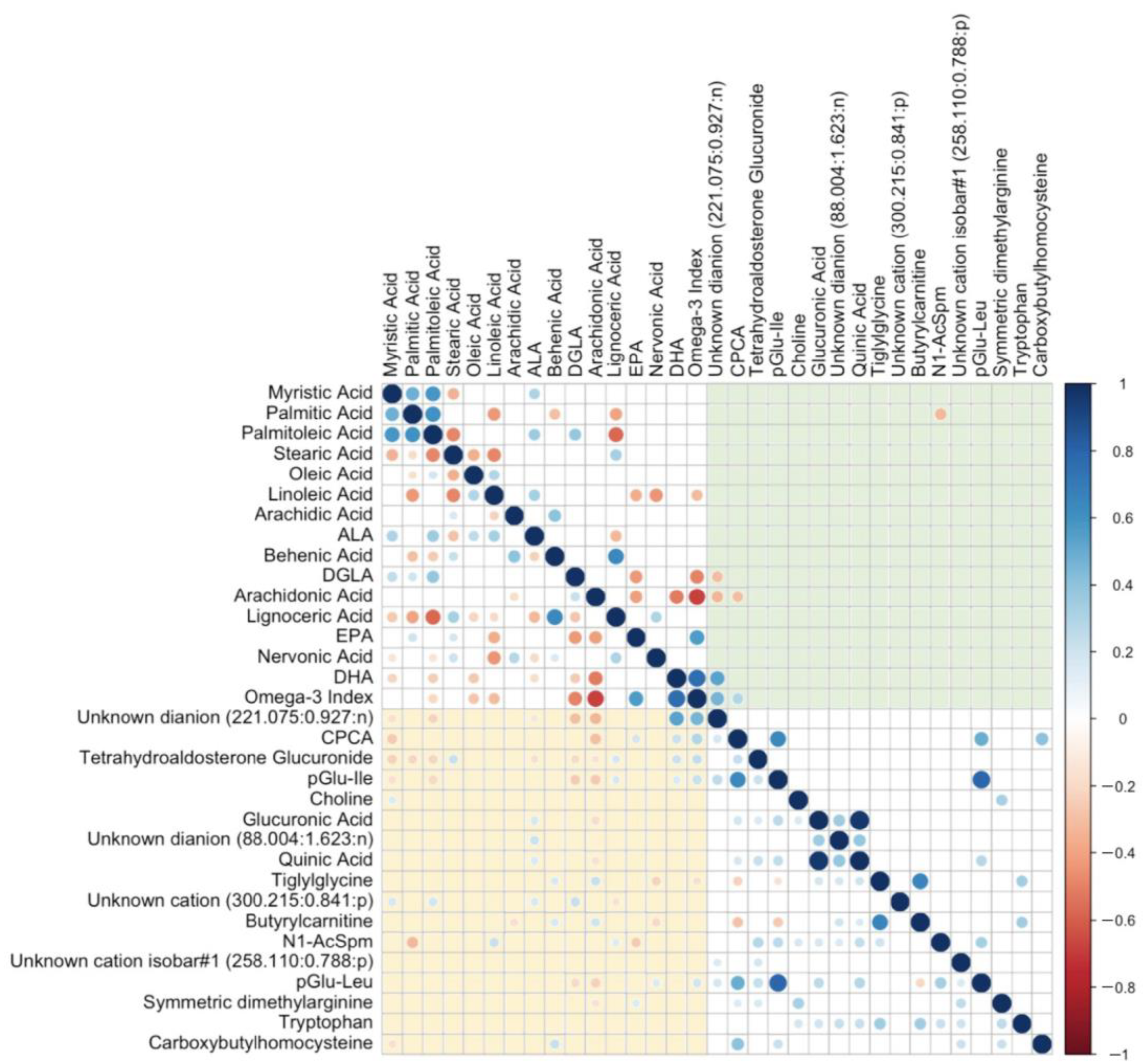
2.5.4. Correlation Plots
3. Results
3.1. Patient Characteristics
3.2. Baseline Participant Data Analysis in the Three Supplement Groups
3.3. Blood Marker and Erythrocyte Fatty Acid Analyses
3.4. Identification of Urinary Metabolites Associated with EPA and/or DHA Supplementation
3.4.1. Distinguishing Supplementation Groups with Unsupervised and Supervised Clustering Approaches
3.4.2. Individual Urinary Metabolite Changes in Response to Supplementation over Time
3.4.3. Urinary Metabolites Associated with the O3I
3.5. Urinary Metabolites Predict Supplement Groups and the O3I
3.6. Increases in Urinary CPCA Are Associated with n3-LCPUFA Erythrocyte Content
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Calder, P.C. Omega-3 Fatty Acids and Inflammatory Processes. Nutrients 2010, 2, 355–374. [Google Scholar] [CrossRef] [PubMed]
- Mozaffarian, D.; Wu, J.H.Y. Omega-3 Fatty Acids and Cardiovascular Disease: Effects on Risk Factors, Molecular Pathways, and Clinical Events. J. Am. Coll. Cardiol. 2011, 58, 2047–2067. [Google Scholar] [CrossRef]
- Abdelhamid, A.S.; Brown, T.J.; Brainard, J.S.; Biswas, P.; Thorpe, G.C.; Moore, H.J.; Deane, K.H.; Summerbell, C.D.; Worthington, H.V.; Song, F.; et al. Omega-3 Fatty Acids for the Primary and Secondary Prevention of Cardiovascular Disease. Cochrane Database Syst. Rev. 2020, 3, CD003177. [Google Scholar] [CrossRef] [PubMed]
- Reimers, A.; Ljung, H. The Emerging Role of Omega-3 Fatty Acids as a Therapeutic Option in Neuropsychiatric Disorders. Ther. Adv. Psychopharmacol. 2019, 9, 204512531985890. [Google Scholar] [CrossRef]
- Harris, W.S.; Tintle, N.L.; Imamura, F.; Qian, F.; Korat, A.V.A.; Marklund, M.; Djoussé, L.; Bassett, J.K.; Carmichael, P.H.; Chen, Y.Y.; et al. Blood N-3 Fatty Acid Levels and Total and Cause-Specific Mortality from 17 Prospective Studies. Nat. Commun. 2021, 12, 2329. [Google Scholar] [CrossRef]
- Burdge, G.C.; Wootton, S.A. Conversion of α-Linolenic Acid to Eicosapentaenoic, Docosapentaenoic and Docosahexaenoic Acids in Young Women. Br. J. Nutr. 2002, 88, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Plourde, M.; Cunnane, S.C. Extremely Limited Synthesis of Long Chain Polyunsaturates in Adults: Implications for Their Dietary Essentiality and Use as Supplements. Appl. Physiol. Nutr. Metab. 2007, 32, 619–634. [Google Scholar] [CrossRef] [PubMed]
- Kipnis, V.; Midthune, D.; Freedman, L.; Bingham, S.; Day, N.E.; Riboli, E.; Ferrari, P.; Carroll, R.J. Bias in Dietary-Report Instruments and Its Implications for Nutritional Epidemiology. Public Health Nutr. 2002, 5, 915–923. [Google Scholar] [CrossRef]
- Poslusna, K.; Ruprich, J.; De Vries, J.H.M.; Jakubikova, M.; Van’T Veer, P. Misreporting of Energy and Micronutrient Intake Estimated by Food Records and 24hour Recalls, Control and Adjustment Methods in Practice. Br. J. Nutr. 2009, 101, S73–S85. [Google Scholar] [CrossRef]
- von Schacky, C. Omega-3 Index and Cardiovascular Health. Nutrients 2014, 6, 799–814. [Google Scholar] [CrossRef]
- Cholewski, M.; Tomczykowa, M.; Tomczyk, M. A Comprehensive Review of Chemistry, Sources and Bioavailability of Omega-3 Fatty Acids. Nutrients 2018, 10, 1662. [Google Scholar] [CrossRef] [PubMed]
- Tu, W.C.; Mühlhäusler, B.S.; Yelland, L.N.; Gibson, R.A. Correlations between Blood and Tissue Omega-3 LCPUFA Status Following Dietary ALA Intervention in Rats. Prostaglandins Leukot. Essent. Fat. Acids 2013, 88, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Harris, W.S.; Von Schacky, C. The Omega-3 Index: A New Risk Factor for Death from Coronary Heart Disease? Prev. Med. 2004, 39, 212–220. [Google Scholar] [CrossRef] [PubMed]
- McLenon, J.; Rogers, M.A.M. The Fear of Needles: A Systematic Review and Meta-Analysis. J. Adv. Nurs. 2019, 75, 30–42. [Google Scholar] [CrossRef]
- Mcmurtry, C.M.; Taddio, A.; Noel, M.; Antony, M.M.; Rieder, M.J.; Robson, K.; Uleryk, E.; Votta, E. Exposure-Based Interventions for the Management of Individuals with High Levels of Needle Fear across the Lifespan: A Clinical Practice Guideline and Call for Further Research. Cogn. Behav. Ther. 2016, 45, 217–235. [Google Scholar] [CrossRef]
- Dempsey, M.; Rockwell, M.S.; Wentz, L.M. The Influence of Dietary and Supplemental Omega-3 Fatty Acids on the Omega-3 Index: A Scoping Review. Front. Nutr. 2023, 10, 1072653. [Google Scholar] [CrossRef]
- Rafiq, T.; Azab, S.M.; Teo, K.K.; Thabane, L.; Anand, S.S.; Morrison, K.M.; De Souza, R.J.; Britz-Mckibbin, P. Nutritional Metabolomics and the Classification of Dietary Biomarker Candidates: A Critical Review. Adv. Nutr. 2021, 12, 2333–2357. [Google Scholar] [CrossRef]
- O’Gorman, A.; Brennan, L. The Role of Metabolomics in Determination of New Dietary Biomarkers. Proc. Nutr. Soc. 2017, 76, 295–302. [Google Scholar] [CrossRef]
- Gao, Q.; Praticò, G.; Scalbert, A.; Vergères, G.; Kolehmainen, M.; Manach, C.; Brennan, L.; Afman, L.A.; Wishart, D.S.; Andres-Lacueva, C.; et al. A Scheme for a Flexible Classification of Dietary and Health Biomarkers. Genes Nutr. 2017, 12, 1–15. [Google Scholar] [CrossRef]
- Clarke, E.D.; Rollo, M.E.; Collins, C.E.; Haslam, R.L.; Pezdirc, K. Urinary Biomarkers of Dietary Intake: A Review. Nutr. Rev. 2020, 78, 364–381. [Google Scholar] [CrossRef]
- Filipovic, M.G.; Reiner, M.F.; Rittirsch, S.; Irincheeva, I.; Aeschbacher, S.; Grossmann, K.; Risch, M.; Risch, L.; Limacher, A.; Conen, D.; et al. Blood Omega-3 Fatty Acids Are Inversely Associated With Albumin-Creatinine Ratio in Young and Healthy Adults (The Omega-Kid Study). Front. Cardiovasc. Med. 2021, 8, 622619. [Google Scholar] [CrossRef]
- Bigornia, S.J.; Harris, W.S.; Falcón, L.M.; Ordovás, J.M.; Lai, C.Q.; Tucker, K.L. The Omega-3 Index Is Inversely Associated with Depressive Symptoms among Individuals with Elevated Oxidative Stress Biomarkers. J. Nutr. 2016, 146, 758–766. [Google Scholar] [CrossRef] [PubMed]
- Gibson, R.; Lau, C.H.E.; Loo, R.L.; Ebbels, T.M.D.; Chekmeneva, E.; Dyer, A.R.; Miura, K.; Ueshima, H.; Zhao, L.; Daviglus, M.L.; et al. The Association of Fish Consumption and Its Urinary Metabolites with Cardiovascular Risk Factors: The International Study of Macro-/Micronutrients and Blood Pressure (INTERMAP). Am. J. Clin. Nutr. 2020, 111, 280–290. [Google Scholar] [CrossRef]
- Ruan, Y.; Zheng, J.; Ren, Y.; Tang, J.; Li, J.; Li, D. Changes of Urine Metabolites in Response to N-3 Fatty Acid Supplements and Their Correlation with Metabolic Risk Factors in Patients with Type 2 Diabetes. Food Funct. 2019, 10, 2471–2479. [Google Scholar] [CrossRef]
- Lee, J.B.; Notay, K.; Klingel, S.L.; Chabowski, A.; Mutch, D.M.; Millar, P.J. Docosahexaenoic Acid Reduces Resting Blood Pressure but Increases Muscle Sympathetic Outflow Compared with Eicosapentaenoic Acid in Healthy Men and Women. Am. J. Physiol. Hear. Circ. Physiol. 2019, 316, H873–H881. [Google Scholar] [CrossRef]
- Metherel, A.H.; Irfan, M.; Klingel, S.L.; Mutch, D.M.; Bazinet, R.P. Compound-Specific Isotope Analysis Reveals No Retroconversion of DHA to EPA but Substantial Conversion of EPA to DHA Following Supplementation: A Randomized Control Trial. Am. J. Clin. Nutr. 2019, 110, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A Simple Method for the Isolation and Purification of Total Lipides from Animal Tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Mcglory, C.; Gorissen, S.H.M.; Kamal, M.; Bahniwal, R.; Hector, A.J.; Baker, S.K.; Chabowski, A.; Phillips, S.M. Omega-3 Fatty Acid Supplementation Attenuates Skeletal Muscle Disuse Atrophy during Two Weeks of Unilateral Leg Immobilization in Healthy Young Women. FASEB J. 2019, 33, 4586–4597. [Google Scholar] [CrossRef]
- Shanmuganathan, M.; Kroezen, Z.; Gill, B.; Azab, S.; de Souza, R.J.; Teo, K.K.; Atkinson, S.; Subbarao, P.; Desai, D.; Anand, S.S.; et al. The Maternal Serum Metabolome by Multisegment Injection-Capillary Electrophoresis-Mass Spectrometry: A High-Throughput Platform and Standardized Data Workflow for Large-Scale Epidemiological Studies. Nat. Protoc. 2021, 16, 1966–1994. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Pinto-Sanchez, M.I.; Bercik, P.; Britz-McKibbin, P. Metabolomics Reveals Elevated Urinary Excretion of Collagen Degradation and Epithelial Cell Turnover Products in Irritable Bowel Syndrome Patients. Metabolomics 2019, 15, 82. [Google Scholar] [CrossRef]
- Yamamoto, M.; Shanmuganathan, M.; Hart, L.; Pai, N.; Britz-Mckibbin, P. Urinary Metabolites Enable Differential Diagnosis and Therapeutic Monitoring of Pediatric Inflammatory Bowel Disease. Metabolites 2021, 11, 245. [Google Scholar] [CrossRef]
- Nori de Macedo, A.; Mathiaparanam, S.; Brick, L.; Keenan, K.; Gonska, T.; Pedder, L.; Hill, S.; Britz-McKibbin, P. The Sweat Metabolome of Screen-Positive Cystic Fibrosis Infants: Revealing Mechanisms beyond Impaired Chloride Transport. ACS Cent. Sci. 2017, 3, 904–913. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Guo, A.C.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Allen, D.; Tian, S.; Oler, E.; Gautam, V.; Greiner, R.; Metz, T.O.; Wishart, D.S. CFM-ID 4.0—A Web Server for Accurate MS-Based Metabolite Identification. Nucleic Acids Res. 2022, 50, W165–W174. [Google Scholar] [CrossRef] [PubMed]
- DiBattista, A.; McIntosh, N.; Lamoureux, M.; Al-Dirbashi, O.Y.; Chakraborty, P.; Britz-McKibbin, P. Temporal Signal Pattern Recognition in Mass Spectrometry: A Method for Rapid Identification and Accurate Quantification of Biomarkers for Inborn Errors of Metabolism with Quality Assurance. Anal. Chem. 2017, 89, 8112–8121. [Google Scholar] [CrossRef] [PubMed]
- Saoi, M.; Li, A.; McGlory, C.; Stokes, T.; von Allmen, M.T.; Phillips, S.M.; Britz-Mckibbin, P. Metabolic Perturbations from Step Reduction in Older Persons at Risk for Sarcopenia: Plasma Biomarkers of Abrupt Changes in Physical Activity. Metabolites 2019, 9, 134. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular Framework for Processing, Visualizing, and Analyzing Mass Spectrometry-Based Molecular Profile Data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Posit Team. RStudio: Integrated Development Environment for R. Posit Software; PBC: Boston, MA, USA, 2020. [Google Scholar]
- Kassambara, A.; Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses; R Package Version 1.0.7. 2020. Available online: https://CRAN.R-project.org/package=factoextra (accessed on 9 September 2022).
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.A. MixOmics: An R Package for ‘omics Feature Selection and Multiple Data Integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef]
- Kassambara, A. rstatix: Pipe-Friendly Framework for Basic Statistical Tests; R Package Version 1.4.8. 2022. Available online: https://CRAN.R-project.org/package=rstatix (accessed on 22 November 2022).
- Pinheiro, J.; Bates, D.; R Core Team. nlme: Linear and Nonlinear Mixed Effects Models; R Package Version 3.1-157. 2022. Available online: https://CRAN.R-project.org/package=nlme (accessed on 9 September 2022).
- Lenth, R.V. Emmeans: Estimated Marginal Means, Aka Least-Squares Means; R Package Version 1.4.8. 2022. Available online: https://CRAN.R-project.org/package=emmeans (accessed on 9 September 2022).
- Groll, A. GlmmLasso: Variable Selection for Generalized Linear Mixed Models by L1-Penalized Estimation; R Package Version 1.6.2. 2022. Available online: https://CRAN.R-project.org/package=glmmLasso (accessed on 9 September 2022).
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Mueller, M. PROC: An Open-Source Package for R and S+ to Analyze and Compare ROC Curves. BMC Bioinform. 2011, 8, 12–77. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis. Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Wei, T.; Simko, V. R package ‘corrplot’: Visualization of a Correlation Matrix. R Package Version 0.92. 2021. Available online: https://github.com/taiyun/corrplot (accessed on 9 September 2022).
- Revelle, W. psych: Procedures for Psychological, Psychometric, and Personality Research; R Package Version 2.3.9. 2022. Available online: https://CRAN.R-project.org/package=psych (accessed on 9 September 2022).
- Klingel, S.L.; Metherel, A.H.; Irfan, M.; Rajna, A.; Chabowski, A.; Bazinet, R.P.; Mutch, D.M. EPA and DHA Have Divergent Effects on Serum Triglycerides and Lipogenesis, but Similar Effects on Lipoprotein Lipase Activity: A Randomized Controlled Trial. Am. J. Clin. Nutr. 2019, 110, 1502–1509. [Google Scholar] [CrossRef] [PubMed]
- Potischman, N. Biomarkers of Nutritional Exposure and Nutritional Status Biologic and Methodologic Issues for Nutritional Biomarkers 1. J. Nutr. 2003, 133, 875S–880S. [Google Scholar] [CrossRef] [PubMed]
- Flock, M.R.; Skulas-Ray, A.C.; Harris, W.S.; Etherton, T.D.; Fleming, J.A.; Kris-Etherton, P.M. Determinants of Erythrocyte Omega-3 Fatty Acid Content in Response to Fish Oil Supplementation: A Dose-Response Randomized Controlled Trial. J. Am. Heart Assoc. 2013, 2, 20–23. [Google Scholar] [CrossRef] [PubMed]
- Ly, R.; MacIntyre, B.C.; Philips, S.M.; McGlory, C.; Mutch, D.M.; Britz-McKibbin, P. Lipidomic Studies Reveal Specific Circulating Phosphatidylcholines as Surrogate Biomarkers of Omega-3 Index. J. Lipid Res. 2023, 100445. [Google Scholar] [CrossRef] [PubMed]
- Dragsted, L.O.; Gao, Q.; Scalbert, A.; Vergères, G.; Kolehmainen, M.; Manach, C.; Brennan, L.; Afman, L.A.; Wishart, D.S.; Andres Lacueva, C.; et al. Validation of Biomarkers of Food Intake-Critical Assessment of Candidate Biomarkers. Genes Nutr. 2018, 13, 14. [Google Scholar] [CrossRef] [PubMed]
- Vidgren, H.M.; Ågren, J.J.; Schwab, U.; Rissanen, T.; Hänninen, O.; Uusitupa, M.I.J. Incorporation of N-3 Fatty Acids into Plasma Lipid Fractions, and Erythrocyte Membranes and Platelets during Dietary Supplementation with Fish, Fish Oil, and Docosahexaenoic Acid-Rich Oil among Healthy Young Men. Lipids 1997, 32, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Simopoulos, A.P. The Importance of the Ratio of Omega-6/Omega-3 Essential Fatty Acids. Biomed. Pharmacother. 2002, 56, 365–379. [Google Scholar] [CrossRef]
- Bouatra, S.; Aziat, F.; Mandal, R.; Guo, A.C.; Wilson, M.R.; Knox, C.; Bjorndahl, T.C.; Krishnamurthy, R.; Saleem, F.; Liu, P.; et al. The Human Urine Metabolome. PLoS ONE 2013, 8, e73076. [Google Scholar] [CrossRef]
- Zeki, Ö.C.; Eylem, C.C.; Reçber, T.; Kır, S.; Nemutlu, E. Integration of GC–MS and LC–MS for Untargeted Metabolomics Profiling. J. Pharm. Biomed. Anal. 2020, 190, 113509. [Google Scholar] [CrossRef]
- Dunn, W.B.; Ellis, D.I. Metabolomics: Current Analytical Platforms and Methodologies. TrAC Trends Anal. Chem. 2005, 24, 285–294. [Google Scholar] [CrossRef]
- Shanmuganathan, M.; Sarfaraz, M.O.; Kroezen, Z.; Philbrick, H.; Poon, R.; Don-Wauchope, A.; Puglia, M.; Wishart, D.; Britz-McKibbin, P. A Cross-Platform Metabolomics Comparison Identifies Serum Metabolite Signatures of Liver Fibrosis Progression in Chronic Hepatitis C Patients. Front. Mol. Biosci. 2021, 8, 676349. [Google Scholar] [CrossRef]
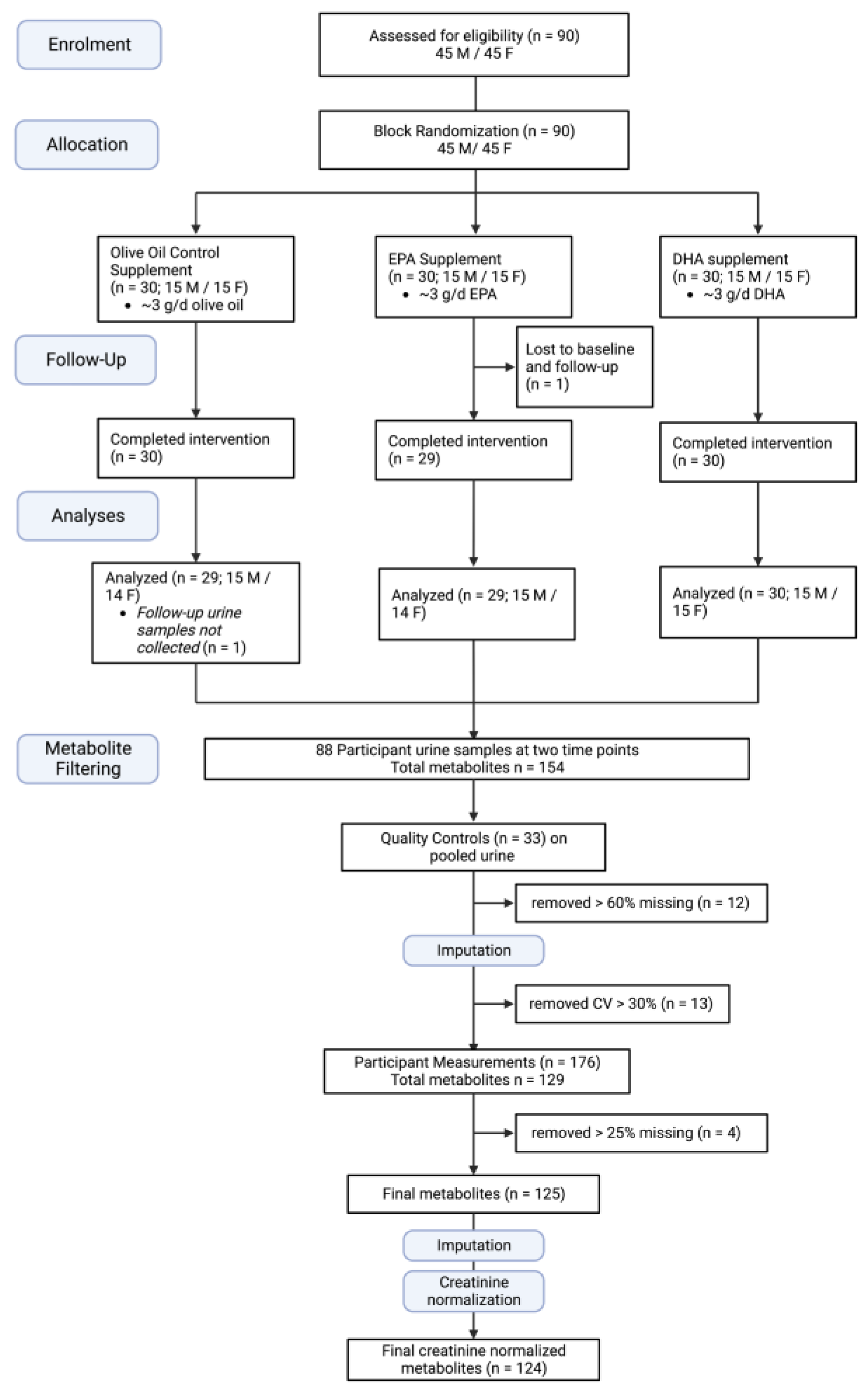
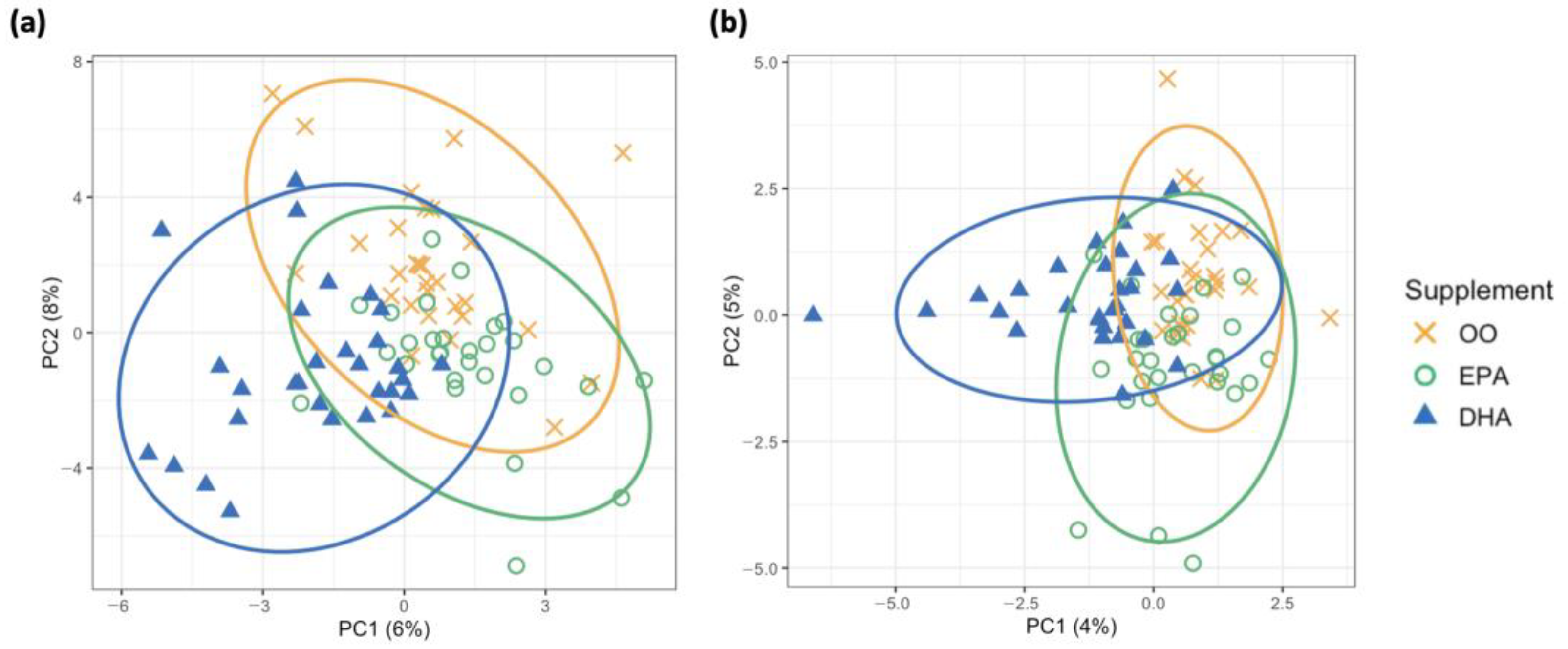
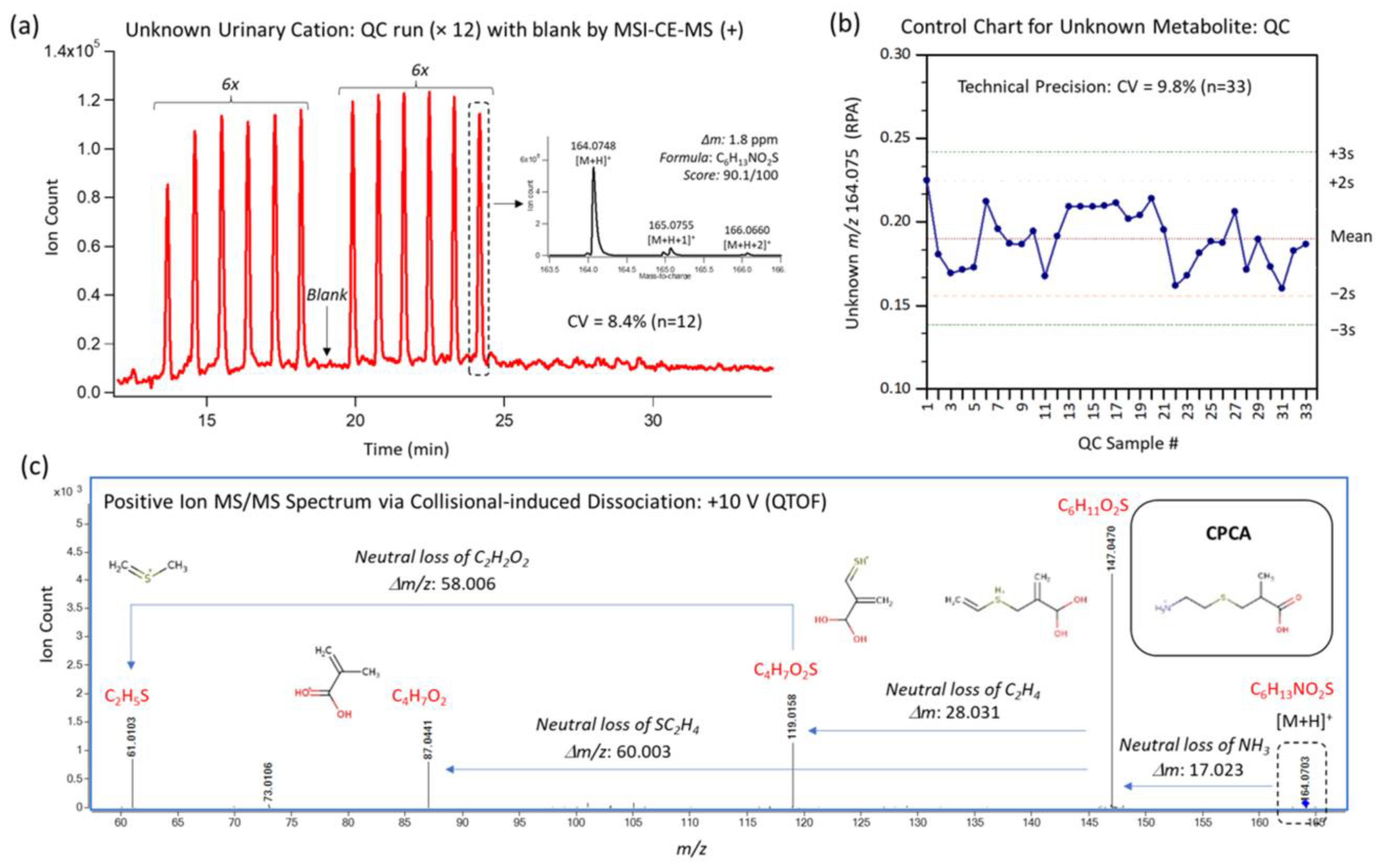

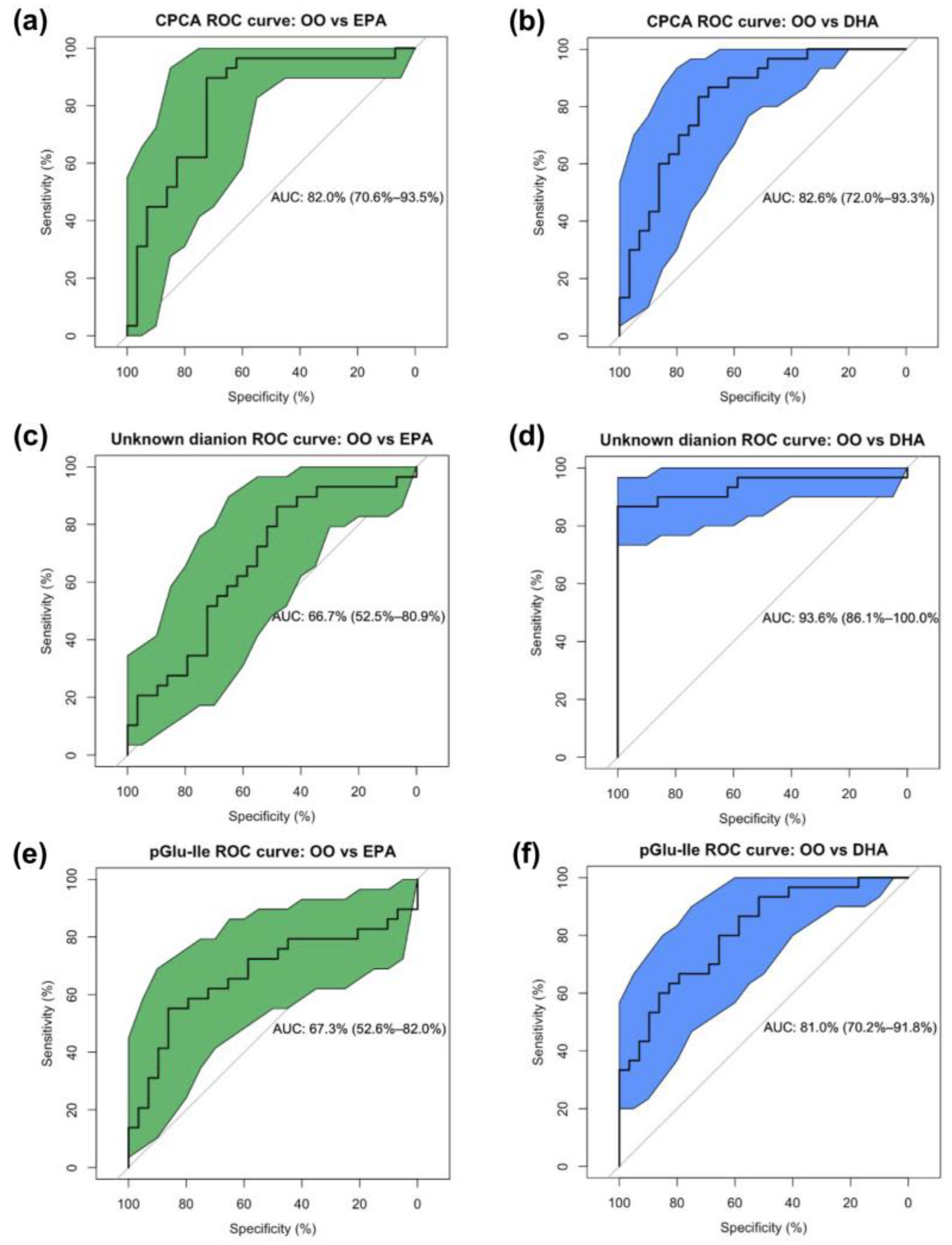
| OO | EPA | DHA | p-Value | |
|---|---|---|---|---|
| M/F, n | 15/15 | 15/14 | 15/15 | |
| Age, y | 21.1 ± 1.9 | 21.4 ± 2.2 | 22.2 ± 2.3 | 0.184 |
| Weight, kg | 69.0 ± 10.9 | 71.0 ± 12.0 | 72.7 ± 14.9 | 0.616 |
| BMI, kg/m2 | 24.1 ± 3.5 | 23.1 ± 2.8 | 23.7 ± 3.4 | 0.613 |
| OO (n = 30) | EPA (n = 29) | DHA (n = 30) | ANOVA | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Baseline | Final | Baseline | Final | Baseline | Final | Pint | Ptreatment | Ptime | |
| Glucose, mmol/L | 4.6 ± 0.3 | 4.7 ± 0.4 | 4.8 ± 0.3 | 4.9 ± 0.3 | 4.7 ± 0.3 | 4.9 ± 0.3 | 0.942 | 0.006 | <0.001 |
| Triglycerides, mmol/L | 0.9 ± 0.4 | 0.9 ± 0.4 a,c | 0.8 ± 0.3 | 0.8 ± 0.3 a,b | 0.8 ± 0.2 | 0.6 ± 0.1 b,$ | 0.049 | 0.023 | 0.028 |
| Cholesterol, mmol/L | 4.4 ± 0.9 | 4.6 ± 0.9 | 4.5 ± 0.7 | 4.5 ± 0.7 | 4.3 ± 0.8 | 4.5 ± 0.9 | 0.400 | 0.809 | 0.108 |
| EPA, ng FA/mg Hb | 27.6 ± 9.4 | 30.6 ± 11.3 c | 27.9 ± 10.7 | 225.5 ± 60.2 a,$ | 25.9 ± 10.0 | 68.6 ± 25.2 b,$ | <0.001 | <0.001 | <0.001 |
| DHA, ng FA/mg Hb | 176.4 ± 40.8 | 172.8 ± 40.6 b | 161.4 ± 39.6 | 148.5 ± 43.6 b | 150.4 ± 32.7 | 401.8 ± 76.3 a,$ | <0.001 | <0.001 | <0.001 |
| EPA, relative % | 0.5 ± 0.2 | 0.5 ± 0.2 c | 0.5 ± 0.2 | 3.9 ± 1.1 a,$ | 0.5 ± 0.2 | 1.2 ± 0.4 b,$ | <0.001 | <0.001 | <0.001 |
| DHA, relative % | 3.2 ± 0.7 | 3.0 ± 0.5 b | 3.0 ± 0.6 | 2.5 ± 0.5 b | 2.9 ± 0.5 | 7.1 ± 1.0 a,$ | <0.001 | <0.001 | <0.001 |
| O3I | 3.7 ± 0.8 | 3.5 ± 0.6 c | 3.5 ± 0.7 | 6.5 ± 1.2 b$ | 3.4 ± 0.6 | 8.3 ± 1.2 a$ | <0.001 | <0.001 | <0.001 |
| Test | sPLS-DA | ANOVA | LASSO | ROC Curves Group at 12 Weeks | ROC Curves Low vs. High O3I 2 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Outcome | Supplement Group (Δ) | Supplement Group × Time | Pairwise (vs. OO) at 12 Weeks | Pairwise (Change vs. Baseline) | O3I | EPA + DHA | AUC (%) | AUC (%) | AUC (%) | |
| Metabolite 1 | m/z:RMT:mode | Pint | OO vs. EPA | OO vs. DHA | <4% vs. >8% O3I | |||||
| Unknown dianion [M − 2H]2− | 221.075:0.927:n | DHA | <0.001 * | ↑DHA | ↑DHA | ↑ | ↑ | 66.7 | 93.6 | 89.4 |
| S-Carboxypropylcysteamine (CPCA) | 164.074:0.613:p | DHA | <0.001 | ↑EPA/ ↑DHA | ↑DHA | ↑ | ↑ | 82.0 | 82.6 | 73.1 |
| Tetrahydroaldosterone glucuronide | 539.249:0.472:n | DHA | 0.019 | ↑ | ↑ | 59.1 | 68.0 | 65.4 | ||
| Pyroglutamylisoleucine (pGlu-Ile) | 241.120:0.653:n | DHA | <0.001 * | ↑EPA/ ↑DHA | 67.3 | 81.0 | 64.7 | |||
| Choline | 104.108:0.333:p | EPA | 55.9 | 53.6 | 57.4 | |||||
| Glucuronic acid | 193.035:0.772:n | EPA | 0.014 | 62.7 | 60.8 | 62.3 | ||||
| Unknown dianion [M − 2H]2− | 88.004:1.622:n | EPA | 49.3 | 47.6 | 51.5 | |||||
| Quinic acid | 191.056:0.798:n | EPA | 0.035 | 55.1 | 59.5 | 62.6 | ||||
| Tiglylglycine | 156.066:0.843:n | EPA | <0.001 | ↓DHA | ↓DHA | 69.4 | 71.3 | 60.6 | ||
| Unknown cation | 300.215:0.841:p | EPA | <0.004 | 52.3 | 68.2 | 66.3 | ||||
| O-Butyrylcarnitine | 232.155:0.718:p | 0.039 | 64.1 | 64.8 | 51.1 | |||||
| N1-Accetylspermidine (N1-AcSpm) | 188.176:0.259:p | 0.040 | ↓ | ↓ | 70.5 | 55.3 | 50.7 | |||
| Unknown cation isobar#2 | 258.110:0.788:p | EPA | 0.049 | 49.6 | 59.0 | 57.2 | ||||
| Pyroglutamylleucine (pGlu-Leu) | 241.120:0.662:n | 0.002 * | ↑EPA/ ↑DHA | 70.9 | 75.9 | 65.4 | ||||
| Symmetric dimethylarginine | 203.151:0.478:p | ↑ | 57.6 | 56.0 | 44.8 | |||||
| Tryptophan | 205.098:0.899:p | ↑ | 68.1 | 54.7 | 63.5 | |||||
| Carboxybutylhomocysteine | 222.080:0.762:p | ↓ | 57.4 | 51.6 | 48.4 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
MacIntyre, B.C.; Shanmuganathan, M.; Klingel, S.L.; Kroezen, Z.; Helmeczi, E.; Seoh, N.-Y.; Martinez, V.; Chabowski, A.; Feng, Z.; Britz-McKibbin, P.; et al. Urinary Metabolite Profiling to Non-Invasively Monitor the Omega-3 Index: An Exploratory Secondary Analysis of a Randomized Clinical Trial in Young Adults. Metabolites 2023, 13, 1071. https://doi.org/10.3390/metabo13101071
MacIntyre BC, Shanmuganathan M, Klingel SL, Kroezen Z, Helmeczi E, Seoh N-Y, Martinez V, Chabowski A, Feng Z, Britz-McKibbin P, et al. Urinary Metabolite Profiling to Non-Invasively Monitor the Omega-3 Index: An Exploratory Secondary Analysis of a Randomized Clinical Trial in Young Adults. Metabolites. 2023; 13(10):1071. https://doi.org/10.3390/metabo13101071
Chicago/Turabian StyleMacIntyre, Brittany C., Meera Shanmuganathan, Shannon L. Klingel, Zachary Kroezen, Erick Helmeczi, Na-Yung Seoh, Vanessa Martinez, Adrian Chabowski, Zeny Feng, Philip Britz-McKibbin, and et al. 2023. "Urinary Metabolite Profiling to Non-Invasively Monitor the Omega-3 Index: An Exploratory Secondary Analysis of a Randomized Clinical Trial in Young Adults" Metabolites 13, no. 10: 1071. https://doi.org/10.3390/metabo13101071
APA StyleMacIntyre, B. C., Shanmuganathan, M., Klingel, S. L., Kroezen, Z., Helmeczi, E., Seoh, N.-Y., Martinez, V., Chabowski, A., Feng, Z., Britz-McKibbin, P., & Mutch, D. M. (2023). Urinary Metabolite Profiling to Non-Invasively Monitor the Omega-3 Index: An Exploratory Secondary Analysis of a Randomized Clinical Trial in Young Adults. Metabolites, 13(10), 1071. https://doi.org/10.3390/metabo13101071







